SUMMARY
Mycobacterium marinum, a bacterium found in freshwater and saltwater, can infect persons with direct exposure to fish or aquariums. During December 2013, the New York City Department of Health and Mental Hygiene learned of four suspected or confirmed M. marinum skin or soft tissue infections (SSTIs) among persons who purchased whole fish from Chinese markets. Ninety-eight case-patients with non-tuberculous mycobacteria (NTM) SSTIs were identified with onset June 2013–March 2014. Of these, 77 (79%) were female. The median age was 62 years (range 30–91). Whole genome sequencing of clinical isolates revealed two main clusters and marked genetic diversity. Environmental samples from distributors yielded NTM though not M. marinum. We compared 56 case-patients with 185 control subjects who shopped in Chinese markets, frequency-matched by age group and sex. Risk factors for infection included skin injury to the finger or hand (odds ratio [OR]: 15·5; 95% confidence interval [CI]: 6·9–37·3), hand injury while preparing fish or seafood (OR 8·3; 95% CI 3·8–19·1), and purchasing tilapia (OR 3·6; 95% CI 1·1–13·9) or whiting (OR 2·7; 95% CI 1·1–6·6). A definitive environmental outbreak source was not identified.
Key words: Mycobacteria, outbreaks, skin infections, soft tissue infections
INTRODUCTION
On December 4, 2013, the New York City (NYC) Department of Health and Mental Hygiene (DOHMH) was notified by an infectious disease physician of four cases of skin or soft tissue infection (SSTI) involving nodules of the hand or arm among women of Chinese ancestry who had handled fish from NYC markets. Patients were evaluated and diagnosed during multiple weeks in late 2013. A skin biopsy from one of the patients yielded an isolate with acid-fast bacilli. Because of skin nodules and history of handling fish, the physician suspected Mycobacterium marinum infection and was concerned an outbreak was occurring. Other than handling fish and Chinese ethnicity, no epidemiological links among the four patients were immediately identified. On January 13, 2014, the physician recontacted DOHMH to report that the acid-fast isolate had been identified as M. marinum and reported two additional cases, bringing the total to six. In January, the US Centers for Disease Control and Prevention (CDC) notified DOHMH about a NYC pathologist who had reported an unusual increase in possible non-tuberculous mycobacteria (NTM) SSTIs and sent biopsies to CDC for laboratory confirmation. DOHMH contacted the pathologist who reported nine additional cases of possible M. marinum infection. These patients had handled fish from Chinese food markets in Brooklyn or Manhattan.
M. marinum is an intermediately growing NTM found in freshwater or saltwater and can cause disease in humans and in fish [1–5]. Human M. marinum infection can cause SSTI with nodules, plaques, or ulcers after a median incubation period of 21 days [6] and can progress to infections of deep tissue requiring surgical management [7]. Handling fish and exposure to fish tanks are known risk factors for M. marinum infection, and minor skin trauma before infection is often reported [8]. Outbreaks of M. marinum infection have been reported among fish farm workers in China and farmers in Micronesia [9, 10]. Other NTM can cause infection with clinical findings similar to M. marinum [11], and >150 NTM species inhabiting surface waters, soils, and potable water systems have been described [5]. Routine bacterial, mycobacterial, and fungal cultures are needed to differentiate NTM from other causes of similarly appearing SSTI (e.g., sporotrichosis and nocardiosis).
The clinical, diagnostic, and pathological features identified in this NTM SSTI outbreak among 29 patients were previously described [12]. Pathological specimens with suppurative granulomas were highly suggestive of NTM SSTI, and both immunohistochemistry staining and polymerase chain reaction (PCR) were found to enhance the yield of laboratory diagnosis [12]. We summarize the public health response to the outbreak, including results of a case–control study to assess risk factors for infection.
METHODS
Case ascertainment and interviews
NTM infections are not reportable to DOHMH unless a cluster of three or more cases or an outbreak is suspected [13]. To actively identify additional cases, DOHMH distributed a citywide health alert on March 5, 2014, notifying clinical providers about the outbreak, recommending that skin biopsy of suspicious lesions be sent for culture and histological staining for acid-fast bacilli, and asking providers to report possible cases to DOHMH [14]. In cooperation with a local Chinese-American medical society, DOHMH held a press conference for Chinese-language news media. The medical society also distributed Chinese translations of the health alert and patient information sheets to its professional membership. DOHMH staff called primary care providers and dermatologists in areas with Chinese immigrant populations in Brooklyn, Manhattan, and Queens to inquire about additional cases. DOHMH staff identified providers through lists maintained by DOHMH and the medical society as serving Chinese communities, and through advertisements for dermatologists and hand surgeons in Chinese-language newspapers. DOHMH staff also contacted NYC pathology laboratories and requested reports from skin biopsies or surgical specimens with suppurative granulomas or granulomatous dermatitis. DOHMH staff also requested that clinical specimens from possible patients be sent to CDC for non-specific mycobacteria immunohistochemical staining (i.e., reacts with tuberculous and non-tuberculous mycobacteria) and PCR testing.
A suspected case was defined as an SSTI with symptom onset during June 1, 2013 to May 31, 2014 in a person who handled fish purchased from a NYC Chinese market (‘market’). A probable case required histopathology reporting granulomas. A confirmed case required either a positive culture or detection by PCR of DNA of M. marinum group species (M. marinum, M. ulcerans, M. liflandii, M. pseudoshottsii, or M. shottsii) or other NTM from a clinical specimen.
A standardized questionnaire was used to interview case-patients. Questions included symptom onset date, types of seafood, frozen fish, or fresh fish (i.e., whole fish alive in a tank or bucket or whole fish on ice) purchased during the 2 months before illness onset, markets where fish or seafood were purchased, history of skin injury (cut, scratch, or other injury) to the finger or hand before or during fish or seafood handling, glove use, aquarium exposure, and gardening.
Environmental investigation
With a New York State (NYS) Department of Agriculture and Markets representative, DOHMH staff visited 10 markets. Market selection was based on association with multiple patients or having a patient who identified the market as the only market patronized. We used a semi-structured questionnaire and interviewed store owners to identify types of fresh fish sold, distributors that supplied fish, and tank cleaning practices. We visited select distributors to collect samples of fish, tank biofilm, and water. Biofilm and water samples were sent to the DOHMH Public Health Laboratory (PHL) and fish samples to the US Department of Agriculture (USDA) National Veterinary Services Laboratories (NVSL).
Laboratory testing
As described previously [12], formalin-fixed paraffin embedded (FFPE) tissues from skin biopsies were tested at CDC with immunohistochemistry, a Mycobacteria genus nested PCR targeting the 16S rRNA gene, and direct sequencing of PCR amplicons for confirmation and speciation.
DOHMH requested all patient NTM isolates from clinical laboratories. Pulsed-field gel electrophoresis (PFGE) of clinical NTM isolates using DNA digestion enzyme XbaI was done at PHL according to standard methods [15] with slight modifications. Whole genome sequencing (WGS) of NTM clinical isolates was conducted at NVSL, using 2 × 250 paired-end chemistry and the Nextera® XT Library Preparation Kit (Illumina, San Diego, California) on a MiSeq® instrument (Illumina, San Diego, California). Because of the diversity in the genomes, a reference independent analysis was selected. ABySS was used to de novo assemble the genomes into contigs. Contigs were verified by blasting them against the National Center for Biotechnology Information (NCBI) nucleotide database, and, if needed, the Mycobacterium sp. contigs were isolated and used in kSNP to create phylogenetic trees and a matrix table, and an unrooted, maximum likelihood phylogenetic tree for the sequenced M. marinum isolates was constructed [16, 17]. For comparison, other M. marinum sequences were included in the analysis, including sequences recovered from different animal species throughout North America at the NVSL, two apparently non-outbreak isolates from the time of the NYC outbreak, an unrelated isolate from the NYS Department of Health PHL, and two NCBI RefSeq M. marinum sequences accessions (NC_010612 and ANPL01) [18].
Biofilm and water samples from distributors were cultured for mycobacteria at PHL, using an N-acetyl-L-cysteine–sodium hydroxide (NALC-NaOH) decontamination procedure, followed by phosphate buffer neutralization, spin, decant, culture in a mycobacterial growth indicator tube at 37 °C, and incubation on Löwenstein–Jensen media at 30 °C. NALC was used for its mucolytic properties and to disrupt biofilms produced by mycobacteria. Fish sampled from tanks were cultured at NVSL for M. marinum, using a 2% NaOH decontamination procedure with media incubation at both 28 and 37 °C.
Case–control study
DOHMH conducted a case–control study to assess risk factors for development of NTM SSTI among persons handling fish or seafood purchased from Chinese markets. Patients meeting the confirmed or probable case definition with symptom onset during June–December 2013 were eligible for inclusion if they responded to questions about fish purchasing practices.
During May 2014, DOHMH staff approached potential controls while they shopped in markets named by patients. Markets in Manhattan or Brooklyn were selected if a patient reported shopping only at that market; or, to be consistent with the distribution of markets named by patients, every Brooklyn market at which at least four patients reported shopping, or every Manhattan market at which at least two patients reported shopping. Markets were excluded if they did not sell fresh or frozen fish, sold fish from only one tank (concerns of limited market size), or were located in Queens (where only 14 (14%) patients reported purchasing fish or seafood).
Interviewers spoke English and at least one of three Chinese languages (Cantonese, Fujianese, or Mandarin). DOHMH created a roster detailing the age group and sex distribution of controls needed for recruitment to frequency match to case-patients with a 3·3:1 ratio. We used 3·3 instead of 3·0 to improve the chances that, given the recruitment challenges (controls solicited at different markets, fluctuating numbers of patients being identified), we would have at least three controls per case-patient. This design was expected to have ⩾80% power to test the hypothesis that a skin injury to the finger or hand was associated with infection (assuming 36 case-patients, 53% exposure among case-patients and ⩽25% exposure among controls, and α = 0·05) [19].
Potential controls were eligible for inclusion if they were NYC residents ages 30 years (minimum age of patients) or older and handled fresh fish from a Chinese market during August–September 2013. Controls were ineligible if they reported having an infection or skin problem of the finger, hand, or arm after June 2013. Interviewers were instructed to approach all potential controls who appeared to meet any of the roster categories (for sex and age) in which enrollment quotas had not been met. To facilitate control interviews, interviewers carried a sheet displaying pictures of fish types with names in English and Chinese characters.
Reference periods selected depended on exposure. For questions relating to cuts or injuries, the reference period for patients was 2 months before illness onset. Although patients might have been asked about incidents that occurred months before the interview, DOHMH suspected patients would likely remember and report cuts or injuries that were temporally associated with their infections. In contrast, for controls, the reference period relating to cuts or injuries was 2 months before interview. This was to obtain a baseline prevalence of cuts and injuries in the population at-risk while attempting to minimize recall bias. For questions relating to purchasing specific types of fish and wearing gloves, the reference period for patients was the 2 months before illness. However, the reference period for controls was August–September 2013 because the majority of patients had illness onset September–November 2013, suggesting exposure to NTM during August–September 2013.
Descriptive and statistical analyses were performed by using SAS® 9·2 (SAS Institute, Incorporated, Cary, North Carolina). Odds ratios (ORs) and 95% confidence intervals (CIs) for potential risk factors were calculated by using multivariable logistic regression and the mid-P exact method [20]. Median unbiased estimates of the OR were reported when warranted. A separate model was created to assess each potential risk factor, adjusting for age group (30–49 years, 50–59 years, 60–69 years, and ⩾70 years), sex, and language of interview (Cantonese, Mandarin or Fujianese, or other). Language was included in models as a potential marker for fish purchasing and preparation practices that might vary across regions in China. Mandarin and Fujianese were combined because some interviewers spoke both languages and the exact Chinese dialect used during the interview was not documented.
The primary analysis included confirmed and probable cases. As a sensitivity analysis, all potential risk factors were assessed when restricting to confirmed cases only. To control for possible seasonal variation in availability of types of fresh fish purchased, we performed a second sensitivity analysis restricted to confirmed cases reporting symptom onset during September–October 2013. Statistical significance was determined as P < 0·05. The DOHMH Institutional Review Board reviewed study objectives and methods and determined the activity to be public health surveillance and non-research.
RESULTS
Case ascertainment and interviews
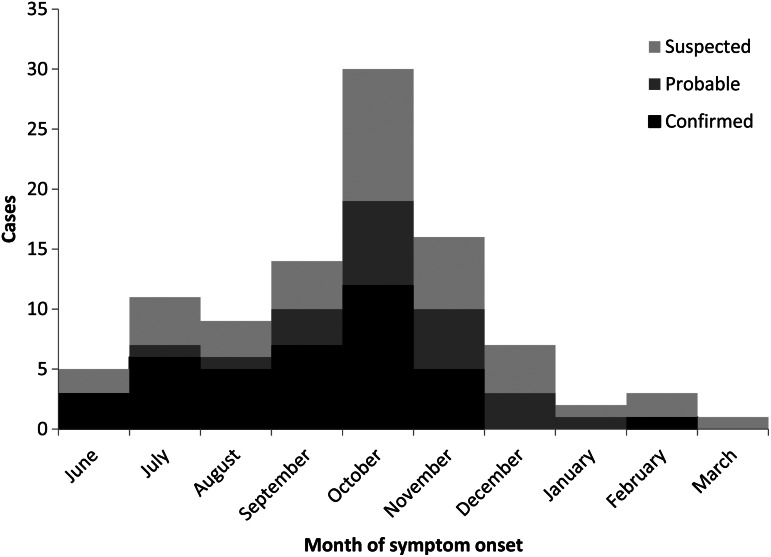
During June 2013–March 2014, we identified 98 cases, including 39 (40%) confirmed, 21 (21%) probable, and 38 (39%) suspected cases (Fig. 1). Sixty (61%) patients reported symptom onset during September–November 2013. Table 1 displays patient demographic characteristics. Seventy-seven (79%) were female; median age was 62 years (range 30–91); 88 (89%) spoke Cantonese, Mandarin, or Fujianese as their primary language; and 59 (60%) lived in Brooklyn, 18 (18%) in Manhattan, 13 (13%) in Queens, and six (6%) outside NYC. Twenty-five (26%) reported working outside the home; reported occupation categories included home health (n = 8) and restaurant or fish butcher (n = 6).
Fig. 1.
Epidemic curve of case-patients with non-tuberculous mycobacterium skin or soft tissue infection after reported fish handling – New York City, 2013–2014.
Table 1.
Demographic characteristics among persons with non-tuberculous mycobacterial skin or soft tissue infection after reported fish handling – New York City, 2013–2014 (N = 98)
| No. | % | |
|---|---|---|
| Female | 77 | 79 |
| Age (years) | ||
| 30–49 | 12 | 12 |
| 50–59 | 28 | 29 |
| 60–69 | 30 | 31 |
| ⩾70 | 28 | 29 |
| Primary language of interview | ||
| Cantonese | 66 | 67 |
| Mandarin or Fujianese | 22 | 22 |
| English | 4 | 4 |
| Russian | 4 | 4 |
| Spanish | 2 | 2 |
| Residence | ||
| Bronx | 0 | 0 |
| Brooklyn | 59 | 60 |
| Manhattan | 18 | 18 |
| Queens | 13 | 13 |
| Staten Island | 2 | 2 |
| Outside of NYC | 6 | 6 |
| Work outside home | 25 | 26 |
| Administrative | 1 | 1 |
| Home health | 8 | 8 |
| Restaurant or fish butcher | 6 | 6 |
| Other | 9 | 9 |
| Unknown | 1 | 1 |
Of 98 case-patients, 90 (92%) completed a standardized interview. Of those completing the interview, during the 2 months before symptom onset, 88 (98%) reported buying fresh fish, 41 (46%) bought frozen fish, 43 (48%) bought fresh seafood, and nine (10%) bought fresh frogs. Approximately 50 markets in Brooklyn, Manhattan, and Queens were mentioned. Sixty-one (68%) case-patients reported having a skin injury to the finger or hand during the 2 months before symptom onset, 10 (11%) reported having a home aquarium, and four (4%) reported having skin contact with aquarium water. Twenty-five (28%) reported gardening. There were no deaths.
Environmental investigation
Store owners reported purchasing fresh fish daily from at least one of four local distributors, and fish were typically sold within 24 h of arrival. Owners reported tanks were cleaned as needed with water and without chemicals. No owner recalled increased numbers of fish appearing sick since June 2013. Of these four distributors, two were visited, distributors A and B. Both reported only selling to NYC markets or restaurants and delivering live fish daily. They also reported that fish were purchased from fisheries in multiple states (ranging from Michigan to North Carolina) and left distributor tanks within 3 days of arrival. Neither distributor reported increased numbers of fish appearing sick since June 2013.
Laboratory testing
Of 98 patients, 39 (40%) had laboratory-confirmed infections. Nineteen were identified with mycobacterial culture of skin biopsy specimens. The remaining 20 were from patients without known mycobacterial culture results and diagnosed by PCR and sequencing of amplicons from FFPE tissue specimens.
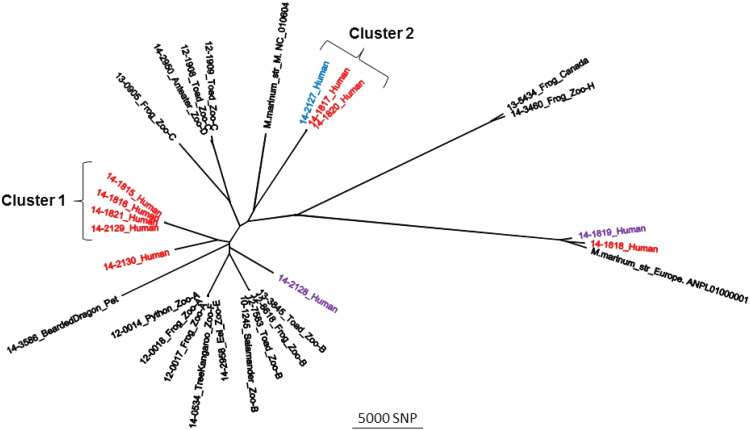
Figure 2 shows the WGS results in an unrooted maximum likelihood phylogenetic tree with eight outbreak isolates in red, one possible outbreak isolate in blue, two unrelated NYS non-outbreak isolates in purple, and 18 USDA veterinary isolates in black. Six of eight M. marinum isolates recovered from outbreak submissions clustered into two groups with intragroup variation of isolates within each of these two clusters of <40 single-nucleotide polymorphisms (SNPs) (see Supplementary material). Of the two remaining outbreak isolates, one (14–2130_Human) varied from isolates in cluster 1 or 2 by at least 8000 SNPs, suggesting a distant common ancestor. The other isolate (14–1818_Human) varied from the isolates in cluster 1 or 2 by at least 25 000 SNPs. Of interest, one non-outbreak isolate (14–2127_Human) varied from the cluster 2 isolates by only 24 SNPs. Lastly, one non-outbreak NYS human isolate (14–1819_Human), an outbreak isolate (14–1818_Human), and a veterinary isolate varied from each other by at least 3000 SNPs. All isolates were deposited in the NCBI Short Read Archive. The assembly statistics, NCBI accession numbers, and a matrix table detailing core SNP differences are included as Supplementary materials.
Fig. 2.
Maximum likelihood phylogenetic tree developed in kSNP using core single-nucleotide polymorphisms (SNPs). The Mycobacterium marinum isolates associated with the 2013–2014 New York City fish handling outbreak are highlighted in red. Two isolates recovered from patients not associated with this outbreak are highlighted in purple, and one isolate originally not considered an outbreak case is highlighted in blue (see text for details). Also included are two reference strains, ‘M’ and ‘Europe’, and various USDA (US Department of Agriculture) isolates recovered from animals (see Supplemental matrix table). Clusters 1 and 2 have intracluster variation of no more than 40 SNPs (see Supplemental matrix table).
PFGE analysis of nine NYC outbreak isolates and three non-outbreak isolates revealed four distinct PFGE patterns. The first included a cluster of six isolates: five had an indistinguishable PFGE pattern (including WGS cluster 1 isolates 14–1815_Human, 14–1816_Human, 14–1821_Human, 14–2129_Human, and a clinical isolate that was not sequenced). The PFGE of the fifth isolate (14–2130_Human) differed from the other four by two bands and initially was considered related. The second PFGE pattern included two outbreak isolates (corresponding to 14–1817_Human and 14–1820_Human) and one non-outbreak isolate (14–2127) with indistinguishable PFGEs. The third PFGE pattern included an outbreak and non-outbreak isolate (14–1818_Human and 14–1819_Human) with indistinguishable PFGEs, and a fourth PFGE pattern (14–2128_Human) was unique. Table 2 compares PFGE and WGS results. PFGE results were concordant with WGS clusters; however, the converse was not found. PFGE did not always distinguish isolates that differed by as much as 3000 SNPs (e.g., 14–1818_Human and 14–1819_Human) (Table 2).
Table 2.
Comparison of WGS and pulsed-field gel electrophoresis results among Mycobacterium marinum outbreak and non-outbreak human isolates – New York City, 2013–2014 (N = 12)
| Isolate name* | Outbreak associated | WGS† cluster | PFGE‡ pattern | WGS and PFGE results discordant |
|---|---|---|---|---|
| 14–1815_Human | Yes | 1 | 1 | |
| 14–1816_Human | Yes | 1 | 1 | |
| 14–1821_Human | Yes | 1 | 1 | |
| 14–2129_Human | Yes | 1 | 1 | |
| 14_Human | Yes | N/A§ | 1 | |
| 14–2130_Human | Yes | Unique¶ | 1b | Yes |
| 14–1817_Human | Yes | 2 | 2 | |
| 14–1820_Human | Yes | 2 | 2 | |
| 14–2127_Human | No | 2 | 2 | |
| 14–1818_Human | Yes | Unique | 3 | Yes |
| 14–1819_Human | No | Unique | 3 | Yes |
| 14–2128_Human | No | Unique | 4 |
DOHMH collected environmental samples for testing from distributor A (three tank biofilm and water samples and 11 live fish) and distributor B (two tank biofilm and water samples and seven live fish). All biofilm samples grew non-M. marinum NTM. M. arupense, a rare human pathogen associated with tenosynovitis [21], was identified from internal organs of three of six tilapia from distributor A and three of three tilapia from distributor B. M. szulgai, a rare human pathogen associated with SSTI [22], was found in one tilapia sample from distributor B. Of six striped bass samples (three each from two distributors), all were negative for NTM.
Case–control study
Of 60 patients meeting the confirmed and probable case definition, 56 (93%) reported symptom onset during June–December 2013, had complete interview data, and were eligible for inclusion. Of 477 potential controls approached at 15 markets, 287 (60%) declined to participate, and five (1%) were excluded because of incomplete questionnaires, leaving 185 (39%) included in analysis.
Interviews were conducted in Cantonese for 44 (79%) case-patients and 123 (66%) controls, Mandarin or Fujianese for eight (14%) case-patients and 52 (28%) controls, and in another language for four (7%) case-patients and 10 (5%) controls. In a model restricting to Chinese speakers and adjusting for age group and sex, case-patients had higher odds of speaking Cantonese than controls (OR 2·4, 95% CI 1·0–5·8).
Table 3 displays the reported frequency of possible risk factors for NTM SSTI related to purchasing or handling fish. Adjusting for age group, sex, and interview language, case-patients (confirmed and probable) and controls had similar odds of purchasing frozen fish, fresh fish, or fresh seafood or wearing waterproof gloves. However, case-patients had higher odds of having had any skin injury to the finger or hand (OR 15·5; 95% CI 6·9–37·3) or having had an injury while preparing fish or seafood (OR 8·3; 95% CI 3·8–19·1). These findings were similar in the sensitivity analysis restricting to confirmed case-patients.
Table 3.
Potential risk factors for non-tuberculous mycobacteria skin or soft tissue infection after reported fish handling – New York City, 2013–2014
| Case-patients (N = 56) | |||||
|---|---|---|---|---|---|
| Control subjects (N = 185) | Confirmed and probable (N = 56) | Confirmed only (N = 36) | |||
| Potential risk factor | No. (%) | No. (%) | OR* (95% CI) | No. (%) | OR* (95% CI) |
| Purchased frozen fish | 84/181 (46) | 26/52 (50) | 1·2 (0·6–2·3) | 17/34 (50) | 1·2 (0·6–2·5) |
| Purchased fresh fish | 181/185 (98) | 56/56 (100) | 1·9 (0·3–∞)† | 36/36 (100) | 1·2 (0·2–∞)† |
| Purchased fresh seafood | 101/171 (59) | 22/55 (40) | 0·5 (0·3–1·0) | 14/36 (39) | 0·5 (0·2–1·1) |
| Removed gills or scales | 73/179 (41) | 26/54 (48) | 1·3 (0·7–2·4) | 18/34 (53) | 1·5 (0·7–3·2) |
| Wore waterproof gloves | 32/170 (19) | 12/54 (22) | 0·8 (0·4–1·8) | 7/35 (20) | 0·9 (0·4–2·4) |
| Had any skin injury to the finger or hand | 25/176 (14) | 32/46 (70) | 15·5 (6·9–37·3)‡ | 21/32 (66) | 11·8 (4·8–30·9)‡ |
| Had injury while preparing fish or seafood | 22/180 (12) | 22/42 (52) | 8·3 (3·8–19·1)‡ | 16/28 (57) | 9·3 (3·7–24·3)‡ |
OR, odds ratio; CI, confidence interval.
Adjusted for age group, sex, and language of interview.
Median unbiased estimate of the OR.
Denotes P < 0·05.
Table 4 displays the most commonly reported fresh fish types purchased. Overall, case-patients (confirmed and probable) had greater odds than controls of purchasing whiting (OR 2·7; 95% CI 1·1–6·6). This association was similar in the sensitivity analysis restricting to confirmed case-patients (OR 3·6; 95% CI 1·4–9·5). In the second sensitivity analysis restricting to case-patients with symptom onset during September–October 2013, case-patients did not have increased odds of having reported purchasing whiting (OR 1·2; 95% CI 0·2–5·6), but had increased odds of purchasing tilapia (OR 3·6; 95% CI 1·1–13·9). Of confirmed case-patients with illness onset during September–October 2013, 79% reported having purchased tilapia, by far the highest percentage of any fish type (Table 4).
Table 4.
Reported fresh fish types purchased by cases-patients with non-tuberculous mycobacteria skin or soft tissue infection and controls – New York City, 2013–2014
| Case-patients | |||||||
|---|---|---|---|---|---|---|---|
| Controls (N = 185) | Confirmed and probable (N = 56) | Confirmed (N = 36) | Confirmed, restricted to onset September–October (N = 19) | ||||
| Fresh fish purchased | No. (%) | No. (%) | OR* (95% CI) | No. (%) | OR* (95% CI) | No. (%) | OR* (95% CI) |
| Bass | 110 (60) | 29 (52) | 0·7 (0·4–1·3) | 16 (44) | 0·5 (0·2–1·1) | 8 (42) | 0·5 (0·2–1·2) |
| Buffalo fish | 74 (40) | 27 (48) | 1·3 (0·7–2·6) | 18 (50) | 1·3 (0·6–2·9) | 9 (47) | 1·1 (0·4–2·9) |
| Carp | 16 (9) | 4 (7) | 0·8 (0·2–2·4) | 4 (11) | 1·2 (0·3–4·1) | 2 (11) | 1·2 (0·2–5·8) |
| Chame | 14 (8) | 5 (9) | 1·4 (0·4–4·3) | 2 (6) | 0·9 (0·1–4·1) | 1 (5) | 1·0 (0·0–7·9) |
| Flounder | 21 (11) | 5 (9) | 0·8 (0·3–2·1) | 2 (6) | 0·4 (0·1–1·8) | 1 (5) | 0·4 (0·0–2·3) |
| Perch | 29 (16) | 9 (16) | 1·7 (0·7–4·1) | 5 (9) | 1·5 (0·5–4·3) | 2 (11) | 0·9 (0·1–4·5) |
| Pomfret | 21 (11) | 5 (9) | 0·9 (0·3–2·4) | 4 (11) | 1·1 (0·3–3·4) | 0 (0) | 0·4 (0·0–1·8) |
| Salmon | 55 (30) | 16 (29) | 0·8 (0·4–1·6) | 8 (22) | 0·6 (0·2–1·3) | 5 (26) | 0·6 (0·2–1·9) |
| Snapper | 30 (16) | 10 (18) | 1·0 (0·4–2·2) | 6 (17) | 0·9 (0·3–2·2) | 5 (26) | 1·5 (0·4–4·4) |
| Sole | 24 (13) | 9 (16) | 1·2 (0·5–2·8) | 7 (19) | 1·5 (0·5–3·9) | 2 (11) | 0·6 (0·1–2·8) |
| Tilapia | 96 (52) | 34 (61) | 1·4 (0·7–2·8) | 25 (69) | 2·0 (0·9–4·8) | 15 (79) | 3·6 (1·1–13·9)† |
| Whiting | 15 (8) | 11 (20) | 2·7 (1·1–6·6)† | 9 (25) | 3·6 (1·4–9·5)† | 2 (11) | 1·2 (0·2–5·6) |
| Yellow fish | 30 (16) | 9 (16) | 0·9 (0·4–2·2) | 7 (19) | 1·1 (0·4–2·9) | 3 (16) | 0·9 (0·2–3·3) |
OR, odds ratio; CI, confidence interval.
Adjusted for age group, sex, and language of interview.
Denotes P < 0·05.
DISCUSSION
This report summarizes the first recognized NTM SSTI outbreak in NYC and the largest reported outbreak in the USA among persons handling fish during food preparation. Of 98 case-patients, the majority were women older than 50 years who spoke Cantonese, Mandarin, or Fujianese and had prepared fish purchased from a Chinese market. The distribution of reported symptom onsets indicated the majority of exposures occurred during late summer or early fall 2013; however, WGS and PFGE results suggested that at least one outbreak-related case occurred as early as May 2013. The grouping of six of eight M. marinum outbreak isolates into two WGS clusters suggests that at least two distinct strains were involved.
In the case–control study, case-patients had increased odds of reporting both a prior skin injury to the finger or hand and of reporting injury during fish preparation, as has been reported during previous NTM outbreaks [6, 9]. Overall, case-patients had increased odds of having purchased whiting; however, this result was not significant when restricted to case-patients with symptom onset most closely corresponding to the purchasing time period asked of controls (September–October 2013). The majority of case-patients who reported purchasing whiting had symptom onset during early summer 2013, preceding the outbreak peak in October. Because case-patients were asked about fish purchasing 2 months before symptom onset and controls were asked about fish purchasing during August–September 2013, seasonal availability of whiting might have influenced results. In the second sensitivity analysis restricting to case-patients reporting symptom onset September–October 2013 (for greatest comparability with controls), tilapia was the only fish type substantially associated with infection. Tilapia is a popular freshwater fish with a spiny dorsal fin grown by aquaculture and has been associated with other bacterial infections among women of Chinese ethnicity after hand injuries during its preparation [23]. To prevent future NTM SSTI infections, persons handling fresh whole fish should consider wearing puncture-resistant gloves or avoiding preparing fish or seafood while having an open skin injury on the finger or hand.
The mechanism by which fish became contaminated could not be determined because of the delay between exposure and environmental investigation. Given the number of markets involved, any contamination likely occurred upstream in the distribution chain. Testing of biofilm, water, and fish samples from two distributors did not yield M. marinum, but other NTM were cultured, indicating that this food distribution chain was susceptible to mycobacterial contamination. Environmental conditions favorable to NTM growth might have been prevalent during summer and fall 2013, but transient, and no longer detectable when the environmental investigation occurred.
WGS revealed marked genetic diversity among available clinical isolates. Because isolates were sequenced from only 10% of cases, the WGS results provided an incomplete description of the outbreak isolates. However, the genetic diversity observed suggests multiple introductions of M. marinum into the distribution chain. This might have occurred if fish species from different geographical areas were comingled in the same holding tank upstream in the distribution chain at a time when environmental conditions were particularly favorable for mycobacterial growth. However, because some of the isolates did not have genetic relatedness to prevalent clusters (i.e., 14–1818_Human and 14–2130_Human), some of the reported NTM SSTIs might not have been related to this outbreak and instead reflected baseline incidence, only detected because of increased awareness associated with the outbreak.
This study had several limitations. First, visits to markets and fish distributors were conducted months after the majority of patients reported symptom onset. Therefore, any fish that might have been contaminated would have been sold or consumed, any conditions that facilitated mycobacterial growth resolved, and any contaminated distributor facilities possibly cleaned and remediated.
Second, latency between NTM exposure and SSTI created inherent outbreak investigation challenges. When DOHMH first identified the outbreak, some patients had experienced symptoms for months, possibly introducing recall bias regarding fish purchased and markets visited. Both case-patients and controls might not have accurately remembered fish purchasing habits or markets visited months before interview, and purchasing multiple fish types at different markets might have led to non-differential misclassification bias. Also, case-patients might have been more likely than controls to recall a skin injury that occurred during food preparation.
Third, the association between having a home aquarium or skin contact with aquarium water and developing an SSTI was not assessed. A known risk factor for M. marinum SSTI, only 10 (11%) of case-patients reported aquarium exposure. Therefore, these exposures were thought to have low overall explanatory value for this outbreak and not assessed in the case–control study.
Fourth, questionnaire differences might have introduced reporting bias in data collected regarding fish purchasing. The case questionnaire was developed for hypothesis generation and included open-ended questions, thus providing more opportunities for case-patients than controls to mention any particular fish type. In contrast, controls might have been prompted to mention a specific fish because they were shown a reference sheet with fish pictures and names. Additionally, reported fish types were not always clearly defined. For example, study participants were asked about purchasing fresh bass, without distinguishing between striped bass and sea bass. Grouping fish types might have masked differences in fish purchasing habits.
Lastly, the environmental investigation was constrained because inspecting all markets and distributors was infeasible, and no city, state, or federal agency had regulatory oversight of live fish delivered to markets and sold to consumers from tanks. Accordingly, a trace-back investigation, as typically occurs when other food items are implicated in outbreaks, was impossible. Without clearly delineated regulatory authorities, future outbreak investigations might be similarly hindered.
This investigation highlighted multiple challenges inherent to investigating an atypical foodborne disease outbreak. Unlike reportable gastrointestinal foodborne diseases, NYC providers are not required to report NTM infections unless they are part of a known or suspected outbreak. Moreover, sensitive diagnostic testing for this disease required either full-thickness skin biopsy or surgical exploration, and clinical specimens needed special handling if mycobacterial culture was attempted. Certain clinicians might have been unfamiliar with these diagnostic concerns, and outbreak-associated infections might have been undiagnosed and unreported. Additionally, although this was the largest such outbreak observed in NYC, the baseline burden of NTM SSTI infections is unknown and might be greater than we realize.
Our investigation yielded several recommendations for future NTM outbreak responses. We recommend investigators consider collecting data to permit analysis by container type (e.g., in a tank vs. on ice), specific fish type (e.g., striped bass vs. sea bass), and source (e.g., freshwater vs. saltwater). Additionally, we recommend investigators consider prioritizing the collection of clinical isolates to be tested with WGS because clustering might guide a trace-back investigation. When more NTM sequences associated with human infections are identified, WGS might be more effective in distinguishing strains or identifying geographic links. As feasible, investigators should also identify and investigate all potential distributors that supply local fish markets. Finally, on the basis of findings from this limited investigation, evaluation of regulatory oversight of the interstate sale of live fish might be warranted.
ACKNOWLEDGEMENTS
The authors thank Sung Hong Chan, Kai Lai Poon, Karen Wong, Alice Yeung, and Wendy Zhu for conducting control interviews, Ann Afordi, Mike Antwi, Sharon Balter, Candacy Browne, Alyssa Chase, Melissa Corcino, Danny Fong, Annie Fine, Elaine Kung, Marcelle Layton, Sam Lee, Natasha McIntosh, Giselle Merizalde, and Sarah Taimur for investigation support, Michael King for editorial support, and Julu Bhatnagar, Dianna Blau, Yvette Francis-Morris, Jana Ritter, Sherif Zaki, the NCEZID Infectious Diseases Pathology Branch, and the DOHMH PHL Mycobacteriology Unit for laboratory support. This investigation received no specific grant from any funding agency, commercial, or not-for-profit sectors.
DISCLAIMER
The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the US Department of Agriculture.
Supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268817001066.
click here to view supplementary material
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Toranzo A, Magarinos B, Romalde JL. A review of the main bacterial fish diseases in mariculture systems. Aquaculture 2005; 246: 37–61. [Google Scholar]
- 2.Swaim LE, et al. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infection and Immunity 2006; 74(11): 6108–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swift S, Cohen H. Granulomas of the skin due to Mycobacterium balnei after abrasions from a fish tank. The New England Journal of Medicine 1962; 267: 1244–1246. [DOI] [PubMed] [Google Scholar]
- 4.Decostere A, Hermans K, Haesebrouck F. Piscine mycobacteriosis: a literature review covering the agent and the disease it causes in fish and humans. Veterinary Microbiology 2004; 99(3–4): 159–166. [DOI] [PubMed] [Google Scholar]
- 5.Falkinham JO III. Environmental sources of nontuberculous mycobacteria. Clinics in Chest Medicine 2015; 36(1): 35–41. [DOI] [PubMed] [Google Scholar]
- 6.Jernigan JA, Farr BM. Incubation period and sources of exposure for cutaneous Mycobacterium marinum infection: case report and review of the literature. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America 2000; 31(2): 439–443. [DOI] [PubMed] [Google Scholar]
- 7.Cheung JP, et al. Mycobacterium marinum infection of the hand and wrist. Journal of Orthopaedic Surgery (Hong Kong) 2012; 20(2): 214–218. [DOI] [PubMed] [Google Scholar]
- 8.Kullavanijaya P, Sirimachan S, Bhuddhavudhikrai P. Mycobacterium marinum cutaneous infections acquired from occupations and hobbies. International Journal of Dermatology 1993; 32(7): 504–507. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, et al. Outbreak of a cutaneous Mycobacterium marinum infection in Jiangsu Haian, China. Diagnostic Microbiology and Infectious Disease 2011; 71(3): 267–272. [DOI] [PubMed] [Google Scholar]
- 10.Lillis JV, et al. Sequelae of World War II: an outbreak of chronic cutaneous nontuberculous mycobacterial infection among Satowanese islanders. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America 2009; 48(11): 1541–1546. [DOI] [PubMed] [Google Scholar]
- 11.Gluckman SJ. Mycobacterium marinum. Clinics in Dermatology 1995; 13(3): 273–276. [DOI] [PubMed] [Google Scholar]
- 12.Sia TY, et al. Clinical and pathological evaluation of Mycobacterium marinum group skin infections associated with fish markets in New York City. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America 2016; 62(5): 590–595. [DOI] [PubMed] [Google Scholar]
- 13.New York City Health Code. Article 11, Reportable Diseases and Conditions (http://www1.nyc.gov/assets/doh/downloads/pdf/about/healthcode/health-code-article11.pdf). Accessed 2 May 2017.
- 14.New York City Department of Health and Mental Hygiene. 2014. Alert #6. Outbreak of Mycobacterium Infections after Preparing Live or Raw Fish or Seafood.
- 15.Ostland VE, et al. Biochemical, molecular, and virulence characteristics of select Mycobacterium marinum isolates in hybrid striped bass Morone chrysops × M. saxatilis and zebrafish Danio rerio. Diseases of Aquatic Organisms 2008; 79(2): 107–118. [DOI] [PubMed] [Google Scholar]
- 16.Gardner SN, Slezak T, Hall BG. kSNP3·0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 2015; 31(17): 2877–2878. [DOI] [PubMed] [Google Scholar]
- 17.Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Research 2009; 19(6): 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatusova T, et al. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Research 2015; 43(7): 3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.PASS 13 Power Analysis and Sample Size Software (2014). Kaysville, Utah, USA: NCSS, LLC. ncss.com/software/pass
- 20.Agresti A. Exact inference for categorical data: recent advances and continuing controversies. Statistics in Medicine 2001; 20(17–18): 2709–2722. [DOI] [PubMed] [Google Scholar]
- 21.Lopez FK, Miley M, Taiwo B. Mycobacterium arupense as an emerging cause of tenosynovitis. Emerging Infectious Diseases 2016; 22(3): 559–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu T, et al. Successful treatment using clarithromycin for a cutaneous lesion caused by Mycobacterium szulgai. British Journal of Dermatology 2000; 142(4): 838–840. [DOI] [PubMed] [Google Scholar]
- 23.U.S. Centers for Disease Control and Prevention. Invasive infection with Streptococcus iniae–Ontario, 1995–1996. MMWR Morbidity and Mortality Weekly Report 1996; 45(30): 650–653. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268817001066.
click here to view supplementary material