Abstract
Accurate knowledge of pathogen incubation period is essential to inform public health policies and implement interventions that contribute to the reduction of burden of disease. The incubation period distribution of campylobacteriosis is currently unknown with several sources reporting different times. Variation in the distribution could be expected due to host, transmission vehicle, and organism characteristics, however, the extent of this variation and influencing factors are unclear. The authors have undertaken a systematic review of published literature of outbreak studies with well-defined point source exposures and human experimental studies to estimate the distribution of incubation period and also identify and explain the variation in the distribution between studies. We tested for heterogeneity using I2 and Kolmogorov–Smirnov tests, regressed incubation period against possible explanatory factors, and used hierarchical clustering analysis to define subgroups of studies without evidence of heterogeneity. The mean incubation period of subgroups ranged from 2·5 to 4·3 days. We observed variation in the distribution of incubation period between studies that was not due to chance. A significant association between the mean incubation period and age distribution was observed with outbreaks involving only children reporting an incubation of 1·29 days longer when compared with outbreaks involving other age groups.
Key words: Bacterial infections, campylobacter, food-borne zoonoses, gastrointestinal infections, outbreaks
INTRODUCTION
Campylobacteriosis is a zoonotic infection caused by a non-spore-forming Gram-negative bacteria [1]. The most common species reported in human diseases are Campylobacter jejuni and Campylobacter coli [2]. In humans, the main route of transmission of Campylobacter is foodborne. Infection occurs following ingestion of undercooked meat and meat products as well as raw or contaminated milk and milk products. Infection can also follow contact with contaminated animals. Person-to-person transmission is rare but can happen. Abdominal cramps and diarrhoea are the most commonly reported symptoms. Non-specific symptoms that can also occur include headache, chills, fever and muscle pain. The duration of illness is usually about a week, with the severity declining after 24–48 h; however, 20% of cases may have a relapse [3, 4].
According to the World Health Organization (WHO), Campylobacter sp. caused 96 million cases of foodborne illness worldwide in 2010 [5]. It is the most commonly reported zoonosis in the European Union accounting for 45·2 cases per 100 000 people [6, 7]. In the UK, there are approximately 9·3 undiagnosed cases in the community for every case reported to the national surveillance system [8], and an estimated 280 000 cases reported each year resulting in over 100 deaths [1, 9].
A large proportion of reported cases are sporadic, however, outbreaks of campylobacteriosis have been reported with foodborne [10, 11] and non-foodborne [12, 13] sources identified. In the UK, 114 outbreaks were reported between 1992 and 2009, affecting a total of 2676 [14]. Outbreak investigation contributes to the reduction of the burden of disease by identifying the source of infection and informing public health strategies and policies. An effective outbreak investigation requires understanding of certain parameters of the infecting pathogen such as the expected incubation period distribution.
Incubation period, which is the time between infection and onset of clinical symptoms, is also important for surveillance and implementation of appropriate public health interventions. In epidemiological studies, incubation period can be used to estimate the period of exposure, identify and exclude travel-related cases, distinguish secondary cases, and formulate a hypothesis [15]. It can help in diagnosing possible cases in the absence of microbiological diagnosis [16] and also offers insights into clinical and public health practices [15]. Essential to an outbreak investigation is constructing a case definition where a time restriction, sometimes based on the incubation period, is set to correctly classify cases as being part of the outbreak under investigation [17].
As a result of certain factors such as infectious dose, host factors and possibly, food matrix, the incubation period may vary between individuals. These, among other factors result in a distribution of incubation period. The incubation period distribution of campylobacteriosis is not clearly defined with different times being reported. The National Health Service in England and WHO report 2–5 days [18, 19], while the Public Health Agency of Canada report 1–10 days [20]. Incorrect estimations may result in formulating inaccurate case definitions, wrongly defined exposure times, excluding outbreak cases as sporadic or travel related cases and vice versa [21] and misclassifying cases. It is therefore important to correctly estimate the incubation period distribution of campylobacteriosis to support effective outbreak investigations.
Point source outbreaks and human experimental studies, in which healthy volunteers are infected with Campylobacter in order to study certain characteristics of the organism, provide an avenue to study the distribution of incubation period. Outbreaks are natural experiments and the outcome can be dependent on the effect of influencing factors, whereas, experimental studies occur in a controlled environment, with less unknown variation as a predetermined dose is administered, and characteristics of participants are screened to ensure similarities.
This study systematically reviewed literature for outbreaks with well-defined point source exposures and human experimental studies. Reported individual patient incubation periods and summary estimates of the distribution of incubation period were extracted and analysed with the aim of describing the distribution of incubation period, identifying any variation in the distribution between outbreaks above expectation by chance, and attempting to explain any variation identified.
METHODS
Research questions and modified PICO elements
Our research questions were:
What is the distribution of incubation period and the average (mean and median) incubation period of Campylobacter in humans?
-
Is there heterogeneity between the reported incubations times amongst studies?
Can any observe variation be explained?
What factors are affecting the distribution of incubation periods?
Population studied/participants – Laboratory confirmed cases of Campylobacter spp. that form part of an outbreak or experimental infection. Probable cases of campylobacter based on clinical symptoms and case definitions in the context of outbreaks
Infectious agent – Campylobacter spp. (all subspecies included)
Route of infection – Foodborne and non-foodborne
Outcome – Onset of gastroenteritis as described or defined by the authors (diarrhoea, vomiting, nausea, abdominal cramps, etc.)
Search strategy and selection process
A systematic literature search for peer reviewed publications of observational studies and experimental studies reporting incubation period was carried out on PubMed, Google Scholar and ISI Web of Knowledge. We searched for the following words: ‘Campylobacter’, ‘outbreaks’, ‘experimental’, and ‘humans’, combining common variations of the words to create search strings (Supplementary Appendix 1). The reference lists of identified review papers were also screened to find other relevant studies where incubation period of Campylobacter spp. may have been reported. The search was carried out between 21 January and 17 March 2016 and there was no restriction on the dates of articles returned or on the reported species. Articles in languages other than English were excluded.
Each article went through the selection and/or assessment stage, which was done in the following phases:
Screening of titles and abstracts for articles with human campylobacteriosis;
Screening of full text for reporting of incubation period data;
Review of full text to assess quality of incubation period data reported;
Further review of full text to assess exposure times and identify outbreaks with confirmed point source exposures.
The quality assessment undertaken in our review focused on assessing the quality of the incubation period data reported based on a set of criteria developed by one of us (JIH) and not the quality of the overall study. This was done because many of the studies did not necessarily set out to study incubation period, but rather to report on the process of an outbreak investigation or provide evidence on the source of infection in an outbreak. This method of quality assessment enabled us to effectively evaluate the quality of incubation period data reported and the accuracy of the estimation. The set of criteria and corresponding components are listed in Table 1 and a scoring system was used to assess the reported data. Two reviewers were involved in the quality assessment stage, and where there was a difference in opinions, discussions were held until a consensus was reached.
Table 1.
Checklist for assessing incubation period data reported by individual studies (adapted from Hawker et al.)
| Criteria | Component |
|---|---|
| Exposure |
|
| Diagnosis |
|
| Accuracy of measurement |
|
| Ascertainment of bias |
|
Data extraction
Data were extracted from the studies using a pre-determined format (Table 2). General information on the published article, the study characteristics, as well as specific information on the outbreak or experiment, including attack rate and exposure, pathogen and patient characteristics, which might influence incubation time were extracted from each study according to a predetermined format. The outcome information to be measured was quantitative which was available as summary or raw data. All studies reported at least one summary statistic of the incubation period distribution as a mean, median, mode or range. The unit of measurement was in days, and where this was reported in hours, we converted to days.
Table 2.
Details of data extracted from the studies
| Section | Information to be collected |
|---|---|
| General information |
|
| Study characteristics |
|
| Pathogen characteristics |
|
| Outcome data/results |
|
| Other outcome data |
|
| Factors that could affect incubation period |
|
| Any other relevant information |
|
Some studies reported raw incubation period for individual cases either as an epidemic curve or a summary table. Where an epidemic curve was provided, the raw incubation period data were extracted using WebPlotDigitizer version 3.10, which is a free web-based data extraction tool [22]. If a summary table was provided instead, the raw data were also extracted. Where both summary and raw data were provided, the raw data were used for analysis.
Descriptive analyses
Frequencies and percentages were calculated to summarise all studies according to the characteristics identified including: study design (observational or experimental), study type (cohort or case–control study), year of study, Campylobacter species, setting of outbreaks, age description of cases, mode of transmission and food vehicle, where applicable.
Using the extracted raw incubation data, histograms of reported incubation periods of individual cases were plotted to re-create the epidemic curves of the outbreaks. All epidemic curves were plotted using a uniform x-axis indicating the incubation period from 0 to 15 days and above, and an individual y-axis indicating the number of cases involved in each outbreak, which varied according to the graph.
Statistical analyses
The raw incubation period distributions extracted from relevant studies were used to test for heterogeneity in the reported data and describe the pattern of heterogeneity, while the summary statistics calculated from these and extracted summary statistics for outbreaks without individual patient data were used to identify factors that may explain heterogeneity. Statistical analyses were carried out using statistical software R version 3.2.3 (2015-12-10) – ‘Wooden Christmas Tree’ [23].
Testing for heterogeneity
We tested for heterogeneity across studies by deriving the value of I2. A P-value of less than 0·05 from the chi-square test provided statistical evidence of heterogeneity and using the Cochran suggested threshold [24] we interpreted the value of I2 to determine the magnitude of heterogeneity.
We also performed a two sample Kolmogorov–Smirnov test (KS test) to compare the cumulative distributions between the studies. We applied a bootstrapped version of the function with repeat sampling conducted 10 000 times in order to derive P-values that will provide improved coverage due to potential ties in the data comparisons. A small P-value indicated that the incubation period distributions are different, and the null hypothesis was rejected. We compared the resulting P-values to confirm if any variation observed was due to chance by calculating the proportion of P-values below 0·05. The probability of obtaining at least the observed proportion of P-values less than 0·05 was calculated, and if it was <0·01, this provided statistical evidence for variation in incubation time distribution.
Identifying factors that explain heterogeneity
In order to examine if the incubation period was influenced by the outbreak characteristics, we performed a linear mixed effect (random and fixed effects) analysis using the individual incubation period data provided as the dependent variable and the outbreak characteristics as the explanatory variables. We applied a square root transformation to the incubation period to reduce skewness of the data. Outbreak characteristics with sufficient information were included in a full multivariable model. Likelihood ratio tests were used as a means of attaining P-values by comparing the full model to an alternative model, which excluded the variable of interest. A final model was developed by excluding variables without statistical significant association with incubation period (P < 0·1).
So as to allow the inclusion of studies reporting only summary data (mean), we further performed a linear regression analysis. The effect of the explanatory variables on the mean incubation period was estimated by using a univariate model. Where statistical support for an association was observed (P < 0·1), a multivariate model was built, which included the associated variables at that threshold to test for confounding.
Due to insufficient information, organism species was excluded as an explanatory variable in both analyses. The significance level for the final models was chosen to be 5%.
Identifying subgroups of studies for analysis
In the presence of statistically significant heterogeneity, we explored the data using subgroup analyses. However, rather than randomly allocating studies to subgroups, we employed hierarchical cluster analysis to identify subgroups of studies that can be combined. The bootstrapped KS test was used to create a hierarchical cluster to show a graphical representation of how the studies grouped together in terms of their dissimilarities. We subtracted the P-values from one to generate a dissimilarity matrix showing the distances between the samples. The cluster analysis algorithm used was the complete linkage method. The output was a dendrogram showing compact visualisation of the dissimilarity matrix.
In order to reduce the likelihood of observing one significant result due to chance or making a type 1 error, we made pragmatic adjustments to the significance level (0·05) by dividing it by the number of studies included in the KS test which was 30. We then subtracted the adjusted P-value from 1 (1 − α) to derive a cut-off point from which studies without evidence of heterogeneity can be defined within separate clusters. These clusters refer to subgroups of studies that do not have evidence of heterogeneity between them and can be combined for meta-analysis.
Subgroup analyses
We pooled the raw incubation data of studies within a subgroup to create a single dataset for each subgroup, and derived the following summary statistics:
Number of studies included in a subgroup;
Total number of cases (sum of cases in all studies included in a subgroup);
Mean and median incubation period of cases within a subgroup;
Standard deviation (s.d.), variance, skew and kurtosis of incubation period of cases within a subgroup.
The mean attack rate of the studies within a subgroup was also calculated.
A forest plot showing the distribution of the mean incubation period and the corresponding 95% confidence interval (CI) was created. Studies without raw data (eight studies) were allocated to subgroups based on their reported mean and included in the forest plot; however, without a CI as this could not be derived.
Risk of bias
We tested our data for ‘small study-effect’ using a funnel plot to visually examine the relationship between small sample sizes and incubation period.
RESULTS
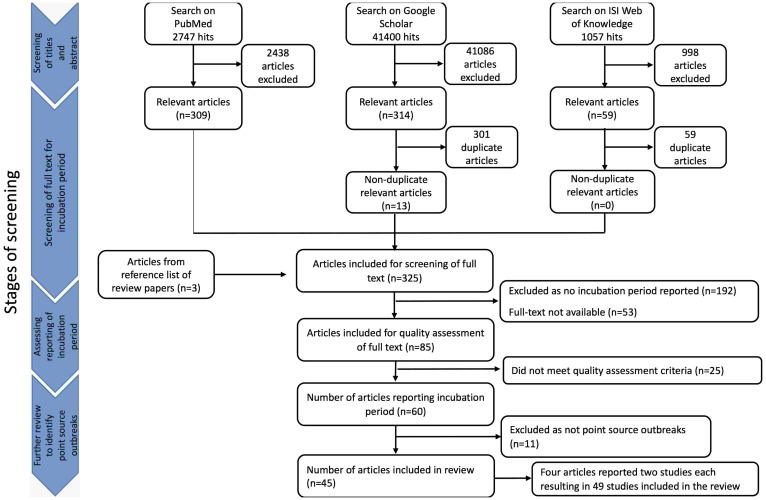
A total of 45 204 search results were retrieved from the three databases and the titles and abstracts were screened for relevance. Exclusion of articles considered irrelevant resulted in 682 articles, and after removing duplicates, 322 articles remained. An additional three articles were identified from searching through the reference list of review papers, resulting in 325 articles available for full text screening for incubation period data. Excluding articles that did not report incubation period and articles that did not meet the quality assessment criteria resulted in 60 articles remaining. These articles were further reviewed to ensure that the reported outbreaks were point source and the reported incubation period was accurate. Excluding outbreaks that were not point source (Supplementary Appendix 2), 45 articles were included in the review (Fig. 1). Four articles reported on two studies each bringing the number of studies included in the review up to 49 (Supplementary Appendix 3). Of these, we were able to extract raw data from 30 studies.
Fig. 1.
Flowchart of study selection process.
Characteristics of studies included in the review
C. jejuni was the most commonly reported species accounting for 75·5% of included studies. Forty-five per cent of the studies were published in year 2000 or later, and 81·6% were carried out in developed countries of Europe and North America (Table 3). Four studies were experimental and the remainder were epidemiological studies undertaken during outbreak investigations to identify the source of infection. Forty-six per cent of these (21/45) were retrospective cohort studies and 29% were descriptive studies.
Table 3.
Characteristics of studies included in review
| N | % | |
|---|---|---|
| Total number of studies | 49 | |
| Year of study | ||
| Before year 2000 | 19 | 38·8 |
| 2000 and later | 22 | 44·9 |
| Unknown | 8 | 16·3 |
| Region of study | ||
| Europe | 20 | 40·8 |
| North America | 20 | 40·8 |
| Australia | 6 | 12·2 |
| Asia | 3 | 6·1 |
| Species | ||
| Campylobacter jejuni | 37 | 75·5 |
| Campylobacter coli | 1 | 2·0 |
| C. jejuni and C. coli | 3 | 6·1 |
| C. jejuni and C. fetus | 1 | 2·0 |
| Unknown | 7 | 14·3 |
| Age distribution | ||
| Mixed ages | 7 | 14·3 |
| Children | 15 | 30·6 |
| Adult | 27 | 55·1 |
| Outbreak setting | ||
| Private party | 14 | 28·6 |
| Farm visit/animal contact | 11 | 22·4 |
| Restaurants | 10 | 20·4 |
| Outdoor activity | 5 | 10·2 |
| School | 5 | 10·2 |
| Experimental study | 4 | 8·2 |
| Food vehicle category | ||
| Poultry | 20 | 40·8 |
| Dairy | 14 | 28·6 |
| Water | 1 | 2·0 |
| Other | 7 | 14·3 |
| Unknown | 7 | 14·3 |
The most common reported setting for outbreaks was private parties (14/49; 28·6%), including weddings and conference dinners, followed by farm visits (11/49; 22·4%). Poultry and dairy were the most frequently reported implicated food vehicle accounting for 40·8% (20/49) and 28·6% (14/49), respectively (Table 3). Comparing the food vehicle and setting of the outbreak, 50% of outbreaks caused by poultry dishes occurred at a private party, and 57·1% of outbreaks caused by dairy or dairy products occurred during a farm visit.
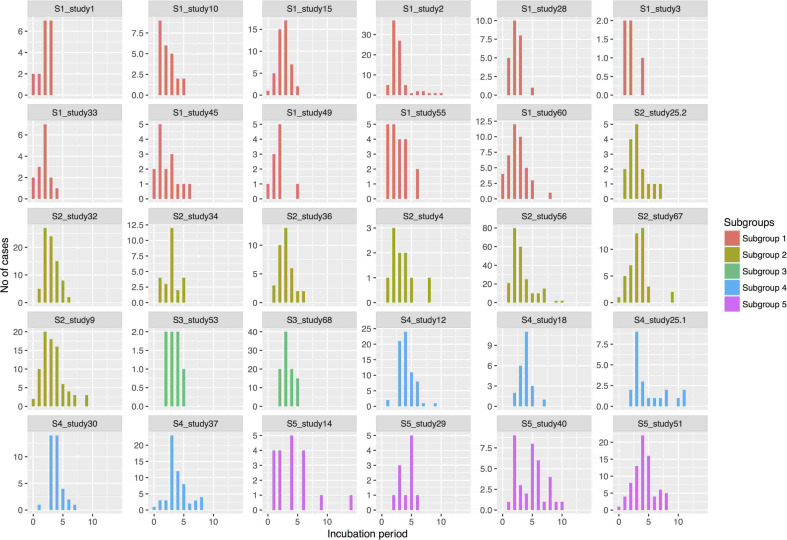
The funnel plot created to test for small study-effect resulted in a symmetric funnel indicating that the size of the study did not have any effect on the reported incubation period (results not shown). From the re-created epidemic curves, we observed a variation in the distribution of incubation period (Fig. 2).
Fig. 2.
Collated epidemic curves re-created from raw data and arranged according to subgroups.
Test of heterogeneity
We calculated that the heterogeneity in the reporting of incubation periods across the different studies was I2 = 72% (P-value for chi-squared ⩽ 0·00001). The proportion of P-values from the KS test that was below 0·05 was >5% ((53%; 231/435). The probability of obtaining the resulting proportion was P<0·00001.
These results indicate a variation in the distribution of incubation periods between studies, which is not due to chance alone.
Factors that may explain heterogeneity
From the linear mixed-effects multivariable analysis and the likelihood ratio tests, age distribution and outbreak setting were significantly associated with incubation period, while food vehicle category showed a weak association with a P-value of 0·08 and met the inclusion criteria into the final model (Table 4). Age distribution and outbreak setting remained significantly associated with incubation period (P < 0·01) in the final model after excluding the non-significant variables (attack rate and year of study) (Table 4).
Table 4.
Linear mixed effect and regression models showing effect of study characteristics on mean incubation period
| Linear mixed effect full model | Linear mixed effect final model | Linear regression univariate analysis | Linear regression multivariable analysis | |||
|---|---|---|---|---|---|---|
| Characteristics | P-value of likelihood ratio test | P-value of likelihood ratio test | Difference in mean incubation period | P-value | Difference in mean incubation period | P-value |
| Attack rate | 0·10 | −0·003 | 0·60 | |||
| Year of study | 0·60 | |||||
| After 2000 | Reference | |||||
| Pre 2000 | 0·19 | 0·57 | ||||
| Age distribution | <0·001 | 0·005 | ||||
| Mixed ages | Reference | Reference | ||||
| Adults | 0·30 | 0·45 | 0·08 | 0·84 | ||
| Children | 1·14 | 0·01 | 1·29 | 0·03 | ||
| Outbreak setting | 0·01 | 0·001 | ||||
| Other | Reference | Reference | ||||
| Farm visit | 0·31 | 0·47 | −0·44 | 0·41 | ||
| Private party | −0·09 | 0·80 | 0·05 | 0·89 | ||
| Restaurant | −0·82 | 0·08 | −0·65 | 0·15 | ||
| School | −0·43 | 0·37 | −0·63 | 0·34 | ||
| Food vehicle category | 0·08 | 0·06 | ||||
| Other | Reference | |||||
| Dairy | −0·03 | 0·95 | ||||
| Poultry | −0·41 | 0·45 | ||||
From the linear regression univariate analysis, age distribution was the only variable with a significant association with the mean incubation period (P < 0·01) with outbreaks involving only children reporting a mean incubation period of 1·14 days longer when compared with mixed outbreaks involving both adults and children. In the final multivariable model also including outbreak setting, as one of the outbreak setting variables had met the inclusion criteria, the association with the mean incubation period remained significant (P < 0·03) with outbreaks involving only children reporting a mean incubation period of 1·29 days longer when compared with mixed outbreaks involving both adults and children (Table 4).
Identifying subgroups of studies
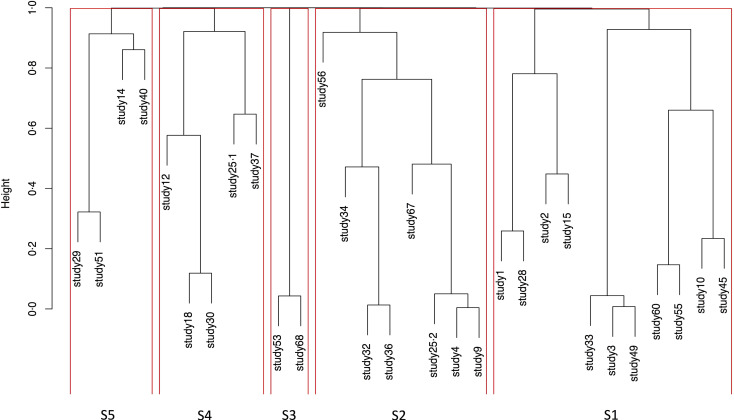
Studies were paired and grouped based on evidence of dissimilarity. Studies found to have the least evidence of dissimilarity between them were paired. Likewise, some studies were not directly paired but attached to other pairs showing that the algorithm could not identify a single study with the least evidence of dissimilarity to them, but instead identified a pair of studies. The resulting output of this cluster analysis is presented as a dendrogram of the dissimilarity matrix (Fig. 3).
Fig. 3.
Dendrogram showing compact visualisation of dissimilarity matrix and identified subgroups.
Following the pragmatic adjustments made to the significance level, the resulting P-value was 0·0017 and the derived cut-off point was 0·9983. Five subgroups were identified using the cut point of 0·9983 to implement the P-value cut point of 0·0017, taking multiple testing into account. These comprised: a subgroup of eleven studies, a subgroup of eight studies and three subgroups of five, four and two studies (Fig. 3).
Summary of subgroup analyses
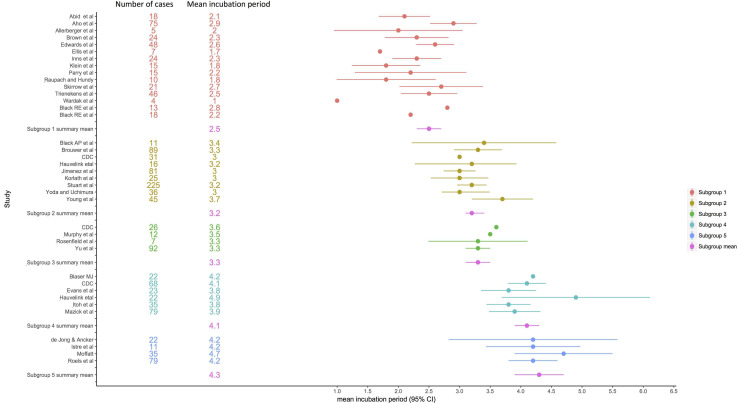
The subgroup containing 11 studies included 302 cases, while the subgroup containing eight studies included 520 cases. The smallest subgroup with two studies also consisted of the lowest number of cases with 102 cases. The mean incubation period of studies in the subgroups varied between 2·5 and 4·3 days (Table 5). There were also substantial differences in the variance, skew and kurtosis between subgroups (Table 5). There was some variation between the studies within subgroups (Fig. 4) albeit not sufficient to evidence difference statistically.
Table 5.
Summary statistics of subgroups
| Frequency | Sum of cases | Attack rate | Median | Mean (95% CI) | Variance | Skew | Kurtosis | |
|---|---|---|---|---|---|---|---|---|
| Subgroup 1 | 11 | 302 | 45·1 | 2 | 2·5 (2·3–2·7) | 2·1 | 1·5 | 4·6 |
| Subgroup 2 | 8 | 520 | 44·4 | 3 | 3·2 (3·1–3·4) | 2·5 | 1·3 | 2·2 |
| Subgroup 3 | 2 | 102 | 26·4 | 3 | 3·3 (3·1–3·5) | 1·0 | 0·3 | −0·9 |
| Subgroup 4 | 5 | 208 | 51·3 | 4 | 4·1 (3·9–4·3) | 2·7 | 1·4 | 3·3 |
| Subgroup 5 | 4 | 145 | 46·4 | 4 | 4·3 (3·9–4·7) | 4·7 | 0·8 | 2·0 |
Fig. 4.
Forest plot showing mean incubation period and 95% CI.
The characteristics of four subgroups were quite similar in terms of the age distribution of cases and food vehicle (Table 6). These four subgroups included outbreaks, which mostly reported poultry as the implicating food vehicle and at least 50% of the outbreaks involved only adults. Food services were reported as an outbreak setting in studies in four subgroups; however, it was the predominant outbreak setting in subgroup 1. The characteristics of subgroup 4 were different with 80% of outbreaks involving only children; dairy products and farm were the most commonly reported food vehicle and outbreak setting, respectively.
Table 6.
Characteristics of studies within subgroups
| Characteristics | Subgroup 1 | Subgroup 2 | Subgroup 3 | Subgroup 4 | Subgroup 5 |
|---|---|---|---|---|---|
| Age | 63% adults | 63% adults | 50% adults | 80% children | 50% adults |
| Food vehicle | 63·6% Poultry | 50% poultry | 100% poultry | 60% dairy | 50% poultry |
| 25% dairy | 20% poultry | ||||
| Setting of outbreaks | 55% Food service | 25% farm | 50% food service | 40% farm | 50% food service |
| 25% school | 50% school | 20% school | 50% school | ||
| 25% food service | |||||
| Severity of illness | 63% | 50% | 50% | 80% | 100% |
| Duration of illness | 0–24 days | 0–20 days | 1–6 days | 0–18 days | 1–9 days |
| Longest incubation period | 10 days | 8 days | 5 days | 11 days | 14 days |
DISCUSSION
Accurate estimations of the period between infection and onset of illness for any infectious disease are essential to support evidence-based interventions in eliminating sources of infection. Our review identified that the reported estimations of the incubation period of campylobacteriosis varied widely, even within subgroups of studies. The results of the I2 and KS tests show that this variation is not due to chance, and there is an underlying pattern of variation. Visual inspection of Figure 2 and the results in Table 5 show that heterogeneity is not only in relation to mean incubation period, but also the shape of the distribution. From both regression analyses, we identified age as a factor that may influence the distribution of incubation period, with reported incubation period in outbreaks affecting children longer than those in mixed age groups. The age structure of cases of campylobacter has changed in recent years with older people becoming increasingly affected [25], and this population shift was reflected in our review where outbreaks investigated after year 2000 mostly involved adults or mixed age groups, while prior to year 2000, more outbreaks involving children were reported.
Furthermore, there appears to be some association between the subgroup characteristics and implicated food vehicle, setting of outbreaks and age of affected cases. However, these differences do not explain all of the variation in distributions of incubation period between subgroups. This may be due to other factors influencing distribution of incubation period that are not evident in the studies or inaccuracy of measurement and reporting. Host characteristics such as underlying medical conditions and immune response [26] as well as dose response have been known to affect infectivity and susceptibility to Salmonella, and may also influence the incubation period of other bacterial infections. These individual patient details have not been provided in the reports, so it is not possible to examine the effect of these factors.
The results of our review might not be generalisable to low- and middle-income countries as majority of included outbreaks and experimental studies took place in high-income countries in Europe and North America. Predisposing factors to campylobacteriosis in low- and middle-income countries, which might also influence incubation period, have been reported to be malnutrition and antimicrobial resistance [27]. A further limitation of the current work is that case definitions varied between studies as authors used different criteria to define cases. The inclusion and exclusion of cases will therefore vary depending on the case definitions used, and this could also affect incubation period. However, all cases were identified at the onset of gastrointestinal symptoms including diarrhoea, vomiting and abdominal cramps, and all were in the context of a known outbreak or experimental study.
Outbreaks that mainly affected children were predominantly caused by consumption of raw milk or raw milk products and exposure was mostly during farm visits. This is similar to the report of Altekruse et al. [28]. The incubation periods of outbreaks involving children were significantly longer than those of outbreaks involving adults or mixed age groups. A review of incubation period of infectious diseases in children reported a similar incubation period to our findings [29].
Our study identified poultry and unpasteurised milk as the most common implicating food vehicles and are known causes of transmission [30, 31]. Studies have identified the presence of virulence genes in both poultry and dairy isolates [32]. However, there is a disparity in the prevalence of Campylobacter in different food products [32], which may result in a variation in acquiring infection as well as incubation period. Also, some type of foods have been known to affect infectivity and thus potentially incubation period of pathogens by being either protective or enabling; an example is fatty food acting as a buffer to protect Salmonella from gastric acid [26].
Infectious dose may have a substantial effect on incubation period distribution, although this may not have varied substantially in the experimental studies included in our review. Studies modelling the dose response of infectious diseases have reported a significant variation in the distribution of incubation period with dose [33, 34]. Human experimental studies of Campylobacter [35] and Salmonella [36] showed a shorter incubation period where the challenge dose was higher. One of the reviewed studies reported a dose–response relationship between the amount of milk consumed and onset of illness and severity, where cases drinking larger amounts of milk had shorter incubation periods and more severe symptoms [10]. A dose–response relationship was also reported in a non-foodborne outbreak involving an outdoor bike race where shorter incubation periods were seen in cases who reported ingesting larger quantities of mud [13]. Another outbreak involving healthy military men who consumed at least 4 litres of untreated surface water during a military training exercise reported no dose–response relationship between the quantity of water consumed and the severity of symptoms [37], however, there was no information on the relationship between ingested dose and incubation period. We were not able to analyse these relationships across the studies due to the lack of individual data related to dose and incubation time.
Host immunity could also influence the incubation period distribution as it determines if an exposure results in illness, and how long the process takes. The development of naturally acquired antibodies in response to a previous infection and the C. jejuni group antigen protects against subsequent illness [35], and may prolong incubation period if illness should occur.
It is worth noting that the bulk of the analyses have been carried out on a subset of studies included in the review from which raw data could be extracted. One problem we encountered in combining results of several studies was the different units of measurement used in reporting. Incubation periods were reported in hours, days or every 2 days. In order to combine the results, we converted all data to days, rounding up or rounding down where necessary. This could result in an over estimation where data were rounded up and an underestimation where data was rounded down and loss of precision for data from some studies. Furthermore, using the online data extraction tool, WebPlotDigitizer, required manual selection of data points, which is open to human error. Separating experimental studies and outbreak reports into relevant subgroups would have been an ideal way of analysing the data; however, there was insufficient information to carry out these analyses, as there were four experimental studies and only two of these reported the mean incubation period.
Exclusion of non-English language articles is appropriate if processing these is inefficient as in our research team and is unlikely to produce bias. Bias would require that non-English papers are associated with different incubation period distributions in outbreaks. However, if there are few eligible studies the translation and inclusion would be warranted. Furthermore, our study population is made up of cases that have been investigated as part of point source outbreaks where incubation period was not the main goal of investigation. This reduces the likelihood of publication bias and selection bias in our study population.
Our results confirm that incubation period in different outbreaks and experiments varied more than can be explained by chance, showed some clustering, and suggested that patient age may contribute to the variation. However, the information provided in the studies was not detailed enough to fully evaluate possible causes for these variations. The ideal data to support identification of factors affecting incubation period would be individual patient data across studies, including information such as underlying conditions, current medications and previous infections. In the absence of access to original individual patient data, reporting of outbreaks could allow better synthesis and meta-regression analysis. Although incubation period is not the main focus of outbreak reports they provide valuable natural experiments to describe incubation period distributions and identify factors affecting this. Increased awareness of the value of this aspect of outbreak reporting can improve the presentation of data to support their use in evidence synthesis.
ACKNOWLEDGEMENTS
The research was funded by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Gastrointestinal Infections at University of Liverpool in partnership with Public Health England (PHE), in collaboration with University of East Anglia, University of Oxford and the Institute of Food Research. Ian Hall is partly funded by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Emergency Preparedness and Response at King's College London and in Modelling Methodology at Imperial, both in partnership with Public Health England (PHE), he is also a Member of NIHR Health Protection Research Units in Emerging and Zoonotic Infections and Gastrointestinal Infections at Liverpool. Adedoyin Awofisayo-Okuyelu is based at University of Oxford. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health or Public Health England.
DISCLAIMER
The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health or Public Health England.
Supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268817001303.
click here to view supplementary material
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Silva J et al. Campylobacter spp. as a foodborne pathogen: a review. Frontiers in Microbiology 2011; 2: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagenaar JA et al. Campylobacter fetus infections in humans: exposure and disease. Clinical Infectious Diseases 2014; 58: 1579–1586. ciu085. doi: 10.1093/cid/ciu085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser MJ et al. Epidemiology of Campylobacter jejuni infections. Epidemiologic Reviews 1983; 5: 157–176. [DOI] [PubMed] [Google Scholar]
- 4.Blaser MJ. Epidemiologic and clinical features of Campylobacter jejuni infections. The Journal of Infectious Diseases 1997; 176(Suppl. 2): S103–S105. [DOI] [PubMed] [Google Scholar]
- 5.Kirk MD et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLOS Med 2015; 12: e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.European Food Safety Authority (EFSA). The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European Union in 2006. EFSA Journal 2007; 6: 130r. [Google Scholar]
- 7.European Food Safety Authority (EFSA). The community summary report on trends and sources of zoonoses and zoonotic agents in the European Union in 2007. EFSA Journal 2009; 7: 223r. [Google Scholar]
- 8.Tam CC et al. Longitudinal study of infectious intestinal disease in the UK (IID2 study): incidence in the community and presenting to general practice. Gut 2012; 61: 69–77; gut.2011.238386. doi: 10.1136/gut.2011.238386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Food Standards Agency. Campylobacter (https://www.food.gov.uk/science/microbiology/campylobacterevidenceprogramme). Accessed 11 July 2016.
- 10.Evans MR et al. A milk-borne campylobacter outbreak following an educational farm visit. Epidemiology and Infection 1996; 117: 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roels TH et al. A foodborne outbreak of Campylobacter jejuni (O:33) infection associated with tuna salad: a rare strain in an unusual vehicle. Epidemiology and Infection 1998; 121: 281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaakoush NO et al. Global epidemiology of Campylobacter infection. Clinical Microbiology Reviews 2015; 28: 687–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stuart TL et al. Campylobacteriosis outbreak associated with ingestion of mud during a mountain bike race. Epidemiology and Infection 2010; 138: 1695–1703. [DOI] [PubMed] [Google Scholar]
- 14.Little CL et al. A recipe for disaster: outbreaks of campylobacteriosis associated with poultry liver pâté in England and Wales. Epidemiology and Infection 2010; 138: 1691–1694. [DOI] [PubMed] [Google Scholar]
- 15.Nishiura H. Early efforts in modelling the incubation period of infectious diseases with an acute course of illness. Emerging Themes in Epidemiology 2007; 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishiura H et al. Incubation period as part of the case definition of severe respiratory illness caused by a novel coronavirus. Euro Surveillance: European Communicable Disease Bulletin 2012; 17: 4–7. [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization. Foodborne disease outbreaks: guidelines for investigation and control (http://www.who.int/foodsafety/publications/foodborne_disease/outbreak_guidelines.pdf). Accessed 24 July 2016.
- 18.NHS Choices. Food poisoning – causes – NHS choices (http://www.nhs.uk/Conditions/Food-poisoning/Pages/Causes.aspx). Accessed 22 July 2016.
- 19.World Health Organization. Campylobacter (http://www.who.int/mediacentre/factsheets/fs255/en/). Accessed 22 July 2016.
- 20.Public Health Agency of Canada. Campylobacter jejuni – Pathogen safety data sheets (http://www.phac-aspc.gc.ca/lab-bio/res/psds-ftss/campylobacter-jejuni-eng.php). Accessed 22 July 2016.
- 21.Horn BJ, Lake RJ. Incubation period for campylobacteriosis and its importance in the estimation of incidence related to travel. Euro Surveillance: European Communicable Disease Bulletin 2013; 18: 17–22. [DOI] [PubMed] [Google Scholar]
- 22.WebPlotDigitizer – Copyright 2010–2016 Ankit Rohatgi (http://arohatgi.info/WebPlotDigitizer/app/). Accessed 11 February 2016.
- 23.R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2015.
- 24.Higgins J, Green S, ed. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 [Updated March 2011]. The Cochrane Collaboration, 2011. Available from www.handbook.cochrane.org. Accessed 22 February 2017. [Google Scholar]
- 25.Nichols GL et al. Campylobacter epidemiology: a descriptive study reviewing 1 million cases in England and Wales between 1989 and 2011. BMJ Open 2012; 2: e001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaser MJ, Newman LS. A review of human salmonellosis: I. Infective dose. Review of Infectious Diseases 1982; 4: 1096–1106. [DOI] [PubMed] [Google Scholar]
- 27.Platts-Mills JA, Kosek M. Update on the burden of Campylobacter in developing countries. Current Opinion in Infectious Diseases 2014; 27: 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altekruse SF et al. Campylobacter jejuni – an emerging foodborne pathogen. Emerging Infectious Diseases 1999; 5: 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.European Centre for Disease Prevention and Control. Systematic Review on the Incubation and Infectiousness/Shedding Period of Communicable Diseases in Children. Stockholm: European Centre for Disease Prevention and Control, 2016. [Google Scholar]
- 30.Bianchini V et al. Prevalence in bulk tank milk and epidemiology of Campylobacter jejuni in dairy herds in Northern Italy. Applied and Environmental Microbiology 2014; 80: 1832–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Sharoud WM. Prevalence and survival of Campylobacter in Egyptian dairy products. Food Research International 2009; 42: 622–626. [Google Scholar]
- 32.Modi S et al. Prevalence of Campylobacter species in milk and milk products, their virulence gene profile and anti-bio gram. Veterinary World 2015; 8: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood RM et al. A dose and time response Markov model for the in-host dynamics of infection with intracellular bacteria following inhalation: with application to Francisella tularensis. Journal of the Royal Society Interface 2014; 11: 20140119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toth DJA et al. Quantitative models of the dose-response and time course of inhalational anthrax in humans. PLoS Pathogens 2013; 9: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Black RE et al. Experimental Campylobacter jejuni Infection in humans. Journal of Infectious Diseases 1988; 157: 472–479. [DOI] [PubMed] [Google Scholar]
- 36.Waddington CS et al. An outpatient, ambulant-design, controlled human infection model using escalating doses of Salmonella typhi challenge delivered in sodium bicarbonate solution. Clinical Infectious Diseases 2014; 58: 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aho M et al. Waterborne outbreak of Campylobacter enteritis after outdoors infantry drill in Utti, Finland. Epidemiology and Infection 1989; 103: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268817001303.
click here to view supplementary material