SUMMARY
Respiratory syncytial virus (RSV) can cause serious respiratory infections, second only to influenza virus. In order to know RSV's genetic changes we examined 4028 respiratory specimens from local hospital outpatients in Gyeonggi Province, South Korea over six consecutive years by real-time one-step RT–PCR; 183 patients were positive for RSV infection. To investigate the specific distribution of RSV genotypes, we performed partial sequencing of the glycoprotein gene. Of the 131 RSV-A specimens sequenced, 61 (43·3%) belonged to the ON1 genotype, 66 (46·8%) were NA1 genotype, 3 (2·1%) were GA5 genotype, and 1 (0·7%) belonged to the GA1 genotype. Of the 31 RSV-B specimens sequenced, 29 were BA9 genotype (87·9%) and 2 were BA10 genotype (6·1%). The most common clinical symptoms were fever, cough, nasal discharge, and phlegm; multiple logistic regression analysis showed that RSV-positive infection on pediatric patients was strongly associated with cough (OR = 2·8, 95% CI 1·6–5·1) and wheezing (OR = 2·8, 95% CI 1·7–4·4). The ON1 genotype was significantly associated with phlegm (OR = 11·8, 95% CI 3·8–46·7), while the NA1 genotype was associated with the pediatric patients’ gender (males, OR = 2·4, 95% CI 1·1–5·4) and presence of chills (OR = 5·1, 95% CI 1·1–27·2). RSV subgroup B was showed association with nasal obstruction (OR = 4·6, 95% CI 1·2–20·0). The majority of respiratory virus coinfections with RSV were human rhinovirus (47·2%). This study contributes to our understanding of the molecular epidemiological characteristics of RSV, which promotes the potential for improving RSV vaccines.
Key words: Clinical symptom, coinfection, genotype, human rhinovirus, respiratory syncytial virus
INTRODUCTION
Human respiratory syncytial virus (RSV) is the second most frequent cause of respiratory infections, and the main cause of bronchiolitis and pneumonia in infants and young children [1]. The incidence of RSV is known to be age-dependent, with an estimated 60% of children infected with RSV in their first year of life [2]. Almost all children are infected by the time they turn 2 years of age [3], whereas fewer infections are detected in older children and adults [4]. Immunity to RSV following recovery from infection is partial and temporary, thus reinfection throughout early childhood is common [5]. Due to the lack of an effective vaccine, substantial health and economic costs are associated with RSV epidemics due to hospitalizations and treatment of RSV infection-related illnesses [6].
RSV is a non-segmented, negative-sense single-strand RNA enveloped virus belonging to the family Paramyxoviridae [7]. There are two major RSV subgroups (A and B) categorized on the basis of antigenic and genetic variability. Each subgroup is further categorized into genotypes based on the nucleotide sequence variation within the second hypervariable region of heavily glycosylated G glycoprotein (G protein). There are 12 genotypes for RSV-A (GA1–7, SAA1, NA1–2, and ON1–2) and 20 genotypes for RSV-B (GB1–4, SAB1–4, URU1–2, and BA1–10) [8, 9]. Depending on the genotypes, there are differences in the attachment G glycoprotein which interacts with host cell receptors [7]. The RSV attachment G glycoprotein is responsible for virus binding to the host cell surface receptor and is a target of human neutralizing antibodies, together with the fusion (F) glycoprotein [3]. The G protein shows the highest genetic and antigenic variability among all the RSV structural proteins. There is abundant evidence of accumulating amino acid changes in its hypervariable regions over time [7, 10], making its study relevant in vaccine development strategies [11].
The dynamics of RSV circulation are further demonstrated with the emergence of new genotypes that spread rapidly worldwide and replace previously circulating genotypes [6]. Such changes were shown after the emergence of the BA genotype [12]. Among the RSV subgroup B, almost all strains detected after 2005 belong to the BA genotype [13]. Coincidently, a similar duplication event in RSV subgroup A led to the emergence of the ON1 genotype [14], and the importance of epidemiological prevalence has also been reported for the ON1 genotype [15, 16]. It is assumed that these specific genetic characteristics provide a selective evolutionary advantage for these viruses [17].
RSV coinfection with other respiratory viruses has been previously studied, in addition to its simple genetic characterization and annual distribution. The relevance to clinical severity in itself and coinfection with other respiratory viruses has also been investigated. For example, several studies revealed that the human rhinovirus (hRV) is the most common virus that coinfects with RSV. A greater number of viruses are not necessarily synonymous with greater disease severity [18], and coinfection occurs frequently with RSV and hRV, as well as RSV and human bocavirus (hBoV). Overall, viral coinfection does not present greater severity, but can present mixed clinical features [19]. However, some studies showed contradictory results. Children coinfected with RSV and other viruses presented more frequently with pneumonia than those with single RSV infection [20]. A previous study showed that during coinfections, one virus could block another simply by being the first to infect the available host cells [21].
In this study, we investigated the molecular epidemiologic characteristics of RSV and its coinfection with other respiratory viruses in Gyeonggi Province in South Korea.
METHODS
Subjects and data collection
This study was conducted by the Korea National Institute of Health (KNIH) as part of the Korea Influenza and Respiratory Viruses Surveillance System (KINRESS). As part of the KINRESS, we collected a total of 4028 respiratory specimens from local hospitals, which are primary hospitals that first level of health system in Gyeonggi Province, South Korea from 2009 to 2014. Epidemiologic data and respiratory specimens from all patients with influenza-like illnesses were sent from local hospitals to the Gyeonggi Province Institute of Health and Environment (GIHE). Swab specimens were immediately placed at 4 °C in 2-ml cryovials containing 1·5 ml of cold viral transport medium (VTM; Difco, USA), and sent to the laboratory. Upon receipt, the specimens were processed immediately for virus detection, identification, and characterization. Aliquots of samples were also stored at −80 °C for additional analysis. Case reports and laboratory analysis data were entered into a KINRESS database (the Korea National Institute of Health, South Korea). The samples were collected for respiratory virus diagnosis, and written informed consent was obtained from the patients, their parents or legal guardians [15]. This study was approved by the Korea National Institute of Health Institutional Review Boards (Approval Nos. 2010-03EXP-1-R, 2011–06EXP-01-C, 2012-08EXP-06-3C, 2013-08EXP-03-5C and 2014-08EXP-08-6C-A).
Virus detection and subsequent molecular analysis
Viral RNA was extracted from 140 µl of each respiratory specimen using QIAamp Viral RNA Mini Kits (Qiagen GmbH, Hilden, Germany). Seven Multiplex-PCR assays were performed for screening 16 respiratory viral pathogens including RSV, influenza (IFV)-A and B, adenovirus (ADV), hBoV, parainfluenza virus (PIV) 1–3, human metapneumovirus (hMPV), human coronaviruses (hCoV) (229E, OC43, NL63), and hRV [15]. Confirmed RSV-positive samples were further screened for subgroup (A/B) [15] and genotyped by amplifying and sequencing the second hypervariable region of the G protein. The G gene-specific primer set for sequence analysis was as follows: forward primer G(151–173) F: CTGGCAATGATAATCTCAACTTC and reverse primer F(3–22) R: CAACTCCATTGTTATTTGCC [22]. The cDNA was prepared using the viral RNA extraction method employed by the routine respiratory virus test. The reaction mixture contained 5 µl of RNA, which was mixed with a final concentration of 10 mM dNTPs, 20 µM random primer, 1× RT buffer, 200 U of Superscript III reverse transcriptase (Invitrogen, CA, USA), 40 U of RNase-OUT RNase inhibitor (Invitrogen), 25 mM MgCl2, 0·1 mM dithiothreitol (DTT), and RNase-free water in a final volume of 20 µl. The mixture was then incubated at 25 °C for 5 min, 50 °C for 60 min, and 72 °C for 5 min to terminate cDNA synthesis. Next, 5 µl of cDNA were added to a PCR mixture containing 1 µl of SP-Taq DNA polymerase (2·5 U/μl) (Cosmo Genetech, Seoul, South Korea), 36 µl of distilled water, 5 µl of 10× PCR buffer, 1 µl of 10 mM dNTPs, and 1 µl each of the forward and reverse primers (both 20 µM) for the G gene. Primary denaturation was conducted at 95 °C for 10 min, which was followed by 35 cycles of PCR where each cycle comprised denaturation for 40 s at 95 °C, annealing for 40 s at 54 °C, and elongation for 1 min at 72 °C, with a final extension cycle of 5 min at 72 °C. The PCR products were separated by electrophoresis using 1% agarose gel and visualized using 1× SYBR Safe DNA Gel Stain (Invitrogen) [15]. BigDye Terminator ver.3·1 (Applied Biosystem, Foster City, CA, USA) was utilized for the sequencing reaction, and nucleotide sequence analysis was performed using a 3730 DNA Analyzer (Applied Biosystem). Multiple nucleotide sequences were aligned and edited with ClustalW ver.1·8 software. Phylogenetic analysis was executed using the neighbor-joining method with a bootstrap value of 1000 replicates for testing statistical significance of the tree topology using MEGA ver.6·06 software [23]. The representative 14 sequences were submitted to Genbank and assigned accession numbers of KY773693-KY773706. Network analysis was visualized using Cytoscape ver.2·6·1 software.
Statistical analysis
The categorical variables were compared using the two-tailed Chi-squared test and multivariate analysis, and multiple logistic regression analysis was applied to estimate the odds ratio (OR) and 95% confidence interval (CI). Statistical analyses were performed using the R.3·0·1 tool. P-values <0.05 were considered to indicate statistical significance.
RESULTS
Prevalence of respiratory viral cases
Among 4028 analyzed samples, 1635 samples were positive for respiratory viral infections of various types (Table 1). The annual percentage of cases that was positive for respiratory infections were 18·0% (75/416) in 2009, 26·9% (179/665) in 2010, 48·8% (335/686) in 2011, 51·7% (474/917) in 2012, 47·1% (345/732) in 2013, and 37·1% (227/612) in 2014. Most of these infections were due to IFV (overall: 22·8%, type A: 14·3%, type B: 8·5%), hRV (14·2%), and ADV (10·0%). RSV viral infection was detected in ~4·5% (n = 183/4028) of the patients enrolled.
Table 1.
Annual incidence of all respiratory infection cases in Gyeonggi Province from 2009 to 2014
| Overall (n, %) | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | |
|---|---|---|---|---|---|---|---|
| Total enrolled patients | 4028 | 416 | 665 | 686 | 917 | 732 | 612 |
| Positive respiratory infections | 1635 (40·6) | 75 (18·0) | 179 (6·9) | 335 (48·8) | 474 (51·7) | 345 (47·1) | 227 (37·1) |
| ADV infection | 402 (10·0) | 5 (1·2) | 54 (8·1) | 53 (7·7) | 102 (11·1) | 157 (21·4) | 31 (5·1) |
| hBoV infection | 106 (2·6) | 1 (0·2) | 5 (0·8) | 34 (5·0) | 28 (3·1) | 26 (3·6) | 12 (2·0) |
| PIV infection | 194 (4·8) | 1 (0·2) | 1 (0·2) | 28 (4·1) | 69 (7·5) | 50 (6·8) | 45 (7·4) |
| RSV infection | 183 (4·5) | 10 (2·4) | 16 (2·4) | 57 (8·3) | 65 (7·1) | 21 (2·9) | 14 (2·3) |
| RSV subgroup (A/B) | |||||||
| RSV-A | 141 (3·5) | 9 (2·2) | 1 (0·2) | 52 (7·6) | 60 (6·5) | 9 (1·2) | 10 (1·6) |
| RSV-B | 33 (0·8) | 0 | 8 (1·2) | 5 (0·7) | 4 (0·4) | 12 (1·6) | 4 (0·7) |
| RSV-A and B | 1 (0·0) | 0 | 0 | 0 | 1 (0·1) | 0 | 0 |
| hCoV infection | 112 (2·8) | 12 (2·9) | 13 (2·0) | 19 (2·8) | 25 (2·7) | 21 (2·9) | 22 (3·6) |
| hMPV infection | 40 (1·0) | – | 6 (0·9) | 7 (1·0) | 3 (0·3) | 7 (1·0) | 16 (2·6) |
| hRV infection | 573 (14·2) | 45 (10·8) | 60 (9·0) | 137 (20·0) | 181 (19·7) | 63 (8·6) | 87 (14·2) |
| IFV infection | 920 (22·8) | 91 (21·9) | 217 (32·6) | 62 (9·0) | 333 (36·3) | 86 (11·7) | 131 (21·4) |
Bold values are target virus value (RSV) in our study.
Epidemiological and clinical characteristics of patients with viral respiratory infections
The epidemiological and clinical characteristics of patients with viral respiratory infections are summarized in Tables 2 and 3; 50·4% of the patients with viral infections were female (PIV (50·5%), hCoV (54·5%), hMPV (57·5%), and IFV (52·9%)). Among the respiratory infections, hBoV and RSV infected the youngest patients (median age = 2 years), whereas IFV was more prevalent in older children (median age = 7 years) than any other viruses. Clinical symptoms such as fever (overall = 81·3%: pediatric patients, 72·7% vs. adult, 8·5%), cough (overall = 66·5%: pediatric patients, 58·5% vs. adult, 8·0%), nasal discharge (overall = 59·1%: pediatric patients, 52·2% vs. adult, 6·8%), and phlegm (overall = 47·8%: pediatric patients, 43·8% vs. adult, 4·0%) were frequently identified among the infected patients. However, several patients might have had conditions such as febrile seizure, and stomachache that were not listed as clinical symptoms. The most common pre-existing conditions among the patients were asthma (overall = 0·7%: pediatric patients, 0·6% vs. adult, 0·1%) and hypertension (overall = 0·9%: pediatric patients, 0% vs. adult, 0·9%). For example, patients infected with IFV had a high prevalence of asthma (n = 8, 30·8%) and hypertension (n = 8, 25·8%); additionally, these patients showed a high prevalence of antibiotic usage (140/210, 66·7%). Pediatric patients mean below 19 years old and adult means over 20 years old.
Table 2.
Demographic and clinical characteristics of all patients with respiratory infections from 2009 to 2014
| Demographic information (n = 4028) | Clinical information (n = 3478) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female/male | Female (%) | Median age (years) | Age group | Clinical symptoms | |||||||||||
| 0–2 years | 3–6 years | 7–19 years | 20–49 years | 50–64 years | Over 65 years | Fever | Cough | Sore throat | Chill | Headache | Muscular pain | ||||
| All patients (n = 4028,%) |
1997/1967 | 50·4 | 5 | 980 (24·3) | 1412 (35·1) | 913 (22·7) | 432 (10·7) | 163 (4·0) | 83 (2·1) | 2827 (81·3) | 2314 (66·5) | 1403 (40·3) | 1086 (31·2) | 1013 (29·1) | 786 (22·6) |
| ADV (n = 402) | 172/227 | 43·1 | 3 | 120 (29·9) | 229 (57·0) | 48 (11·9) | 1 (0·2) | 2 (0·5) | 0 | 337 (83·8) | 272 (67·7) | 148 (40·3) | 106 (26·4) | 72 (17·9) | 66 (16·4) |
| hBoV (n = 106) | 48/58 | 45·3 | 2 | 60 (56·6) | 36 (34·0) | 8 (7·5) | 1 (0·9) | 1 (0·9) | 0 | 90 (84·9) | 76 (71·7) | 27 (25·5) | 23 (21·7) | 13 (12·3) | 11 (10·4) |
| PIV (n = 194) | 97/95 | 50·5 | 3 | 80 (41·2) | 96 (49·5) | 10 (5·2) | 2 (1·0) | 3 (1·5) | 1 (0·5) | 172 (88·7) | 152 (78·4) | 56 (28·9) | 26 (13·4) | 14 (7·2) | 18 (9·3) |
| RSV (n = 183) | 81/102 | 44·3 | 2 | 116 (63·4) | 58 (31·7) | 6 (3·3) | 1 (0·5) | 1 (0·5) | 1 (0·5) | 159 (86·9) | 154 (84·2) | 37 (20·2) | 32 (17·5) | 17 (9·3) | 7 (3·8) |
| hCoV (n = 112) | 54/45 | 54·5 | 4 | 38 (33·9) | 36 (32·1) | 20 (17·9) | 10 (8·9) | 3 (2·7) | 5 (4·5) | 85 (75·9) | 73 (65·2) | 38 (33·9) | 30 (26·8) | 22 (19·6) | 20 (17·9) |
| hMPV (n = 40) | 23/17 | 57·5 | 3 | 13 (32·5) | 22 (55·0) | 4 (10·0) | 0 | 0 | 1 (2·5) | 31 (77·5) | 29 (72·5) | 13 (32·5) | 11 (27·5) | 3 (7·5) | 5 (12·5) |
| hRV (n = 573) | 273/289 | 48·6 | 3 | 211 (36·8) | 211 (36·8) | 82 (14·3) | 52 (9·1) | 10 (1·7) | 5 (0·9) | 388 (67·7) | 345 (60·2) | 161 (28·1) | 109 (19·0) | 82 (14·3) | 69 (12·0) |
| IFV (n = 920) | 474/422 | 52·9 | 7 | 59 (6·4) | 59 (6·4) | 359 (39·0) | 108 (11·7) | 25 (2·7) | 15 (1·6) | 681 (74·0) | 566 (61·5) | 362 (39·3) | 351 (38·2) | 36 139·2) | 268 (29·1) |
Clinical conditions including febrile seizure and stomachache.
Case was defined based WHO surveillance [43].
Case with acute respiratory illness except influenza.
Table 3.
Clinical characteristics of all patients with stratified by specific age group from 2009 to 2014
| Age group | Clinical information (n = 3478) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Clinical symptoms | ||||||||||
| Fever | Cough | Sore throat | Chill | Headache | Muscular pain | Nasal discharge | Nasal obstruction | Hoarseness | Wheezing | |
| Total (%) | 2827 (81·3) | 2314 (66·5) | 1403 (40·3) | 1086 (31·2) | 1013 (29·1) | 786 (22·6) | 2056 (59·1) | 1398 (40·2) | 473 (13·6) | 183 (5·3) |
| 0–2 years | 1163 (33·4) | 978 (28·1) | 347 (10·0) | 248 (7·1) | 81 (2·3) | 107 (3·1) | 944 (27·1) | 683 (19·6) | 242 (7·0) | 102 (2·9) |
| 3–6 years | 849 (24·4) | 678 (19·5) | 381 (11·0) | 284 (8·2) | 253 (7·3) | 201 (5·8) | 588 (16·9) | 386 (11·1) | 109 (3·1) | 63 (1·8) |
| 7–19 years | 517 (14·9) | 380 (10·9) | 350 (10·1) | 287 (8·3) | 364 (10·5) | 213 (6·1) | 284 (8·2) | 188 (5·4) | 52 (1·5) | 11 (0·3) |
| 20–49 years | 218 (6·3) | 183 (5·3) | 226 (6·5) | 199 (5·7) | 224 (6·4) | 196 (5·6) | 162 (4·7) | 103 (3·0) | 50 (1·4) | 3 (0·1) |
| 50–64 years | 52 (1·5) | 64 (1·8) | 69 (2·0) | 47 (1·4) | 58 (1·7) | 46 (1·3) | 46 (1·3) | 23 (0·7) | 14 (0·4) | 3 (0·1) |
| Over 65 years | 26 (0·7) | 30 (0·9) | 29 (0·8) | 19 (0·5) | 32 (0·9) | 23 (0·7) | 28 (0·8) | 14 (0·4) | 6 (0·2) | 0 |
Annual incidence of RSV and RSV subgroups in patients with respiratory viruses
Among 4028 samples, 1635 samples had positive results for respiratory infections; of these 183 (4·5%) were RSV-positive, as confirmed by multiplex-PCR assays. Using real-time one-step RT–PCR to distinguish between RSV subgroups A and B, we categorized 141 RSV-positive samples as subgroup A (80·6%), 33 as subgroup B (18·8%), and 1 as RSV-A/B coinfection (0·6%). RSV-A was the major subgroup between January 2009 and November 2014, with January 2010 to December 2010 as the only exception. RSV-B (n = 8, 88·9%) was found in the majority of RSV cases from January 2013 to December 2013, while it was detected at only a slightly higher rate (n = 12, 57·1%) than RSV-A infections (n = 9, 42·9%). Most cases of RSV-A (n = 112, 79·4%) were detected from January 2011 to December 2012, whereas most cases of RSV-B (n = 12, 36·4%) were detected from January 2013 to December 2013.
Phylogenetic analysis of RSV and annual distribution of RSV subgroup A and B genotypes
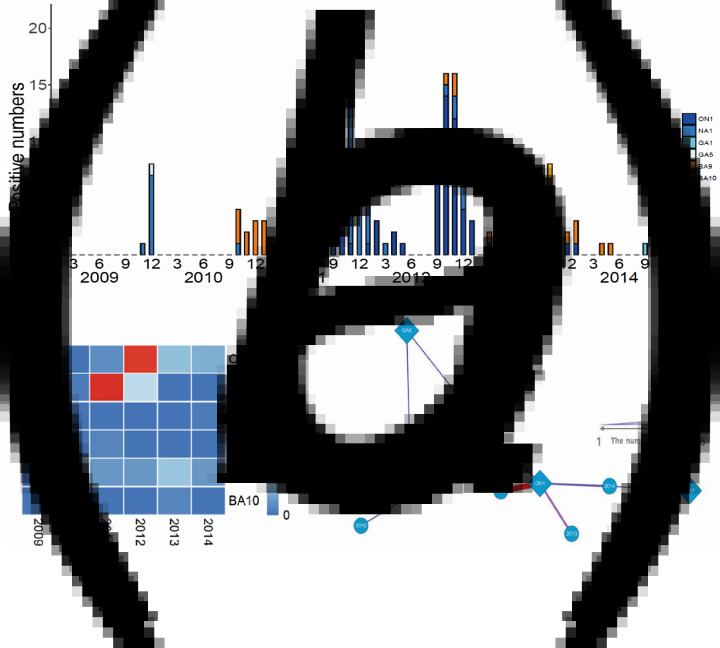
We conducted viral genotyping based on the nucleic acid sequence of the second hypervariable region of the G protein. The target region was sequenced and aligned using MEGA ver.6·06. This phylogenetic analysis included 40 representative RSV-A isolates, 14 RSV-A reference sequences, 31 RSV-B isolates, and 26 RSV-B reference sequences. Of the RSV subgroup A (n = 141) and RSV subgroup B (n = 33) samples, 131 RSV-A (92·9%) and 31 RSV-B (93·9%) were sequenced successfully. RSV-A samples clustered in the genotypes of ON1 (n = 61, 43·3%), NA1 (n = 66, 46·8%), GA1 (n = 1, 0·7%), and GA5 (n = 3, 2·1%), whereas RSV-B samples clustered in the genotypes of BA9 (n = 29, 87·9%) and BA10 (n = 2, 6·1%) (Fig. 1). The ON1 and NA1 genotypes (total: n = 127, 96·9%) constituted the majority of RSV-A genotypes, while the BA9 genotype (n = 29, 87·9%) constituted the majority of RSV-B genotypes. In contrast, GA1 and GA5 genotypes (n = 4, 3·1%) in RSV-A, and BA10 genotype (n = 2, 6·1%) in RSV-B were observed less frequently. From 2009 to 2012, the NA1 genotype was consistently identified in the study subjects. The ON1 genotype was newly detected from 2011 to 2014. Between 2011 and 2012, both ON1 and NA1 genotypes were identified; however, the NA1 genotype was not identified from 2013 to 2014. These results suggest that the ON1 genotype of RSV newly emerged and was first isolated in December 2011. Then, it gradually dominated and replaced the NA1 genotype. For the RSV-B genotype, BA9 was observed continuously, except in 2009. There were no other notable genetic changes detected in RSV-B genotypes during the study period (Table 4, Fig. 2).
Fig. 1.
Annual distribution of RSV genotypes in Gyeonggi Province from 2009 to 2014. (a) Seasonal distribution of RSV genotypes. (b, c) Heatmap and network analyses show the relationship between isolation year and RSV genotype.
Table 4.
Annual incidence of infections with all respiratory viruses, RSV, RSV subgroups (A/B) and genotypes in Gyeonggi Province from 2009 to 2014
| Overall (n, %) | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | |
|---|---|---|---|---|---|---|---|
| Overall patients | 4028 | 416 | 665 | 686 | 917 | 732 | 612 |
| Positive respiratory infections | 1635 (40·6) | 75 (18·0) | 179 (26·9) | 335 (48·8) | 474 (51·7) | 345 (47·1) | 227 (37·1) |
| RSV positive infection | 183 (11·3) | 10 (13·3) | 16 (8·9) | 57 (17·0) | 65 (13·9) | 21 (6·1) | 14 (6·2) |
| RSV subgroup (A/B) (n = 175) | |||||||
| RSV-A-positive infection | 141 (80·6) | 9 (100) | 1 (11·1) | 52 (91·2) | 60 (90·9) | 9 (42·9) | 10 (71·4) |
| RSV-B-positive infection | 33 (18·9) | 0 | 8 (88·9) | 5 (8·8) | 4 (6·1) | 12 (57·1) | 4 (28·6) |
| RSV-A and B coinfection | 1 (0·6) | 0 | 0 | 0 | 1 (1·5) | 0 | 0 |
| Total | 175 | 9 | 9 | 57 | 66 | 21 | 14 |
| RSV-A genotype (n = 131) | |||||||
| ON1 genotype | 61 (43·3) | 0 | 0 | 3 (5·8) | 44 (73·3) | 8 (88·9) | 6 (60·0) |
| NA1 genotype | 66 (46·8) | 8 (88·9) | 1 (100) | 45 (86·5) | 12 (20·0) | 0 | 0 |
| GA1 genotype | 1 (0·7) | 0 | 0 | 0 | 0 | 0 | 1 (10·0) |
| GA5 genotype | 3 (2·1) | 1 (11·1) | 0 | 2 (3·8) | 0 | 0 | 0 |
| Not classified | 10 (7·1) | 0 | 0 | 2 (3·8) | 4 (6·7) | 1 (11·1) | 3 (30·0) |
| Total | 141 | 9 | 1 | 52 | 60 | 9 | 10 |
| RSV-B genotype (n = 31) | |||||||
| BA9 genotype | 29 (87·9) | 0 | 8 (100) | 4 (80·0) | 4 (100) | 9 (75·0) | 4 (100) |
| BA10 genotype | 2 (6·1) | 0 | 0 | 0 | 0 | 2 (16·7) | 0 |
| Not classified | 2 (6·1) | 0 | 0 | 1 (20·0) | 0 | 1 (8·3) | 0 |
| Total | 33 | 0 | 8 | 5 | 4 | 12 | 4 |
Fig. 2.
Phylogenetic analysis of the RSV-positive samples using partial G protein sequence based on nucleotide sequences of (a) RSV subgroup A (b) RSV subgroup B. ( ) Indicates HRSV-A genotype, and (
) Indicates HRSV-A genotype, and ( ) indicates RSV-B genotype.
) indicates RSV-B genotype.
General information and clinical characteristics of patients with RSV subgroups and genotypes
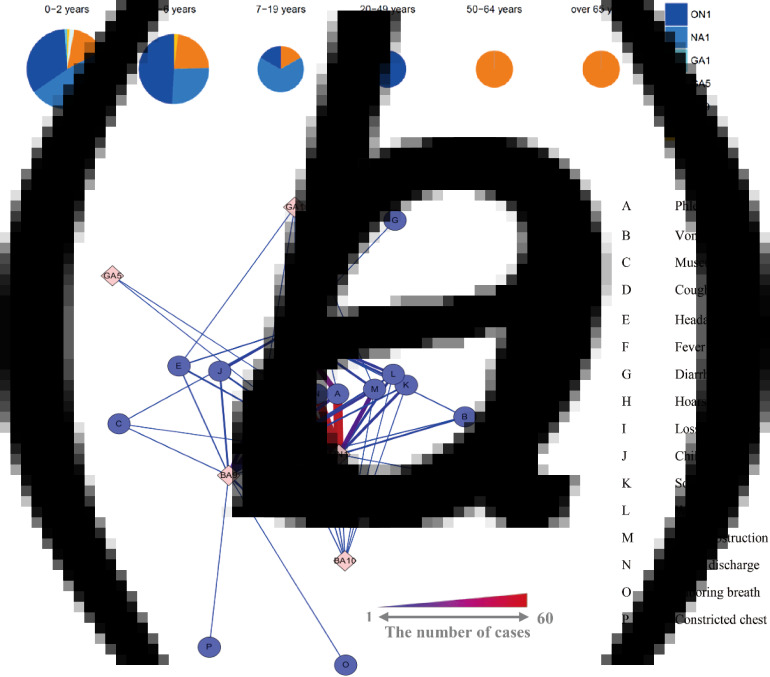
Table 5 summarizes the demographic and clinical characteristics of patients associated with RSV subgroups and genotypes. The ON1 genotype (n = 31, 50·8%), RSV subgroup B (n = 19, 55·9%), BA9 genotype (n = 15, 51·7%), and BA10 genotype (n = 2, 100%) had more females than males. All RSV subgroups and genotypes were associated with the young patient group (median age of below 3 years) and the GA5 genotype with the youngest group (median age of 1·0 years). Most of the cases were 0–2 years of age (n = 116, 63·4%); however, two older patients (1 in the 50–65 year range and 1 >65 years) were infected with RSV subgroup B (BA9 genotype) (Fig. 3a). We performed network analysis to address the correlation between RSV genotype and clinical symptoms (Fig. 3b). Fever, cough, nasal discharge, and phlegm were the most common clinical symptoms; however, the ON1 genotype was associated with slightly more nasal discharge and phlegm than the NA1 genotype. Subjects infected with the BA9 genotype (RSV-B) had labored breathing, which is generally regarded as a serious symptom (Table 5).
Table 5.
Demographic and clinical characteristics of patients in the RSV, RSV subgroups (A/B), and RSV genotype groups from 2009 to 2014
| Demographic information (n = 183) | Clinical information (n = 183) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female/male | Female (%) | Median age (years) | Age group | Clinical symptoms | ||||||||
| 0–2 years | 3–6 years | 7–19 years | 20–49 years | 50–64 years | Over 65 years | Fever | Cough | Sore throat | ||||
| RSV (n = 183) | 81/102 | 44·3 | 2·0 | 116 (63·4) | 58 (31·7) | 6 (3·3) | 1 (0·5) | 1 (0·5) | 1 (0·5) | 159 (86·9) | 154 (84·2) | 37 (20·2) |
| RSV-A (n = 142) | 59/82 | 41·8 | 2·0 | 94 (66·2) | 42 (29·6) | 5 (3·5) | 1 (0·7) | 0 | 0 | 129 (90·8)) | 125 (88·0) | 30 (21·1) |
| ON1 genotype (n = 61) | 31/29 | 51·7 | 2·0 | 35 (57·4) | 24 (39·3) | 1 (1·6) | 1 (1·6) | 0 | 0 | 60 (98·4) | 57 (93·4) | 14 (23·0) |
| NA1 genotype (n = 66) | 23/43 | 34·8 | 1·5 | 49 (74·2) | 13 (19·4) | 4 (6·0) | 0 | 0 | 0 | 57 (86·4) | 56 (84·8) | 12 (18·2) |
| GA1 genotype (n = 1) | 0/1 | 0 | 2·0 | 1 (100·0) | 0 | 0 | 0 | 0 | 0 | 1 (100·0) | 1 (100·0) | 0 |
| GA5 genotype (n = 3) | 0/3 | 0 | 1·0 | 3 (100·0) | 0 | 0 | 0 | 0 | 0 | 1 (33·3) | 1 (33·3) | 0 |
| RSV-B (n = 34) | 19/15 | 55·0 | 3·0 | 16 (47·1) | 15 (44·1) | 1 (2·9) | 0 | 1 (2·9) | 1 (2·9) | 26 (76·5) | 26 (76·5) | 9 (26·5) |
| BA9 genotype (n = 29) | 15/14 | 51·7 | 2·0 | 15 (51·7) | 11 (37·9) | 1 (3·4) | 0 | 1 (3·4) | 1 (3·4) | 21 (72·4) | 21 (72·4) | 7 (24·1) |
| BA10 genotype (n = 2) | 2/0 | 100 | 3·0 | 1 (50·0) | 1 (50·0) | 0 | 0 | 0 | 0 | 2 (100·0) | 2 (100·0) | 1 (50·0) |
Fig. 3.
Demographic and clinical characteristics of patients with RSV genotypes. (a) RSV genotype incidence rates stratified by patient age. (b) Associations between RSV genotypes and clinical symptoms in patients. Thicker red lines indicate stronger relationships.
Multiple logistic regression analysis of association of clinical characteristics in patients with RSV and RSV subgroups and genotypes
We performed multiple logistic regression analysis to understand the association between clinical features and RSV by dividing into pediatric patients (below 19 years) and adult (over 20 years) as a separate group. Demographical information (age and gender) and 16 clinical symptoms were included in the statistical analysis. OR values >1 indicate a positive association. Clinical symptoms such as cough (OR = 2·8, 95% CI 1·6–5·1), wheezing (OR = 2·8, 95% CI 1·7–4·4), and vomiting (OR = 2·2, 95% CI 1·2–3·9) were significantly higher in RSV-infected groups compared with non-RSV-infected groups on pediatric patients (P < 0·05). No statistically significant ORs were observed in the RSV subgroup A and BA9 genotype. In the ON1 genotype, only phlegm (OR = 11·8, 95% CI 3·8–46·7) was statistically significant (P < 0·05) and the NA1 genotype showed a significant association with gender (males, OR = 2·4, 95% CI 1·1–5·4) and chills (OR = 5·1, 95% CI 1·1–27·1). RSV subgroup B showed a significant association with nasal obstruction (OR = 4·6, 95% CI 1·2–20·0), but no other symptoms showed a significant association (Table 6). However, No statistically significant ORs were observed in the RSV-infected groups compared with non-RSV-infected groups on adult (data not shown) and there are only 3 data on adult in RSV subgroups and genotypes that is too little to analyze.
Table 6.
Multiple logistic regression analysis for RSV, RSV subgroups, and RSV genotypes with clinical symptoms on pediatric patients
| Demographic information and clinical information (Odds ratio:95% CI, P-value) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Clinical symptoms | |||||||||
| Age | Gender | Fever | Cough | Sore throat | Chill | Headache | Muscular pain | Nasal discharge | |
| RSV | |||||||||
| OR | 0·7 (0·6–0·8) | 1·1 (0·8–1·5) | 0·3 (0·2–0·7) | 2·8 (1·6–5·1) | 0·8 (0·5–1·3) | 1·5 (0·9–2·4) | 1·1 (0·6–2·0) | 0·4 (0·1–0·8) | 1·3 (0·9–2·1) |
| P-value | <0·001* | 0·718 | <0·01* | <0·001* | 0·406 | 0·092 | 0·739 | <0·05* | 0·219 |
| RSV-A | |||||||||
| OR | 0·9 (0·7–1·1) | 1·8 (0·7–4·9) | 1·3 (0·2–7·7) | 4·0 (0·8–18·3) | 0·5 (0·1–2·4) | 1·0 (0·3–4·6) | 8·2 (0·9–129) | 0·2 (0·0–2·6) | 1·4 (0·4–4·7) |
| P-value | 0·177 | 0·205 | 0·761 | 0·076 | 0·406 | 0·975 | 0·091 | 0·206 | 0·558 |
| ON1 genotype | |||||||||
| OR | 1·2 (1·0–1·5) | 0·5 (0·2–1·1) | 5·4 (0·4–333·3) | 1·5 (0·3–8·6) | 0·7 (0·2–2·2) | 0·3 (0·1–1·3) | 3·1 (0·6–20·5) | 0·3 (0·0–3·9) | 2·2 (0·8–6·6) |
| P-value | 0·059 | 0·109 | 0·297 | 0·596 | 0·506 | 0·114 | 0·208 | 0·424 | 0·129 |
| NA1 genotype | |||||||||
| OR | 0·8 (0·6–1·0) | 2·4 (1·1–5·4) | 1·5 (0·3–7·5) | 2·4 (0·6–10·8) | 1·7 (0·4–6·8) | 5·1 (1·1–27·1) | 0·4 (0·0–2·2) | 1·0 (0·1–10·7) | 1·0 (0·4–2·5) |
| P-value | <0·05* | <0·05* | 0·623 | 0·218 | 0·480 | <0·05* | 0·301 | 0·971 | 0·917 |
| RSV-B | |||||||||
| OR | 1·2 (0·9–1·4) | 0·5 (0·2–1·4) | 0·8 (0·1–5·0) | 0·3 (0·1–1·2) | 1·8 (0·4–7·7) | 1·0 (0·2–4·0) | 0·1 (0·0–1·1) | 5·1 (0·4–70·4) | 0·7 (0·2–2·3) |
| P-value | 0·177 | 0·205 | 0·761 | 0·076 | 0·406 | 0·975 | 0·091 | 0·206 | 0·558 |
| BA9 genotype | |||||||||
| OR | 1·1 (0·9–1·4) | 0·7 (0·3–1·8) | 0·8 (0·1–5·2) | 0·3 (0·1–1·3) | 1·3 (0·3–6·0) | 1·2 (0·3–5·0) | 0·2 (0·0–1·8) | 4·5 (0·3–64·1) | 0·6 (0·2–2·0) |
| P-value | 0·232 | 0·453 | 0·761 | 0·083 | 0·701 | 0·802 | 0·175 | 0·254 | 0·396 |
Statistically significant values are in bold.
Statistically significant.
ND [not determined]: missing data.
ND [not determined]: one data point.
Coinfection with RSV and other respiratory viruses
We investigated simultaneous infections with RSV and other respiratory viruses using a multiplex-PCR assay. Among all tested respiratory viruses, hRV (n = 25, 47·2%) and ADV (n = 12, 22·6%) were detected most frequently with RSV. Most coinfections were detected from January 2011 to December 2012 (n = 45, 84·9%) proportionate with RSV-positive cases. Among all RSV-positive cases (n = 183), RSV single-infection was observed with the most frequency (n = 137, 74·5%), followed by mono-coinfections (n = 39, 21·2%) and dual-coinfections (n = 7, 3·8%) with RSV. Due to the number of cases in our study, only hRV coinfection with RSV (n = 21) could be evaluated with RSV single-infection (n = 138) in demographic and clinical comparisons between patients with RSV single- and coinfections. In RSV single-infections, fever (n = 117, 85·4%), cough (n = 115, 83·9%), nasal discharge (n = 100, 73·0%), and phlegm (n = 88, 64·2%) were the major clinical symptoms. In cases with hRV and RSV coinfection, fever (n = 20, 95·2%), cough (n = 18, 85·7%), nasal discharge (n = 17, 81·0%), and phlegm (n = 17, 81·0%) were the major clinical symptoms (Table 7). We performed a two-tailed χ2 test to test the statistical difference between two independent groups [23]. No significant association was detected between single (n = 137) and overall coinfection (n = 46) or between single (n = 137) and RSV and hRV coinfection (n = 21) (P > 0·05) (data not shown).
Table 7.
Demographic and clinical characteristics of patients with RSV coinfection
| Demographic information (n = 183) | Clinical information (n = 183) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female/male | Female (%) | Median age (years) | Age group | Clinical symptoms | ||||||||
| 0–2 years | 3–6 years | 7–19 years | 20–49 years | 50–64 years | Over 65 years | Fever | Cough | Sore throat | ||||
| RSV single infection (n = 137) | 60/77 | 43·8 | 3·5 | 85 (62·0) | 44 (32·1) | 5 (3·6) | 1 (0·7) | 1 (0·7) | 1 (0·7) | 117 (85·4) | 115 (83·9) | 30 (21·9) |
| RSV + ADV (n = 7) | 5/2 | 71·4 | 1·7 | 6 (85·7) | 1 (14·3) | 0 | 0 | 0 | 0 | 6 (85·7) | 6 (85·7) | 1 (14·3) |
| RSV + PIV1 (n = 3) | 2/1 | 66·7 | 2·3 | 2 (66·7) | 1 (33·3) | 0 | 0 | 0 | 0 | 3 (100·0) | 3 (100·0) | 1 (33·3) |
| RSV + hRV (n = 21) | 11/10 | 52·4 | 2·0 | 15 (71·4) | 6 (28·6) | 0 | 0 | 0 | 0 | 20 (95·2) | 18 (85·7) | 3 (14·3) |
| RSV + IFV (n = 2) | 0/2 | 0 | 6·5 | 0 | 1 (50·0) | 1 (50·0) | 0 | 0 | 0 | 2 (100·0) | 1 (50·0) | 1 (50·0) |
| RSV + hCoV (n = 1) | 0/1 | 0 | 4·0 | 0 | 1 (100·0) | 0 | 0 | 0 | 0 | 1 (100·0) | 1 (100·0) | 1 (100·0) |
| RSV + hBoV (n = 5) | 1/4 | 20·0 | 1·8 | 4 (80·0) | 1 (20·0) | 0 | 0 | 0 | 0 | 3 (60·0) | 3 (60·0) | 0 |
| RSV + ADV + hBoV (n = 2) | 1/1 | 50·0 | 1·5 | 2 (100·0) | 0 | 0 | 0 | 0 | 0 | 2 (100·0) | 2 (100·0) | 0 |
| RSV + ADV + hCoV (n = 1) | 0/1 | 0 | 4·0 | 0 | 1 (100·0) | 0 | 0 | 0 | 0 | 1 (100·0) | 1 (100·0) | 0 |
| RSV + ADV + hRV (n = 2) | 0/2 | 0 | 1·5 | 2 (100·0) | 0 | 0 | 0 | 0 | 0 | 2 (100·0) | 2 (100·0) | 0 |
| RSV + hCoV + hRV (n = 1) | – | – | 3·0 | 0 | 1 (100·0) | 0 | 0 | 0 | 0 | 1 (100·0) | 1 (100·0) | 0 |
| RSV + hMPV + hRV (n = 1) | 1/0 | 100·0 | 3·0 | 0 | 1 (100·0) | 0 | 0 | 0 | 0 | 1 (100·0) | 1 (100·0) | 0 |
DISCUSSION
In this study, we investigated the distributions of all respiratory viruses, the RSV subgroups, and the coinfections of RSV with other respiratory viruses using real-time one-step RT–PCR. The RSV genotypes were determined by sequencing the second hypervariable region of the G protein, and the results were correlated with the demographic and clinical characteristics from consented clinical information from patients.
RSV evolved and changed continuously after an RSV presumed outbreak reported in 1941 [24]. For six consecutive years in Gyeonggi Province located in South Korea, RSV-A and B cocirculated; RSV-A predominated during most seasons, but RSV-B predominated in 2010 and 2013 (Table 4). This tendency was consistent with the results from previous studies in northern Italy [25] and Japan [26], but differed somewhat from the results from other countries in East Asia [8, 27, 28]. The difference appears to be due to various geographic regions and periods. Consistent with previous reports, higher proportions of infected patients were male [8, 25, 28], and most RSV infections were detected in patients <2 years of age [29]. In terms of seasonality, RSV epidemics commonly span the months from October to March, peaking between November and January [30], and our study showed an analogous pattern. However, different seasonal patterns were noted in tropical regions such as Vietnam, with peaks in the hot and dry season (July to September), suggesting a role of climate parameters such as temperature and relative humidity [23].
After the first discovery of the ON1 genotype in Ontario, Canada in 2010 [14], the ON1 genotype was reported in many other countries, including Japan [31], China [32], Germany [17], Italy [16], India [33], and South Africa [34]. In our study, the ON1 genotype emerged in October 2011. In South Korea, the first ON1 genotype was discovered in August 2011 [35], which was the next season after Eshaghi et al. first discovered the novel ON1 genotype in Canada [14, 15]. Other previous studies in South Korea [15, 35] used clinical samples collected from the Korean Influenza and Respiratory Viruses Surveillance System, which was incorporated nationwide with the Institute of Health and Environment. Therefore, the first ON1 genotype discovered in August 2011 may be from clinical specimens collected from another province in South Korea. From 2009 to 2010, most RSV subgroup A infections were clustered in the NA1 genotype; from 2011 to 2012, the NA1 genotype and ON1 genotypes coexisted but demonstrated opposite predominance: 2011 showed genotype distributions of 92% NA1 and 6% ON1, whereas 2012 showed distributions of 21% NA1 and 79% ON1. Furthermore, no cases of NA1 genotype (subgroup A) were detected from January 2013 to November 2014. Based on the pattern of disease epidemics, we surmise that the NA1 genotype (ancestor of ON1) was replaced with the ON1 genotype during 2011 to 2012. Such substitutions were observed in studies conducted in other countries [23, 25].
The results of many studies [14, 36] demonstrate that the main characteristic of the ON1 genotype is the presence of a 72-nucleotide duplication insertion in the G protein, and this characteristic may have a positive evolutionary effect on RSV. For example, genetic variations may play a crucial biological role for enhancing the efficiency of viral attachment to the cell receptors or for faster viral replication during pathogenesis [23], which may elicit strong resistance to herd immunity [27]. The BA genotype of RSV-B gained a 60-nucleotide duplication insertion in the G protein, a change that occurred since the BA genotype was first isolated in 1999 (Buenos Aires, Argentina) [12]. The GA1 genotype was newly isolated in September 2014, but reports showed that it was isolated previously in 2009. In addition, isolation of the GA1 genotype was reported in Senegal, Africa in 2015 [36] (Table 4).
Demographic and clinical information is important to complement the limitations of advanced molecular technology, as it can take several days to identify a respiratory virus genotype in an infected patient. Therefore, the relevance of clinical information and RSV-positive infections has been investigated continuously through various studies [16, 23, 25, 28, 36]. We performed two analyses with respect to the clinical relevance of RSV-positive infections. We first performed a network analysis using simple frequencies, and then conducted a multiple logistic regression analysis. In RSV-infected patients, clinical symptoms such as fever, cough, nasal discharge, and phlegm were major signs. Serious symptoms such as labored breathing (n = 1) and constricted chest (n = 1) were reported in patients with the BA9 genotype in RSV-B, however, the low frequency prevented a robust analysis (Table 5, Fig. 3b). On the other hand, multiple logistic regression analysis on pediatric patients showed that RSV-positive infections were associated with a 2·8-fold increased risk of cough and wheezing. Furthermore, the ON1 genotype was associated with an 11·8-fold increased risk of phlegm, and the NA1 genotype was associated with a 5·1-fold increased risk of chills and RSV subgroup B was associated with a 4·6-fold increased risk of nasal obstruction (Table 6). However, these results had limitations because the demographic and clinical information was not fully recorded in some patients and our study was performed through local hospital in Gyeonggi Province and these hospitals were all primary hospital. Therefore, most of clinical symptoms were mild and so few patients had serious symptoms. Further studies will be necessary to overcome these limitations and understand the exact associations of clinical symptoms and respiratory virus genotypes.
Many studies have reported contradictory results about coinfection and clinical severity between RSV subgroup and genotype [18, 19, 21, 28, 37–40]. hRV was the most commonly detected virus (47·2%) with RSV among all respiratory viruses in Gyeoggi Province (Table 7). These results were consistent with other studies in China, Spain, and Norway [18, 19, 28, 40]. No significant associations were detected between coinfection and the severity of clinical symptoms between RSV single-infection and overall coinfection, and between RSV single-infection and hRV and RSV coinfection (data not shown); these results were consistent with those reported in studies conducted in Vietnam [37], UK [38], and Spain [18, 19]. However, other studies have shown that coinfections with RSV and respiratory viruses such as hRV, hMPV, and PIV type 3 increase clinical severity [39]. Recently, experimental studies were performed to determine the mechanism underlying coinfection and clinical severity. According to Pinky and Dobrovolny [21], one virus can block another during coinfection by being the first to infect the available host cells. hRV, which is a fast-growing virus, reduces the replication of the remaining viruses. The slowest growing virus is suppressed in the presence of another virus, and the effect of viral coinfections on clinical outcomes is no more severe than a single-virus infection, and can even result in a less severe clinical impact. Skjerven et al. [40] reported that viral genomic load shows a positive correlation with disease severity. In other words, clinical severity depends on the viral genomic load rather than coinfection. However, other studies reported no correlation between viral load and clinical symptoms [41, 42]. Further study is warranted to investigate this clinical aspect in future.
We investigated the molecular epidemiological characteristics of RSV in patients with viral respiratory infections. The association between clinical symptoms in patients and the molecular epidemiology of RSV infections will be useful for developing treatment therapies and vaccines against RSV.
ACKNOWLEDGEMENT
This study was supported by the Intramural Research Fund (grant no. 4851-300) of the Korea National Institute of Health.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Borchers AT, et al. Respiratory syncytial virus – a comprehensive review. Clinical Reviews in Allergy & Immunology 2013; 45: 331–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glezen WP, et al. Risk of primary infection and reinfection with respiratory syncytial virus. American Journal of Diseases of Children 1986; 140: 543–546. [DOI] [PubMed] [Google Scholar]
- 3.Sorce LR. Respiratory syncytial virus: from primary care to critical care. Journal of Pediatric Health Care 2009; 23: 101–108. [DOI] [PubMed] [Google Scholar]
- 4.Hogan AB, et al. Exploring the dynamics of respiratory syncytial virus (RSV) transmission in children. Theoretical Population Biology 2016; 110: 78–85. [DOI] [PubMed] [Google Scholar]
- 5.Meng J, et al. An overview of respiratory syncytial virus. PLoS Pathogens 2014; 10: e1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slovic A, et al. A molecular epidemiological study of human respiratory syncytial virus in Croatia, 2011–2014. Infection, Genetics and Evolution 2016; 44: 76–84. [DOI] [PubMed] [Google Scholar]
- 7.Johnson PR, et al. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proceedings of the National Academy of Sciences of the United States of America 1987; 84: 5625–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui G, et al. Genetic variation in attachment glycoprotein genes of human respiratory syncytial virus subgroups A and B in children in recent five consecutive years. PLoS ONE 2013; 8: e75020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano E, et al. Molecular evolution of human respiratory syncytial virus attachment glycoprotein (G) gene of new genotype ON1 and ancestor NA1. Infection, Genetics and Evolution 2014; 28: 183–191. [DOI] [PubMed] [Google Scholar]
- 10.Cane PA, Pringle CR. Evolution of subgroup A respiratory syncytial virus: evidence for progressive accumulation of amino acid changes in the attachment protein. Journal of Virology 1995; 69: 2918–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otieno JR, et al. Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya. Journal of Virology 2016; 90: 4990–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trento A, et al. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. Journal of General Virology 2003; 84: 3115–3120. [DOI] [PubMed] [Google Scholar]
- 13.Trento A, et al. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. Journal of Virology 2010; 84: 7500–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eshaghi A, et al. Genetic variability of human respiratory syncytial virus A strains circulating in Ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS ONE 2012; 7: e32807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y-J, et al. Rapid replacement of human respiratory syncytial virus A with the ON1 genotype having 72 nucleotide duplication in G gene. Infection, Genetics and Evolution 2014; 26: 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierangeli A, et al. Rapid spread of the novel respiratory syncytial virus A ON1 genotype, central Italy, 2011 to 2013. Eurosurveillance 2014; 19: 20843. [DOI] [PubMed] [Google Scholar]
- 17.Prifert C, et al. Novel respiratory syncytial virus a genotype, Germany, 2011–2012. Emerging Infectious Diseases 2013; 19: 1029–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Roig A, et al. Viral coinfection in childhood respiratory tract infections. Archivos de Bronconeumología (English Edition) 2015; 51: 5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvo C, et al. Respiratory syncytial virus coinfections with rhinovirus and human bocavirus in hospitalized children. Medicine 2015; 94: e1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asner SA, et al. Is virus coinfection a predictor of severity in children with viral respiratory infections? Clinical Microbiology and Infection 2015; 21: 264.e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinky L, Dobrovolny HM. Coinfections of the respiratory tract: viral competition for resources. PLoS ONE 2016; 11: e0155589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antoniassi da Silva LH, et al. Genetic variability in the G protein gene of human respiratory syncytial virus isolated from the Campinas metropolitan region, Brazil. Journal of Medical Virology 2008; 80: 1653–1660. [DOI] [PubMed] [Google Scholar]
- 23.Yoshihara K, et al. Association of RSV-A ON1 genotype with increased pediatric acute lower respiratory tract infection in Vietnam. Scientific Reports 2016; 6: 27856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams JM. Primary virus pneumonitis with cytoplasmic inclusion bodies: study of an epidemic involving thirty-two infants, with nine deaths. Journal of the American Medical Association 1941; 116: 925–933. [Google Scholar]
- 25.Esposito S, et al. Characteristics and their clinical relevance of respiratory syncytial virus types and genotypes circulating in Northern Italy in five consecutive winter seasons. PLoS ONE 2015; 10: e0129369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dapat IC, et al. New genotypes within respiratory syncytial virus group B genotype BA in Niigata, Japan. Journal of Clinical Microbiology 2010; 48: 3423–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui G, et al. Rapid replacement of prevailing genotype of human respiratory syncytial virus by genotype ON1 in Beijing, 2012–2014. Infection, Genetics and Evolution 2015; 33: 163–168. [DOI] [PubMed] [Google Scholar]
- 28.Yu X, et al. Human respiratory syncytial virus in children with lower respiratory tract infections or influenza-like illness and its co-infection characteristics with viruses and atypical bacteria in Hangzhou, China. Journal of Clinical Virology 2015; 69: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gimferrer L, et al. Molecular epidemiology and molecular characterization of respiratory syncytial viruses at a tertiary care university hospital in Catalonia (Spain) during the 2013–2014 season. Journal of Clinical Virology 2015; 66: 27–32. [DOI] [PubMed] [Google Scholar]
- 30.Thorburn K. Respiratory syncytial virus–more chimera than chimpanzee? Current Medical Research and Opinion 2016; 32: 699–701. [DOI] [PubMed] [Google Scholar]
- 31.Tsukagoshi H, et al. Genetic analysis of attachment glycoprotein (G) gene in new genotype ON1 of human respiratory syncytial virus detected in Japan. Microbiology and Immunology 2013; 57: 655–659. [DOI] [PubMed] [Google Scholar]
- 32.Cui G, et al. Emerging human respiratory syncytial virus genotype ON1 found in infants with pneumonia in Beijing, China. Emerging Microbes & Infections 2013; 2: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choudhary M, et al. Genetic variability of human respiratory syncytial virus in Pune, Western India. Infection, Genetics and Evolution 2013; 20: 369–377. [DOI] [PubMed] [Google Scholar]
- 34.Valley-Omar Z, et al. Novel respiratory syncytial virus subtype ON1 among children, Cape Town, South Africa, 2012. Emerging Infectious Diseases 2013; 19: 668–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee W-J, et al. Complete genome sequence of human respiratory syncytial virus genotype A with a 72-nucleotide duplication in the attachment protein G gene. Journal of Virology 2012; 86: 13810–13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fall A, et al. Epidemiology and molecular characterization of human respiratory syncytial virus in Senegal after four consecutive years of surveillance, 2012–2015. PLoS ONE 2016; 11: e0157163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran DN, et al. Molecular epidemiology and disease severity of human respiratory syncytial virus in Vietnam. PLoS ONE 2013; 8: e45436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cebey-López M, et al. Does viral co-infection influence the severity of acute respiratory infection in children? PLoS ONE 2016; 11: e0152481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Míguez A, Iftimi A, Montes F. Temporal association between the influenza virus and respiratory syncytial virus (RSV): RSV as a predictor of seasonal influenza. Epidemiology and Infection 2016; 144: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skjerven HO, et al. Virus type and genomic load in acute bronchiolitis: severity and treatment response with inhaled adrenaline. Journal of Infectious Diseases 2016; 213: 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee N, et al. High viral load and respiratory failure in adults hospitalized for respiratory syncytial virus infections. The Journal of Infectious Diseases 2015; 212: 1237–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franz A, et al. Correlation of viral load of respiratory pathogens and co-infections with disease severity in children hospitalized for lower respiratory tract infection. Journal of Clinical Virology 2010; 48: 239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.World Health Organization. WHO Surveillance Case Definitions for ILI and SARI. Geneva: WHO, 2014. [Google Scholar]