Abstract
In Arabidopsis, phytochrome (phy) A, phyB, and cryptochrome 1 (cry1) are representative far‐red, red, and blue light photoreceptors, respectively. Members of the SUPPRESSOR OF PHYA‐105 (SPA) protein family (SPA1–SPA4) form E3 ubiquitin ligase complexes with CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), which mediates the degradation of photomorphogenesis‐promoting factors to desensitize light signaling. SPA2 has been reported to promote seedling etiolation in the dark. However, the unique roles of SPA2 and its three functional domains in suppressing photomorphogenesis under different light conditions are largely unknown. Here, we demonstrate that overexpression of the full‐length or the central coiled‐coil and C‐terminal WD‐repeat domains of SPA2 cause hyper‐etiolation phenotypes under several light conditions. The SPA2 central coiled‐coil and C‐terminal WD‐repeat domains are necessary and sufficient for repressing seedling de‐etiolation, cotyledon unfolding, and promoting hypocotyl negative gravitropism under several light conditions. Furthermore, phyA, phyB, cry1, and COP1 repress protein accumulation or nuclear translocation of SPA2 through direct interactions with its kinase‐like and coiled‐coil domains located in the N‐terminus in response to far‐red, red, and blue light treatments, respectively. Taken together, our results demonstrate that SPA2 functions under multiple light conditions; moreover, light‐activated photoreceptors rapidly suppress SPA2 activity via direct interactions in response to different light treatments.
Keywords: Arabidopsis thaliana , cryptochrome 1, photomorphogenesis, phytochrome A, phytochrome B, SPA2
1. INTRODUCTION
Light and its properties (e.g., wavelength, irradiance, direction, and periodicity) serve as major environmental cues that modulate many aspects of plant growth and development from seed germination to flowering time (Bae & Choi, 2008; Deng & Quail, 1999; Li et al., 2011). Plants monitor light quality, quantity, and duration through multiple photoreceptors (Briggs & Olney, 2001; Christie, 2007; Rizzini et al., 2011). Among these photoreceptors, phytochrome (phy) A predominantly regulates plant far‐red (FR) light responses; phyB through phyE mediates seedling de‐etiolation in red (R) and white (W) light, with phyB playing a major role (Bae & Choi, 2008; Fankhauser & Casal, 2004; Li et al., 2011). Cryptochromes (cry1 and cry2) participate in plant photomorphogenic responses and photoperiodic flowering in response to blue (B, 400–500 nm) light and ultraviolet A (UV‐A, 320–400 nm) light. Cry1 is more efficient in high B light, while cry2 largely functions in low B light (Lin, 2002; Lin & Shalitin, 2005; Whitelam & Halliday, 2007). The UV‐B (290–320 nm) light‐absorbing UV RESISTANCE LOCUS10 (UVR8) protein controls UV‐B‐induced photomorphogenesis and the accumulation of UV‐B‐absorbing flavonols, which may aid in plant tolerance to abiotic and biotic stressors (Rizzini et al., 2011; Tilbrook et al., 2013).
Downstream of the photoreceptors in Arabidopsis light signaling pathways, CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1, a ring finger protein containing WD40 repeats and a coiled‐coil domain), functions as a rate‐limiting repressor of light signaling (Feng & Deng, 2007; Hoecker, 2017; Holm & Deng, 1999; Lau & Deng, 2012; Li et al., 2011; Podolec & Ulm, 2018). SUPPRESSOR OF PHYA‐105 (SPA) genes, which constitute a small family of four members (SPA1–SPA4), encode proteins that contain an N‐terminal kinase‐like domain, a central coiled‐coil domain, and a C‐terminal WD‐repeat domain (Hoecker et al., 1999). COP1 interacts with the SPA proteins to form E3 ubiquitin ligase complexes (Fittinghoff et al., 2006; Laubinger et al., 2004; Laubinger & Hoecker, 2003). These complexes repress light signaling by targeting a group of photomorphogenesis‐promoting factors for ubiquitylation and degradation; the targeted factors include ELONGATED HYPOCOTYL 5 (HY5) (Saijo et al., 2003), LONG HYPOCOTYL IN FAR‐RED 1 (HFR1) (Duek et al., 2004; Jang et al., 2005; Yang, Lin, Hoecker, et al., 2005; Yang, Lin, Sullivan, et al., 2005), LONG AFTER FAR‐RED LIGHT 1 (LAF1) (Seo et al., 2003), PHYTOCHROME RAPIDLY REGULATED 1 (PAR1) (Zhou et al., 2014), phyA (Zhu et al., 2008), and phyB (Jang et al., 2010).
The role of COP1 ubiquitination is conserved from animals to plants. Thus, the four plant‐specific SPA family members may have evolved to fine‐tune COP1 E3 activity (Fittinghoff et al., 2006; Marine, 2012). Although SPA proteins possess similar functional domains, they have overlapping but partially distinct functions in the regulation of plant development (Laubinger et al., 2004). SPA1 suppresses light signaling under R, FR, and B light conditions in a phyA‐dependent manner; it also functions in the dark (Fittinghoff et al., 2006; Hoecker et al., 1998, 1999; Laubinger et al., 2004; Lian et al., 2011; Liu et al., 2011; Sheerin et al., 2015; Yang & Wang, 2006; Zuo et al., 2011). SPA3 and SPA4 promote elongation growth only in adult plants (Fittinghoff et al., 2006; Laubinger et al., 2004; Laubinger & Hoecker, 2003). Unlike SPA1, SPA3 and SPA4 function in both light‐grown seedlings and adult plants. SPA2 has only a limited function under light conditions (Fittinghoff et al., 2006; Laubinger et al., 2004).
The function of SPA1 has been well defined (Chen et al., 2016; Hoecker et al., 1998, 1999; Liu et al., 2011; Lu et al., 2015; Saijo et al., 2003; Yang & Wang, 2006; Zheng et al., 2013; Zuo et al., 2011). SPA1 and SPA2 share about 66% amino acid identity in the C‐terminal WD40‐repeat domain, but considerable differences exist in their N‐terminal kinase‐like (23.5% identity) and central coiled‐coil (31.2% identity) domains (Laubinger & Hoecker, 2003). Moreover, SPA1 and SPA2 strongly differ in their responsiveness to light, while they have indistinguishable activities in the dark (Chen et al., 2016; Laubinger et al., 2004). However, the unique roles of SPA2 and its three functional domains in suppressing photomorphogenesis under different light conditions are largely unknown. In the present study, we found that SPA2 had functions identical to SPA1 under R, B, and W light conditions, but these functions were not conserved under FR light. Overexpression of full‐length SPA2 or the central coiled‐coil and C‐terminal WD‐repeat domains resulted in a hyper‐etiolation phenotype under various light conditions. Thus, the central coiled‐coil and C‐terminal WD‐repeat domains of SPA2 mimic full‐length SPA2 in repressing seedling de‐etiolation and cotyledon unfolding and promoting hypocotyl negative gravitropism, while both the kinase‐like and coiled‐coil domains located in the N‐terminus suppress its protein activity through direct interactions with photoreceptors and COP1. We confirmed that the SPA2 protein is rapidly degraded in response to light; moreover, phyA, phyB, cry1, and COP1 regulate SPA2 protein activity through direct interactions with its N‐terminal kinase‐like and coiled‐coil domains.
2. METHODS
2.1. Plant materials and plant growth conditions
The phyA‐211 (Reed et al., 1994), phyB‐9 (Reed et al., 1993), cry1‐304 (Mockler et al., 1999), cop1‐4 (McNellis et al., 1994), spa1‐100 (Yang, Lin, Hoecker, et al., 2005), spa2‐100 (CS410769), PHYB–GFP (Zheng et al., 2013), Myc–SPA1 (line B1–17), and Myc–SPA1–CT509 (line B15–9) (Yang & Wang, 2006) transgenic plants are the A. thaliana Columbia (Col) ecotype. PHYA–GFP (Kim et al., 2000) and CRY1–OE (Ahmad et al., 1998) are in Wassilewskija (WS) ecotype. PHYA–OE (Boylan & Quail, 1991) is in the Nossen (No) ecotype. Seeds were sterilized and cold treated as described previously (Zheng et al., 2013; Zhou et al., 2014) and then sown on MS plates (Murashige and Skoog medium salts, 1% sucrose, 1% agar, and 0.05% MES [2‐(N‐Morpholino) ethanesulfonic acid], pH 5.7). After exposure to white light for 3 h, the plates were transferred to darkness for 21 h and then to appropriate light conditions for 4 days at 22°C. FR, R, and B light are supplied by the light‐emitting diode (LED) light sources (model E‐30LEDL3; Percival Scientific, Perry, Iowa, USA), with irradiance fluence rates of approximately 1.9, 24.4, and 11.6 μmol·m−2·s−1, respectively, unless otherwise indicated (model HR‐350 Spectrometer; Taiwan Hipoint Co. Kaohsiung). White light was supplied using cool‐white fluorescent lamps. The hypocotyl lengths of at least 30 seedlings from each sample were measured using ImageJ software (http://rsb.info.nih.gov/ij/download.html).
2.2. Plasmids construction
To generate the SPA2 clone, a full‐length SPA2 cDNA fragment was cloned into pEASY–T1 cloning vector (TransGen, Beijing, China) after it was obtained by RT‐PCR using the primer pairs SPA2‐XBE1F and SPA2‐1036XSXR. After pEASY–SPA2 (amino acids 1–1,036) was validated by DNA sequencing, it was used as template to generate various deletion derivatives of SPA2 by PCR cloning. Primer pairs used to produce various deletion derivatives of SPA2 were as follows: SPA2‐XBE1 and SPA2‐566XR (for SPA2‐NT566, amino acids 1–566), SPA2‐XBE1 and SPA2‐703XSXR (for SPA2‐NT703, amino acids 1–703), and SPA2‐565XBEF and SPA2‐1036XSXR (for SPA2‐CT472, amino acids 565–1,036) (Primers see Table S1). These PCR products were cloned into the pEASY–T1 vector to produce their respective pEASY derivative clones. All PCR inserts of the SPA2 deletion derivatives were validated by DNA sequencing.
To generate the Myc–SPA2, Myc–SPA2–NT703, and Myc–SPA2–CT472 transgenic plants, the XhoI‐SpeI fragments from their respective pEASY clones were ligated into the XhoI and XbaI sites of pJIM19–Myc binary vector (Yang, Lin, Sullivan, et al., 2005; Yang & Wang, 2006) to produce pJIM19–Myc–SPA2, pJIM19–Myc–SPA2–NT703, and pJIM19–Myc–SPA2–CT472 under the control of the constitutive CaMV 35S promoter.
To generate the mCherry–SPA2 transgenic plants, PCR products of mCherry using primer pair of mCherry‐F and mCherry‐R were cloned into the pEASY–T1 clone vector. Then, double digestion fragments of KpnI and BamHI from pEASY–mCherry were cloned into the pCAMBIA2300 binary vector (http://www.cambia.org/daisy/cambia/materials/vectors/585.html), resulting in pCAMBIA2300–mCherry. Last, a BamHI‐SpeI fragment of SPA2 from pEASY–SPA2 was cloned into the BamHI‐XbaI site of pCAMBIA2300–mCherry to produce pCAMBIA2300–mCherry–SPA2.
To generate the constructs for yeast two‐hybrid assay of SPA2, its full length or deletion derivatives were released as the EcoRI‐XhoI fragments from their respective pEASYclones and ligated into the corresponding sites of the vector pEG202 or pJG4‐5 (Wang et al., 2001) to produce the translational fusions with the LexA DNA binding domain (BD) or LexA active domain (AD). To generate the BD‐SPA2–dCC, which lacks the coiled‐coil domain and has NT566 (amino acids 1–566) and CT355 (amino acids 682–1,036), two PCR fragments of NT566 and CT355 using primer pair of SPA2‐XBE1 and SPA2‐dCCR, or SPA2‐dCCF and SPA2‐1036XSXR, separately, were purified and mixed together as templates to do the second PCR next. The new PCR products of SPA2–dCC using primer pair of SPA2‐XBE1 and SPA2‐1036XSXR were cloned into TA vector to obtain pEASY–SPA2–dCC.
To generate the pEASY–PHYA, pEASY–PHYB, and pEASY–CRY1 clones, the full length of PHYA, PHYB, and CRY1 was cloned into the pEASY–T1 vector using primer pairs of PHYA‐1BMF and PHYA‐1122XR, PHYB‐1SMF and PHYB‐1172SXR, or CRY1‐1ESF and CRY1‐681XR, separately. To generate the domain deletion constructs of PHYA and CRY1, phyA–CT272 and CRY1–CT192 were obtained by PCR using pEASY–PHYA or pEASY–CRY1 as template with the primer pairs of PHYA‐851MF and PHYA‐1122XR, or CRY1‐490EF and CRY1‐681XR, separately.
To generate constructs for bimolecular fluorescent complimentary (BIFC), PHYA, PHYB, and CRY1 were cloned into pSPYNE–35S (Walter et al., 2004) via double digestion of BamHI‐XhoI, SmaI‐SalI, and SpeI‐BamHI from their pEASY clones, respectively. A BamHI‐XmaI fragment of SPA2 from pEASY–SPA2 was cloned into pSPYCE–35S to produce pSPYCE–SPA2.
To generate the constructs for Co‐IP, SPA2‐NT566, CRY1–CT192, and PHYA–FL were released from corresponding pEASY vectors and then cloned to pJIM19–Myc binary vector (Yang, Lin, Sullivan, et al., 2005; Yang & Wang, 2006), p1305‐Luc‐HA binary vector, and pSYCE‐35S to produce Myc–SPA2–NT566, HA‐CRY1‐CT192, and pSYCE‐PHYA separately.
To generate the constructs for yeast two‐hybrid assay of photoreceptors, the full‐length of phyA, phyB, and cry1 or its deletion derivatives were released as MfeI‐XhoI (for phyA and phyB) or EcoRI‐XhoI (for CRY1) fragments from their respective pEASY clones and ligated into the corresponding sites of the vector pEG202 or pJG4‐5 (Wang et al., 2001) to produce the translational fusions with the LexA DNA binding domain or LexA active domain.
2.3. Plant transformation and selection of the transgenic plants
All the binary constructs of pJIM19–Myc–SPA2, pJIM19–Myc–SPA2–NT703, pJIM19–Myc–SPA2–CT472, pCAMBIA2300–mCherry–SPA2, and pJIM19–Myc–SPA1–NT545 were electroporated into Agrobacterium tumefaciens strain GV3101 and then introduced into Arabidopsis Col‐0 via a floral dip method (Clough & Bent, 1998). Transgenic plants were selected as previous described (Zheng et al., 2013; Zhou et al., 2014). For most experiments, homozygous T3 or T4 transgenic plants with single T‐DNA insertion were used.
2.4. Construction of double mutants
The double mutants phyA‐211/Myc–SPA2, PHYA–OE/Myc–SPA2, phyB‐9/Myc–SPA2, PHYB–GFP/Myc–SPA2, cry1‐304/Myc–SPA2, CRY1–OE/Myc–SPA2, Myc–SPA2/cop1‐4, Myc–SPA2–NT703/cop1‐4, and Myc–SPA2–CT472/cop1‐4, PHYA–GFP/mCherry–SPA2, PHYB–GFP/mCherry–SPA2, and GFP–CRY1/mCherry–SPA2 were derived from genetic crosses of the two respective single parental mutants (or transgenic lines). Putative double mutants were selected in the F2 generation and confirmed in the F3 generation based on the mutant phenotype, or antibiotic selection markers, immunoblot and RT‐PCR analysis.
2.5. Yeast two‐hybrid analysis
The assay system and all the procedures have been described by Serino et al. (1999). Yeast two‐hybrid analysis was performed as previously described (Yang, Lin, Hoecker, et al., 2005; Zheng et al., 2013). Two constructs of AD (activation domain)–SPA2 and AD–CRY1 were able to cause self‐activation; thus, we removed the results.
2.6. Imunoblot analysis
For immunoblotting, Arabidopsis seedlings were grown in different light conditions as indicated in the text. The protein extraction method and procedure described previously by Yang, Lin, Sullivan, et al. (2005) were used for immunoblotting. Myc tagged SPA2 protein, and its domain‐deletion mutant forms were detected with anti‐myc monoclonal antibody (Abmart Biotech, Shanghai, China). Endogenous phyB was detected with anti‐phyB polyclonal antibody (Zheng et al., 2013). These proteins were visualized by incubation with the secondary antibodies (1) goat anti‐rabbit IgG alkaline phosphatase conjugate (1:5,000, A3687; Sigma‐Aldrich, Taufkirchen, Germany) or goat anti‐mouse IgG alkaline phosphatase conjugate (1:5,000, DC05L; Sigma‐Aldrich, Taufkirchen, Germany) in the presence of 5‐bromo‐4‐chloro‐3‐indolylphosphate and nitro blue tetrazolium as substrates or (2) the rabbit and mouse IgG‐HRP (1:5,000, M21003; Abmart Biotech, Shanghai, China) and the Femto‐sig ECL Western Blotting Substrate180–506 (180–506; Tanon Biotech, Shanghai, China) with Chemiluminescence imaging system (Tanon 5200; Tanon Biotech, Shanghai, China).
2.7. Nuclear fractionation
Measurement of nuclear fractions was performed following the protocol in the CeILytic™ PN Plant Nuclei Isolation/Extraction Kit (Sigma‐Aldrich, Saint Louis, Missouri, USA). Anti‐Histone H3 (1:10,000, ab1791) (Abcam, Cambridge, UK) and anti‐HSP90 (1:8000, at‐115) (Santa Cruz, Dallas, Texas, USA) antibodies were used to detect nuclear and cytosolic markers, respectively.
2.8. BIFC assays and fluorescence microscopy
The constructs for BIFC were delivered into leaf cells of tobacco (Nicotiana benthamiana) following the reported procedure (Walter et al., 2004). Briefly, tobacco plants were co‐infiltrated with p19 (Liu et al., 2010) and pSPYCE–SPA2 with either of pSPYNE–PHYA, pSPYNE–PHYB, or pSPYNE–CRY1 in GV3101 and were transferred into the dark for 3 days and then transferred into FR, R, and B light conditions for 8 h. Fluorescence of YFP in the transformed tobacco was imagined using a BX41 fluorescence microscope (Olympus, Tokyo, Japan). To visualize protein co‐localization of mCherry–SPA2 (lines 7‐6) and the phyA–GFP (Kim et al., 2000), phyB–GFP (Zheng et al., 2013), or GFP–CRY1 (Gao et al., 2015) in the nucleus of Arabidopsis cells, GFP or mCherry fluorescence of the PHYA–GFP/mCherry–SPA2, PHYB–GFP/mCherry–SPA2, and GFP–CRY1/mCherry–SPA2 seedlings were mounted on slides and were examined with a BX41 fluorescence microscope (Olympus, Tokyo, Japan). Double mutants of PHYA–GFP/mCherry–SPA2, PHYB–GFP/mCherry–SPA2, and GFP–CRY1/mCherry–SPA2 were grown in the dark for 4 days and then were transferred to FR, R, or Blight for 2, 1, or 1 h, respectively. Nuclei positions were confirmed by 4′,6‐diamidino‐2‐phenylindole (DAPI) staining. Representative images were documented by photography with a DP20 digital camera system (Olympus, Tokyo, Japan). All images were taken from the same region of hypocotyls or roots with identical exposure. For each condition, at least 20 seedlings were observed; a representative image is presented.
2.9. In vivo co‐immunoprecipitation (co‐IP) assay
In vivo Co‐IP assays were performed as previously described, unless otherwise indicated (Zheng et al., 2013). To operate in vivo co‐IP of phyA–GFP by Myc–SPA2–NT566 under FR light, tobacco plant was co‐infiltrated with p19 and either or both of PHYA–GFP and Myc–SPA2‐NT566 in GV3101 and then transferred to FR light for 3 days; leaf extracts were incubated with anti‐Myc‐conjugated agarose (M20012; Shanghai Abmart Biotech, China) under FR light (1.87 μmol·m−2·s−1) for 4 h at 4°C. The pellets were analyzed by immunoblotting with anti‐Myc and anti‐GFP antibodies (Zheng et al., 2013). To perform in vivo co‐IP of endogenous phyB by Myc–SPA2–CT472 under R light, native protein extract was prepared from Myc–SPA2‐CT472 (line 8) seedlings grown in R light for 4 days and incubated with anti‐Myc‐conjugated agarose under R light. To carry out in vivo co‐IP of Myc–SPA2–NT566 by HA–CRY1–CT192, tobacco plant was co‐infiltrated with p19 and either or both of HA–CRY1–CT192 and Myc–SPA2–NT566 in GV3101 and then transferred to B light (20 μmol·m−2·s−1) for 3 days; leaf extracts were incubated with anti‐HA‐conjugated agarose (M20003; Shanghai Abmart Biotech, China) under B light (20 μmol·m−2·s−1) for 4 h at 4°C.
2.10. Quantitative RT‐PCR (qRT‐PCR) Analysis
qRT‐PCR were performed according to a previously reported method (Song et al., 2015; Zhou et al., 2014). The 18S rRNA and Actin 2 was selected as an internal control, and the primers of detect genes used for qRT‐PCR analysis were showed in Supplementary Table S1. All samples were analyzed in three biological replicates. The relative expression levels were calculated using the 2–ΔΔCT method.
2.11. Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SPA2 (At4g11110), SPA1 (At2g46350), PHYA (At1g09570), PHYB (At2g18790), CRY1 (At4g08920), CAB3 (At1g29910), Actin 2 (At3g18780), and Tubulin 2 (At5g62690).
3. RESULTS
3.1. The spa2‐100 mutant enhances the seedling de‐etiolation phenotype of the spa1‐100 mutant in the dark and under various weak light conditions
Previous studies involving the spa1‐3/spa2‐1 double mutant, which was generated from spa1‐3 (RLD ecotype) and spa2‐1 (Columbia ecotype [Col‐0]), showed that SPA2 primarily functions in the dark and has only a limited role in the light (Hoecker et al., 1998; Laubinger et al., 2004). To investigate SPA2 functions during seedling de‐etiolation, we generated a spa1‐100/spa2‐100 double mutant in the Col‐0 ecotype background using the spa1‐100 (Yang, Lin, Hoecker, et al., 2005) and spa2‐100 alleles. The spa2‐100 mutant allele carries two T‐DNA insertions in the sixth intron of the SPA2 gene at 4,008 and 4,036 bp after the presumed start codon (Figures 1a and S1a). RT‐PCR analysis revealed that no SPA2 mRNA was produced in the spa2‐100 mutant (Figure 1b). These results suggest that spa2‐100 is a null allele.
FIGURE 1.
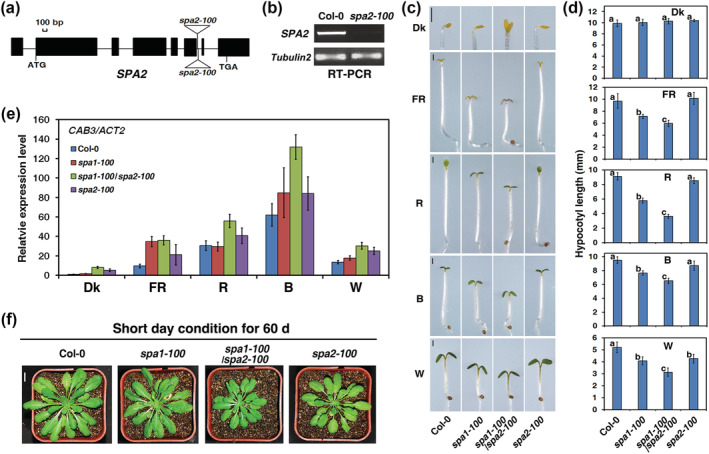
The spa2‐100 mutant enhances the seedling de‐etiolation phenotype conferred by the spa1‐100 mutant in the dark or under various weak light conditions. (a) Diagram of the genomic structure of the SPA2 gene and the T‐DNA insertions (represented by the triangle). Black rectangles represent the exons and lines are introns. (b) RT‐PCR analysis showing that SPA2 mRNA accumulation is abolished in spa2‐100 mutant. Tubulin 2 is shown below as a control. (c) Morphology of the Columbia wild type (Col‐0), spa1‐100, spa2‐100, and spa1‐100/spa2‐100 grown in darkness (Dk), weak far‐red (FR, 0.06 μmol·m−2·s−1), red (R, 4.05 μmol·m−2·s−1), blue (B, 0.17 μmol·m−2·s−1), or white light (W, 7.75 μmol·m−2·s−1) for 4 days. Bars = 1 mm. (d) Quantification of hypocotyl lengths (average of at least 30 seedlings) corresponding to (c). Bars stand for standard deviations. The same lower case letter indicates no significant difference (P < 0.05) between two lines according to the F test, while different lower case letters indicate a significant difference. (e) qRT‐PCR analysis of CAB3 in the wild type (Col‐0), spa1‐100, spa2‐100, and spa1‐100/spa2‐100 in the dark or under FR, R, B, and white light corresponding to (c) and (d). The column shows the mean relative expression of CAB3/Actin2 of three biological repeats. Error bars indicate the standard deviation. (f) Seedling morphology of the wild type (Col‐0), spa1‐100, spa2‐100, and spa1‐100/spa2‐100 under short day (8‐h‐light/16‐h‐dark) condition for 60 days. See also Figure S1
As reported previously for the spa2‐1 mutant (Laubinger et al., 2004), the hypocotyl length of the spa2‐100 mutant was indistinguishable from the hypocotyl length of the wild‐type (WT) in the dark or under FR, R, B, and W light conditions (Figures 1c,d and S1b,c). Although the hypocotyl length of the spa1‐100/spa2‐100 double mutant did not differ from the hypocotyl length of the Col‐0 WT or either of the parental lines in the dark, the seedlings exhibited some features of light‐grown seedlings, such as an open apical hook and unfolded cotyledons (Figures 1c,d and S1b). When the spa1‐100/spa2‐100 double mutant was grown under weak FR (0.06 μmol·m−2·s−1), weak and medium R (4.05 and 24.40 μmol·m−2·s−1, respectively), B (0.17 μmol·m−2·s−1), or weak and medium W light (7.75 and 21.30 μmol·m−2·s−1, respectively) for 4 days, its hypocotyls were significantly shorter than the hypocotyls of Col‐0 WT and the spa1‐100 mutant (Figures 1c,d and S1c). In addition, the spa1‐100/spa2‐100 double mutant displayed increased expression of the CAB3 gene (encoding chlorophyll a/b binding protein 3) in the dark, as well as under various light conditions (Figure 1e). Furthermore, the spa1‐100/spa2‐100 double mutant was smaller than the spa1‐100 and spa2‐100 single mutants under short‐day conditions (Figure 1f). Taken together, these results suggest that spa2‐100 mutant enhances de‐etiolation phenotypes caused by spa1‐100 mutant under darkness and several weak light conditions.
3.2. Overexpression of Myc–SPA2 results in a hyper‐etiolation phenotype under R, B, and W light conditions, but not under FR light
To investigate the functional role of SPA2 in light signaling, we first generated transgenic plants overexpressing c‐Myc epitope‐tagged full‐length SPA2 (Myc–SPA2) driven by the constitutively strong CaMV 35S promoter (Figure S2). Independent homozygous transgenic lines (>30) were obtained for the transgenes, and representative lines were selected for functional analyses. The transcript abundances of two lines of transgenic Myc–SPA2 (lines 15 and 33) were 15.4‐ and 12.6‐fold higher than the transcript abundance of endogenous SPA2 in the Col‐0 WT (Figure S3a,b). The dark‐grown Myc–SPA2 transgenic plants possessed hypocotyl lengths similar to the lengths of WT seedlings (Figure 2a,b). Unlike transgenic Myc–SPA1 plants, which had extremely long hypocotyls under FR light, the Myc–SPA2 transgenic plants unexpectedly displayed hypocotyl lengths similar to the hypocotyl lengths of WT seedlings in response to FR light (Figures 2a,b and S4; Yang & Wang, 2006). Strikingly, under R, B, and W light conditions, overexpression of Myc–SPA2 resulted in a hyper‐etiolation phenotype that was comparable with the phenotype of transgenic Myc–SPA1 plants (Figure 2a,b; Yang & Wang, 2006).
FIGURE 2.
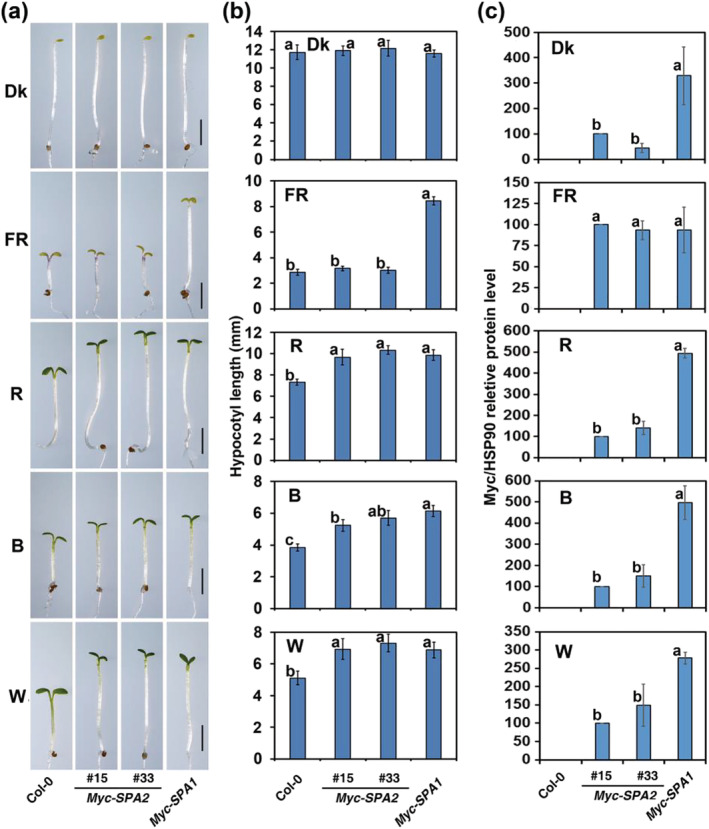
Overexpression of Myc–SPA2 results in hyper‐etiolation under red (R), blue (B), and white (W) light‐conditions, but not under FR light. Morphology (a) and quantification of hypocotyl lengths (average of at least 30 seedlings) (b) of the wild type (Col‐0), Myc–SPA2 (lines 15 and 33), and Myc–SPA1 (lines B1–17; Yang & Wang, 2006) in the dark or under FR (1.9 μmol·m−2·s−1), R (24.4 μmol·m−2·s−1), B (11.6 μmol·m−2·s−1), or W (7.8 μmol·m−2·s−1) for 4 days. Bars = 2 mm. Bars stand for standard deviations. The same lower case letter indicates no significant difference (P < 0.05) between two lines according to the F test, while different lower case letters indicate a significant difference (b). (c) Quantification of relative Myc–SPA2/HSP90 and Myc–SPA1/HSP90 protein levels of Myc–SPA2 and Myc–SPA1 under different light conditions. Seedlings of all the lines were grown as (a). See also Figures S2–S6
Next, we determined the protein abundances of Myc–SPA1 and Myc–SPA2 under different light conditions. Myc–SPA2 accumulated at much lower levels than did Myc–SPA1 in the dark (Figures 2c and S5). Although the Myc–SPA2 seedlings showed no repression of photomorphogenesis under FR, they accumulated similar quantities of Myc‐tagged protein, compared with Myc–SPA1 seedlings. In contrast, the Myc–SPA2 and Myc–SPA1 transgenic plants showed an almost identical level of seedling de‐etiolation, and the Myc–SPA2 seedlings accumulated much less Myc‐tagged protein than did the Myc–SPA1 seedlings under R, B, and W light (Figures 2c and S5).
3.3. SPA2–CT472 (coiled‐coil and WD40‐repeat domains), not SPA2–NT703 (kinase‐like and coiled‐coil domains), mimics full‐length SPA2 in repressing light signaling
A previous study demonstrated that the SPA1 central coiled‐coil and C‐terminal WD40‐repeat domains mimic the function of full‐length SPA1 in repressing light signaling and are sufficient for photomorphogenesis repression (Yang & Wang, 2006). SPA2 also keeps the three function domains as SPA1 (Laubinger et al., 2004). To further investigate the relationships among the three structural domains of SPA2, we generated transgenic plants that overexpressed different versions of genes encoding the SPA2 c‐Myc epitope‐tagged region with the N‐terminal kinase‐like and central coiled‐coil domains (Myc–SPA2–NT703), as well as the region of SPA2 encoding the central coiled‐coil and C‐terminal WD40‐repeat domains (Myc–SPA2–CT472) (Figure S2). Transcript abundances of transgenic Myc–SPA2–NT703 (#19 and #37) or Myc–SPA2–CT472 (#8 and #15) were 6.8–9.1‐fold or 10.2–10.5‐fold higher than the abundance of endogenous SPA2 in the Col‐0 WT (Figure S3c–f).
When grown under various light conditions for 4 days, the transgenic plants overexpressing c‐Myc epitope‐tagged SPA2–CT472 (Myc–SPA2–CT472) (Figure S2) displayed much longer hypocotyls than did plants overexpressing Myc–SPA1–CT509 (encodes SPA1 central coiled‐coil and C‐terminal WD‐repeat domains) (Yang & Wang, 2006), with normal etiolation in the dark (Figure 3a,b). Notably, the hyper‐etiolation phenotype caused by transgenic Myc–SPA2–CT472 in transgenic plants was comparable with the phenotypes of the phyA‐211, phyB‐9, cry1‐304, and phyB‐9 mutants under FR, R, B, and W light conditions, respectively (Figure 3a,b). Like Myc–SPA1–NT545 (encodes SPA1 lacking both the central coiled‐coil domain and the C‐terminal WD‐repeat domain) plants, the hypocotyl length of Myc–SPA2–NT703 (Figure S2) plants was not visibly different from the hypocotyl length of WT plants (Figure 3d,e). The level of Myc‐tagged protein accumulation was twofold to threefold higher in Myc–SPA2–CT472 seedlings than in Myc–SPA1–CT509 seedlings (Figure 3c); there was only a slight difference in Myc‐tagged protein accumulation between Myc–SPA2–NT703 and Myc–SPA1–NT545 seedlings (Figure 3f). These results indicate that SPA2–CT472 is responsible for mediating repressed light signaling, while SPA2–NT703 is not.
FIGURE 3.
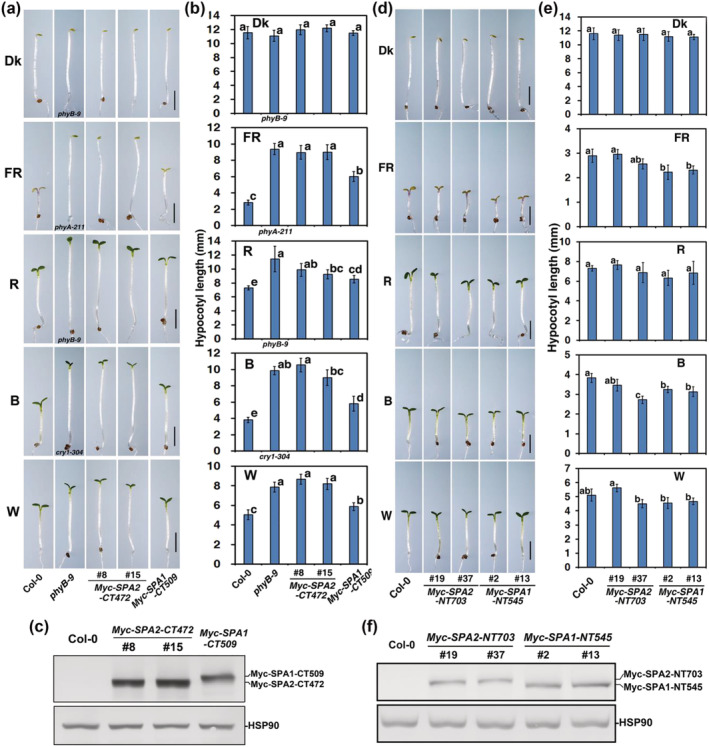
SPA2–CT472, not SPA2–NT703, mimics full‐length SPA2 in repressing light signaling. Morphology (a, d) and quantification of hypocotyl lengths (average of at least 30 seedlings) (b, e) of the wild type (Col‐0), phyB‐9 (in Dk, R, W), phyA‐211 (in FR), cry1‐304 (in B), Myc–SPA2–NT703 (lines 19 and 37) and Myc–SPA1–NT545 (lines 2 and 13), Myc–SPA2–CT472 (lines 8 and 15), and Myc–SPA1–CT509 (line B15–9; Yang & Wang, 2006) in the dark or under FR (1.9 μmol·m−2·s−1), R (24.4 μmol·m−2·s−1), B (11.6 μmol·m−2·s−1), or W (7.8 μmol·m−2·s−1) for 4 days. Bars = 2 mm (a, d). Bars stand for standard deviations. The same lower case letter indicates no significant difference (P < 0.05) between two lines according to the F test, while different lower case letters indicate a significant difference (b, e). Anti‐Myc immunoblot analyses of transgenic (c) Myc–SPA2–NT703 and Myc–SPA1–NT545 and (f) Myc–SPA2–CT472 and Myc–SPA1–CT509. Immunoblots of anti‐HSP90 are shown at the bottom to indicate approximately equal loading. Seedlings were grown in W (7.8 μmol·m−2·s−1) for 4 days. See also Figures S2–S6
Because metabolizable sugars are involved in seedling de‐etiolation (Short, 1999), we investigated whether sugar availability affected de‐etiolation in the transgenic lines. Our results indicated that etiolation of Myc–SPA2 and Myc–SPA2–CT472 seedlings, as well as Myc–SPA1 and Myc–SPA1–CT509 seedlings, was promoted under FR, R, B, and W light conditions despite the absence of additional sugar (Figure S6).
3.4. Myc–SPA2 and Myc–SPA2–CT472 are involved in regulating cotyledon unfolding and hypocotyl negative gravitropism in FR light
To investigate whether the three SPA2 domains are involved in the regulation of cotyledon unfolding, we compared the cotyledon angles of the transgenic lines with the WT in response to different light treatments. All Myc–SPA2, Myc–SPA2‐NT703, Myc–SPA2‐CT472, and WT seedling cotyledons were closed in the dark. After FR, R, or W exposure for 24 h, the angles between the cotyledons of the WT were 136.4°, 154.7°, and 158.8°, whereas the angles of the Myc–SPA2 seedlings (line 15) only reached 66.6%, 51.4%, and 54.2% of the Col‐0 WT, respectively (Figure 4a,b). The cotyledons of the Myc–SPA2–NT703 seedlings (lines 19 and 37) continued to unfold as observed in the Col‐0 WT (Figure 4a,b). Notably, the cotyledons of Myc–SPA2–CT472 (lines 8 and 15) remained folded after exposure to the different light treatments (Figure 4a,b).
FIGURE 4.
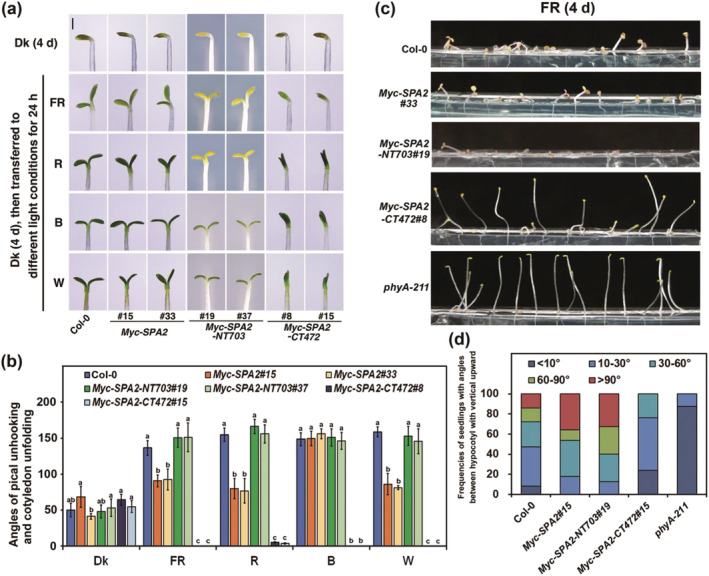
The coiled‐coil and WD‐repeat domains of SPA2 are involved in promotion of cotyledon folding and hypocotyl negative gravitropism response. (a) Comparison of cotyledon folding of Myc–SPA2 (lines 15 and 33) Myc–SPA2–NT703 (lines 19 and 37), and Myc–SPA2–CT472 (lines 8 and 15) with the wild type Col‐0. Seedlings were grown in the dark (Dk) for 4 days and subsequently transferred to FR (1.9 μmol·m−2·s−1), R (24.4 μmol·m−2·s−1), B (11.6 μmol·m−2·s−1), or W (7.8 μmol·m−2·s−1) for 24 h. Bar = 1 mm. (b) Angles of apical unhooking in and cotyledon unfolding in response to FR, R, B, or W light treatments according to (a). Error bars depict the means ± SD (n ≥ 20). Hypocotyl negative gravitropism response of the wild type Col‐0, Myc–SPA2 (line 15), Myc–SPA2–NT703 (line 19), and Myc–SPA2–CT472 (line 8). The same lower case letter indicates no significant difference (P <0.05) between two lines according to the F test, while different lower case letters indicate a significant difference. Seedlings were grown under FR (1.9 μmol·m−2·s−1) for 4 days (c), then the angles between hypocotyl with vertical upward were measured (d)
The angles between the hypocotyl and the vertical axis were measured under FR to detect hypocotyl negative gravitropism; 100% of the phyA‐211 seedlings and 76.2% of Myc–SPA2–CT472 (line 8) seedlings displayed angles <30°, whereas the remaining Myc–SPA2–CT472 seedlings had angles <60° (Figure 4c,d). Unexpectedly, the Myc–SPA2 (line 33) and Myc–SPA2–NT703 (line 19) seedlings showed much more lodging than did Col‐0 WT seedlings. The Myc–SPA2 (line 33) and Myc–SPA2–NT703 (line 19) seedlings with angles ≥90° were 2.58‐ and 2.34‐fold more common than were WT seedlings with angles ≥90° (Figure 4c,d). These observations suggest that the N‐terminal kinase‐like domain of SPA2 is not essential for its repressive function; moreover, both the central coiled‐coil and C‐terminal WD40‐repeat domains are sufficient for repressing light signaling and promoting hypocotyl negative gravitropism.
3.5. The N‐terminal kinase‐like and coiled‐coil domains of SPA2 are responsible for its stability during light signaling
The SPA1 N‐terminal kinase‐like domain is responsible for the degradation of SPA1 (Yang & Wang, 2006). To determine whether the SPA2 N‐terminal kinase‐like domain has a role in regulating the activity of SPA2, we compared the protein levels of Myc–SPA2, Myc–SPA2–NT703 (kinase‐like and coiled‐coil domains), and Myc–SPA2–CT472 (coiled‐coil and WD40‐repeat domains) in seedlings that had been grown in W light. Although all mRNA levels were in the same range (Figure S7a,b), the levels of the Myc–SPA2–CT472 protein in two transgenic lines were approximately 17.2‐ to 29.8‐fold higher than the levels of the Myc–SPA2 protein (line 15), while the levels of two transgenic lines of Myc–SPA2–NT703 only reached 23–47% of the Myc–SPA2 protein (line 15) (Figures 5a,b and S7c). To test whether the low levels of Myc–SPA2 and Myc–SPA2–NT703 in white light‐grown seedlings might be due to their degradation mediated by the 26S proteasome pathway, we tested the effects of a proteasome inhibitor, MG132, on their protein accumulations. Seedling treatment with the proteasome inhibitor MG132 eliminated the effect of light on the degradation of Myc–SPA2 and Myc–SPA2–NT703 proteins, but it did not influence the Myc–SPA2–CT472 protein levels (Figures 5c and S7d).
FIGURE 5.
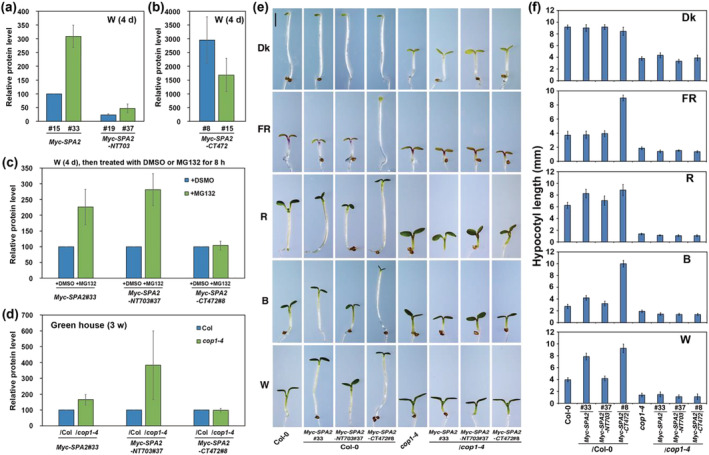
The N‐terminal kinase‐like and coiled‐coil domains of SPA2 are responsible for its stability in light signaling. (a, b) Comparison of relative Myc–SPA2/HSP90, Myc–SPA2–NT703/HSP90, and Myc–SPA2–CT472/HSP90 protein levels in white light. (c) Comparison of relative Myc–SPA2/HSP90, Myc–SPA2–NT703/HSP90, and Myc–SPA2–CT472/HSP90 protein levels after treated with DMSO or MG132. Error bars depict the means ± SD (n = 3). (d) Comparison of relative Myc–SPA2/HSP90, Myc–SPA2–NT703/HSP90, and Myc–SPA2–CT472/HSP90 in wild type (Col‐0) or cop1‐4 mutant background. (e) Morphology of Myc–SPA2 (line 33), Myc–SPA2–NT703 (line 37), and Myc–SPA2–CT472 (line 8) in both the WT (Col‐0) and cop1‐4 mutant backgrounds. Seedlings were grown in the dark (Dk) or under FR (1.9 μmol·m−2·s−1), R (24.4 μmol·m−2·s−1), B (11.6 μmol·m−2·s−1), or W (7.8 μmol·m−2·s−1) for 4 days. Bar = 1 mm. (f) Quantification of hypocotyl lengths of Myc–SPA2 (line 33), Myc–SPA2–NT703 (line 37), and Myc–SPA2–CT472 (line 8) in both the WT (Col‐0) and cop1‐4 mutant backgrounds. See also Figures S7 and S8
Previous studies indicated that SPA2 is degraded through the 26S proteasome and that COP1 is involved (Chen et al., 2015). To investigate whether COP1 regulated SPA2 degradation through N terminal kinase‐like domain, Myc–SPA2, Myc–SPA2–NT703, and Myc–SPA2–CT472 transgenes were separately introduced into the cop1‐4 mutant background to generate double‐homozygous plants of Myc–SPA2/cop1‐4, Myc–SPA2–NT703/cop1‐4, and Myc–SPA2–CT472/cop1‐4. The protein levels of Myc–SPA2 and Myc–SPA2–NT703 were 1.7‐ and 4.7‐fold higher in the cop1‐4 background than in either of their parental lines under W light, while Myc–SPA2–CT472 accumulated at almost the same level in the Col‐0 and cop1‐4 backgrounds (Figures 5d and S7e). As expectedly, the accumulation rates of the Myc–SPA2 protein were 5.2‐, 12.8‐, 18.7‐, and 28.4‐fold higher in the cop1‐4 background than in the Col‐0 WT background in the dark or under FR, R, and B light conditions, respectively (Figure S8a,b). Yeast two hybrid assays showed that COP1 was able to interact with the SPA2 kinase‐like and coiled‐coil domains (Figure S9a). These results demonstrate that the N‐terminus of SPA2 is necessary to target SPA2 protein for degradation through the 26S proteasome pathway using COP1 as E3 ligase.
We also explored whether overexpression of Myc–SPA2 and Myc–SPA2–CT472 could substitute for the loss of COP1 activity. The Myc–SPA2/cop1‐4, Myc–SPA2–NT703/cop1‐4, and Myc–SPA2–CT472/cop1‐4 double homozygous plants displayed a phenotype identical to the phenotype of the cop1‐4 single mutant in the dark and under FR, R, B, and W light (Figure 5e,f). This result suggests that the repressive function of Myc–SPA2 requires functional COP1; the functions of COP1 and SPA2 are not interchangeable.
3.6. PhyA, phyB and cry1 suppress SPA2 activities under R and B light conditions
PhyA, phyB, and cry1 are predominantly responsible for promoting seedling de‐etiolation under FR, R, and B light conditions, respectively. The phyA‐211 (Reed et al., 1994), phyB‐9 (Reed et al., 1993), and cry1‐304 (Mockler et al., 1999) mutants are nonsense alleles of PHYA, PHYB, and CRY1 genes. To investigate whether these photoreceptors regulate SPA2 activities under different light conditions, we introduced Myc–SPA2 (line 33) into the phyA‐211, phyB‐9, and cry1‐304 mutants, as well as plants overexpressing phyA (PHYA–OE) (Boylan & Quail, 1991), GFP‐tagged phyB (PHYB–GFP) (Zheng et al., 2013), or cry1 (CRY1–OE) (Ahmad et al., 1998) backgrounds with genetic crosses; thus, we generated phyA‐211/Myc–SPA2, PHYA–OE/Myc–SPA2, phyB‐9/Myc–SPA2, PHYB–GFP/Myc–SPA2, cry1‐304/Myc–SPA2, and CRY1–OE/Myc–SPA2 plants. The hypocotyl lengths and cotyledon opening rates of these plants were compared with the corresponding characteristics in their parental lines under different light conditions.
Under FR light, the hypocotyl length of phyA‐211/Myc–SPA2 plants was almost equivalent to the hypocotyl length of the phyA‐211 single mutant and much greater than the hypocotyl lengths of the Myc–SPA2 and Col‐0 WT plants (Figure 6a). Similar to the PHYA–OE seedlings, the PHYA–OE/Myc–SPA2 plants displayed distinctly shorter hypocotyls than did Myc–SPA2 and Col‐0 WT seedlings (Figure 6b). Under continuous R light, phyB‐9/Myc–SPA2 plants had longer hypocotyls than did the Myc–SPA2 and phyB‐9 parent plants (Figure 6d), while the hypocotyls of the PHYB–GFP/Myc–SPA2 seedlings were as short as the hypocotyls of PHYB–GFP seedlings (Figure 6e). Under continuous B light, the hypocotyls of the cry1‐304/Myc–SPA2 seedlings were much longer than the hypocotyls of its parent plants cry1‐304 and Myc–SPA2 (Figure 6g), while the CRY1–OE/Myc–SPA2 seedlings displayed hypocotyls comparable with the hypocotyls of the CRY1–OE seedlings (Figure 6h).
FIGURE 6.
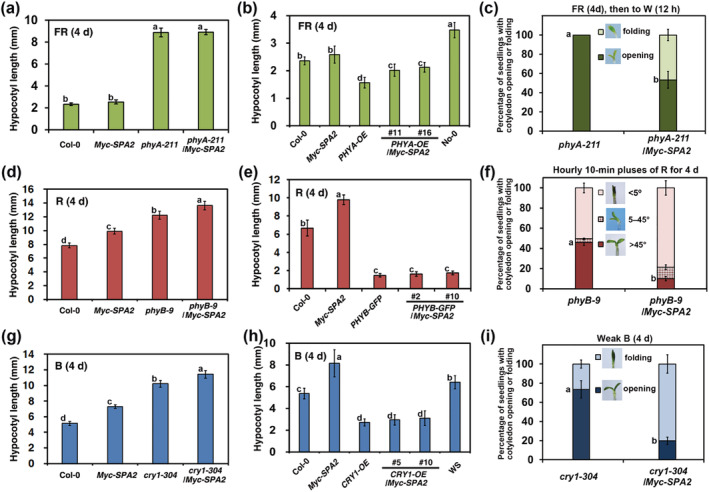
PhyA, phyB, and cry1 suppress SPA2 activities under far‐red (FR), red (R), and blue (B) light conditions. The fluence rates of the FR, R, B, or W were 1.9, 24.4, 11.6, or 7.8 μmol·m−2·s−1, respectively, unless otherwise indicated. Quantification of hypocotyl lengths (average of at least 30 seedlings) of the wild type (Col‐0), Myc–SPA2 (parental line #33), (a) phyA‐211 and phyA‐211/Myc–SPA2; (d) phyB‐9 and phyB‐9/Myc–SPA2; (g) cry1‐304 and cry1‐304/Myc–SPA2; (b) No‐0, PHYA–OE (No‐0 background, parental line, Boylan & Quail, 1991), and PHYA‐OE/Myc–SPA2 (lines 1 and 16); (e) PHYB–GFP (parental line, Zheng et al., 2013), and PHYB–GFP/Myc–SPA2 (lines 2 and 10); (h) CRY1–OE (parental line, Ahmad et al., 1998) and CRY1–OE/Myc–SPA2 (lines 5 and 10) under FR (a, b), R (d, e), or B (g, h) light condition for 4 days, respectively. Morphology and percentage of seedlings with cotyledon opening (c) phyA‐211 and phyA‐211/Myc–SPA2; (f) phyB‐9 and phyB‐9/Myc–SPA2; (i) cry1‐304 and cry1‐304/Myc–SPA2. Seedlings were grown (c) in FR light for 4 days and subsequently transferred to white light for 12 h; (f) in hourly 10‐min pluses of R light for 4 days; (i) in B weak light (0.17 μmol·m−2·s−1) for 4 days. Bars stand for standard deviations
We also evaluated the cotyledon unfolding rates in the double mutants and their parental lines. When grown under FR for 4 days, all Col‐0 and Myc–SPA2 seedlings had open cotyledons, whereas the cotyledons of the phyA‐211 and phyA‐211/Myc–SPA2 seedlings remained folded (data not shown). After they had been transferred to W light for 12 h, 100% of the phyA‐211 mutant seedlings had open cotyledons, while 53.3% of the phyA‐211/Myc–SPA2 seedlings had folded cotyledons (Figure 6c). When subjected to hourly 10‐min pulses of R for 4 d, all Col‐0 and Myc–SPA2 seedlings had open cotyledons (data not shown), but the proportions of phyB‐9 and phyB‐9/Myc–SPA2 seedlings with cotyledon angle <5° were 50.3% and 78.7%, respectively (Figure 6f). Although the cotyledons unfolded in the Col‐0 and Myc–SPA2 seedlings after they had been grown under weak B light (0.17 μmol·m−2·s−1) for 4 days (data not shown), the proportions of cry1‐304 and cry1‐304/MycSPA2 seedlings with folded cotyledons were 26.3% and 79.9%, respectively (Figure 6i). These photoreceptors suppress elongation of the hypocotyl and folding of the cotyledons caused by Myc–SPA2 overexpression in response to different light treatments.
3.7. SPA2 N‐terminal kinase‐like and coiled‐coil domains mediate interactions with phyA, phyB, and cry1 in vitro and in vivo
The above results showed that phyA, phyB, and cry1 promote photomorphogenesis by suppressing SPA2 activity (Figure 6). Previous results demonstrated that phyA, phyB, and cry1 interact with SPA2 to promote photomorphogenesis (Chen et al., 2015; Sheerin et al., 2015). However, the domain responsible for interactions between photoreceptors and SPA2 has been unknown. Here, we employed a yeast two‐hybrid assay to test whether SPA2 interacted with phyA, phyB, and cry1. Our results indicated that the N‐terminal 566‐amino acid fragment of SPA2 (NT566, kinase‐like domain) interacted with full‐length phyA and a C‐terminal 272‐amino acid fragment of phyA (PHYA–CT272, HKRD domain) (Figure 7a). Full‐length phyB interacted with full‐length of SPA2 and SPA2‐NT703 (N‐terminal kinase‐like and coiled‐coil domains) (Figure 7b). A C‐terminal 192‐amino acid fragment of CRY1 (CRY1–CT192, DAS domain) interacted with NT566, NT703, and a deletion derivative of SPA2 that lacked the coiled‐coil domain (dCC) (Figure 7c). These results suggest that the NT566 (kinase‐like domain), NT703 (kinase‐like and coiled‐coil domains), and dCC (kinase‐like and WD repeat domains) of SPA2 are responsible for interacting with phyA (HKRD domain), phyB, and cry1 (DAS domain), respectively.
FIGURE 7.
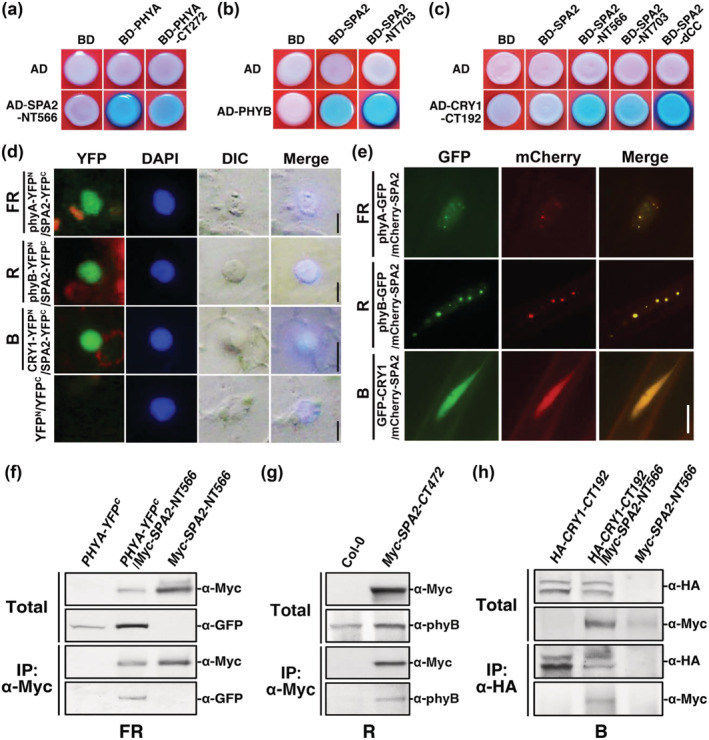
The N‐terminal kinase‐like and coiled‐coil domains of SPA2 mediate interaction with phyA, phyB, and cry1 in vitro and in vivo. The fluence rates of the far‐red (FR), red (R), or blue (B) light were FR (1.9 μmol·m−2·s−1), R (24.4 μmol·m−2·s−1), B (11.6 μmol·m−2·s−1), or W (7.8 μmol·m−2·s−1), respectively, unless otherwise indicated. The interaction between SPA2 with phyA (a), phyB (b), and cry1 (c) in yeast. AD, activation domain; BD, DNA binding domain. (SPA2, full length) Amino acids 1–1,036; (SPA2–NT566, kinase‐like domain) amino acids 1–566; (SPA2–NT703, kinase‐like domain and coiled‐coil domain) amino acids 1–703; (SPA2–dCC, which lacks the coiled‐coil domain) amino acids 1–566 and 682–1,036; (phyA, full length) amino acids 1–1,122; (phyA–CT272) amino acids 851–1,122; (phyB, full length) amino acids 1–1,172; (CRY1‐CT192) amino acids 490–681. (d) Protein co‐localization of SPA2–YFPc and phyA–YFPN, phyB–YFPN or CRY1–YFPN by BIFC assays in the nucleus of tobacco (Nicotiana benthamiana) cells. Bars = 5 μm. (e) Co‐localization of phyA, phyB, cry1 with SPA2 in the nucleus of Arabidopsis cells in response to FR (2 h), R (1 h), or B (1 h) light transitions. Bar = 5 μm. (f) In vivo co‐immunoprecipitation (co‐IP) of phyA–GFP by Myc–SPA2–NT566 in tobacco plant under FR light. (g) In vivo co‐IP of endogenous phyB by Myc–SPA2–CT472 in Arabidopsis under R light. (h) In vivo co‐IP of Myc–SPA2–NT566 by HA–CRY1–CT192 in tobacco plant under B light (20 μmol·m−2·s−1)
Next, we observed interactions between SPA2 and phyA, phyB, or cry1 in living plant cells using the bimolecular fluorescence complementation (BIFC) assay (Walter et al., 2004). Tobacco (Nicotiana benthamiana L.) plants were transfected with the pSPYCE plasmid encoding SPA2‐YFPC and the pSPYNE plasmid encoding phyA–YFPN, phyB–YFPN, or CRY1–YFPN using A. tumefaciens strain GV3101. After the plants had been exposed to FR, R, or B light for 8 h, strong fluorescent signals indicating protein co‐localization were observed by fluorescence microscopy when SPA2–YFPC was co‐expressed with phyA–YFPN, phyB–YFPN, or CRY1–YFPN; in contrast, cells transfected with the empty vector produced no fluorescent signal (Figure 7d).
Then, co‐localization of SPA2 with phyA, phyB, or cry1 was detected in the nucleus using transgenic plants that co‐expressed mCherry–SPA2 and PHYA–GFP, PHYB–GFP, or GFP–CRY1 in response to different light treatments. After growth in the dark for 4 days, the PHYA–GFP/mCherry–SPA2 seedlings were transferred to FR light for 2 h; the GFP fluorescence signal of phyA–GFP or mCherry fluorescence signal of mCherry–SPA2 was then observed. In FR light, the phyA–GFP and mCherry–SPA2 proteins co‐localized in the nucleus at small but clear nuclear bodies (NBs, small yellow dots) (Figure 7e). After a dark‐to‐R light transition for 1 h, phyB–GFP and mCherry–SPA2 in PHYB–GFP/mCherry–SPA2 transgenic seedlings were co‐localized in large and bright NBs (large yellow dots) (Figure 7e). Unlike the PHYA–GFP/mCherry–SPA2 and PHYB–GFP/mCherry–SPA2 seedlings, the seedlings of GFP–CRY1/mCherry–SPA2 displayed only diffuse GFP and mCherry fluorescence signals in the nucleus (Figure 7e, yellow nuclear fluorescence).
We conducted in vivo co‐immunoprecipitation (co‐IP) assays to investigate whether phyA and SPA2 co‐localize in the same protein complex (Liu et al., 2010). Tobacco plants were co‐infiltrated with the gene‐silencing suppressor p19 (Liu et al., 2010) and either or both PHYA–GFP and Myc–SPA2–NT566 using A. tumefaciens strain GV3101 and then transferred to FR light for 3 days. Subsequently, the native protein was extracted from tobacco leaves and subjected to co‐IP using antibodies against the Myc epitope. The Myc‐tagged SPA–NT566 protein was co‐purified with YFPC ‐tagged full‐length phyA (Figure 7f). To examine whether phyB and SPA2 co‐localize in the same protein complex in response to R light, a native protein extract was prepared from seedlings of the Myc–SPA2–CT472 transgenic line that had been grown in R light for 4 days and incubated with anti‐Myc‐conjugated agarose under R light. The Myc‐tagged SPA2–CT472 protein co‐purified with endogenous phyB under R light (Figure 7g). Finally, in vivo co‐IP of Myc–SPA2–NT566 by CRY1–CT192–HA was carried out using tobacco plants that had been co‐infiltrated with p19 and either or both CRY1–CT192–HA and Myc–SPA2–NT566 using A. tumefaciens strain GV3101. After the tobacco seedlings had been transferred to B light for 3 days, the leaf extracts were subjected to co‐IP using an antibody against the HA epitope (Figure 7h). The results of all in vivo co‐IP assays showed that phyA, phyB, and cry1 co‐localize in the same protein complex as SPA2 in FR, R, and B‐light treatments, respectively.
3.8. PhyA, phyB, and cry1 repress SPA2 protein activity in response to different light treatments through different ways
The above results indicated that phyA, phyB, and cry1 are responsible for inhibiting hypocotyl growth and cotyledon unfolding in response to FR, R, and B light conditions, respectively. Next, we investigated how phyA, phyB, and cry1 inhibited SPA2 activity. Previous studies revealed that phyA and phyB are involved in the degradation of nuclear‐localized SPA2 protein under light conditions, while cry1 and cry2 have minimal effects on this process (Chen et al., 2015). We first detected Myc–SPA2 accumulation under continuous darkness and in FR, R, B, and W light. The Myc–SPA2 protein accumulated to much higher levels under R and W light, compared with the dark; much lower levels accumulated under FR and B light (Figure S10a,b). The transcript abundances of transgenic Myc–SPA2 were comparable under the different light conditions (Figure S10c,d) and in the WT, phyA‐211, phyB‐9, and cry1‐304 backgrounds (Figure S10e,f). Therefore, the differences in Myc–SPA2 protein levels in these plants were caused by changes in protein stability in response to different light treatments.
The Myc–SPA2 total protein was slightly degraded in the Myc–SPA2 plants during exposure to FR light, but its levels sharply increased in the phyA‐211/Myc–SPA2 seedlings (Figures 8a and S11a). Although the nuclear Myc–SPA2 protein level was much higher in the phyA‐211 background than in the Col‐0 WT background, it tended to decrease in both the phyA‐211 and WT backgrounds in response to FR treatment (Figures 8b and S11a). In response to R light, the Myc‐SPA2 total protein was gradually degraded in the Myc‐SPA2 seedlings, but its levels increased in the phyB‐9/Myc–SPA2 seedlings (Figures 8c and S11b). The nuclear Myc–SPA2 protein increased much more rapidly in the phyB‐9 background than in the Col‐0 WT background in response to R treatment (Figures 8d and S11b). In response to B light, Myc–SPA2 total protein was gradually degraded in Myc–SPA2 and cry1‐304/Myc–SPA2 plants (Figures 8e and S11c). We also found that accumulation of nuclear Myc–SPA2 protein was faster in cry1‐304/Myc–SPA2 plants than in Myc–SPA2 plants in response to B light (Figures 8f and S11c). Thus, phyA, phyB, and cry1 repressed SPA2 protein activity in different ways. PhyA inhibited SPA2 protein accumulation in the cytoplasm but did not affect translocation of SPA2 into the nucleus in response to FR light (Figures 8a,b and S11a). Although cry1 protein had minimal effects on SPA2 protein abundance, it may have inhibited the translocation of SPA2 to the nucleus in response to B light (Figures 8e,f and S11c). Unlike either phyA or cry1, phyB induced SPA2 protein degradation in the cytoplasm and restrained the translocation of SPA2 into the nucleus (Figures 8c,d and S11b).
FIGURE 8.
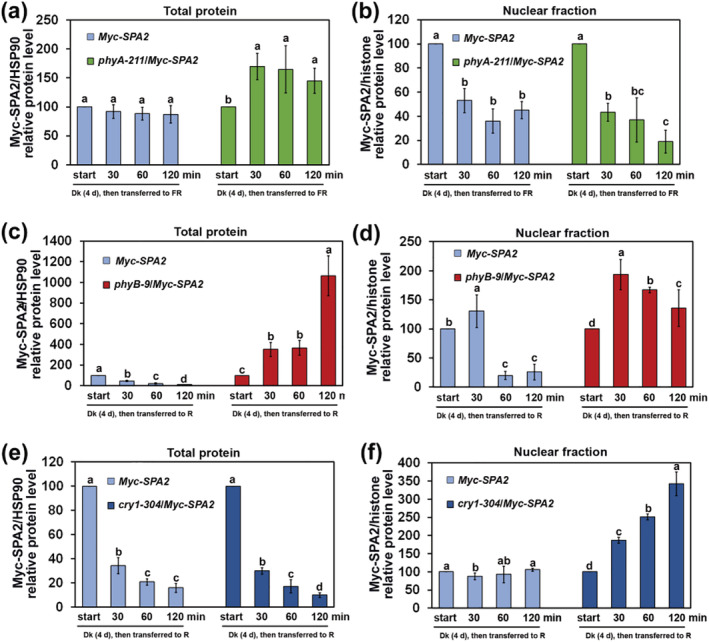
Quantification of relative Myc–SPA2/HSP90 protein levels (a, c, e) in total protein or relative Myc–SPA2/histone protein levels in nuclear fraction (b, d, f) in response to different light‐treatments. Seedlings were grown in the dark (Dk) and subsequently transferred to FR (1.9 μmol·m−2·s−1), R (24.4 μmol·m−2·s−1), and B (11.6 μmol·m−2·s−1) light‐conditions for 30, 60, or 120 min. See also Figure S5 and S6. Error bars depict the means ± SD (n = 3). The same lower case letter indicates no significant difference (P < 0.05) between two cases according to the F test, while different lower case letters indicate a significant difference. See also Figures S9 and S10
4. DISCUSSION
4.1. Similar to SPA1, SPA2 represses Arabidopsis de‐etiolation in the dark and under multiple light conditions
The functions of SPA1 under various light conditions have been well characterized. SPA1 was originally identified as a component that acts negatively in both the phyA‐mediated very‐low‐fluence responses and high‐irradiance responses (Baumgardt et al., 2002; Hoecker et al., 1998, 1999). Then, SPA1 was shown to participate in cry1‐ and cry2‐mediated B light signaling (Holtkotte et al., 2017; Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011). Next, SPA1 was found to act genetically downstream of phyB to promote seedling etiolation under R light (Lu et al., 2015). Furthermore, SPA1 acts downstream of phyB‐mediated repression of FR light signaling to enhance COP1 nuclear activity (Zheng et al., 2013). SPA proteins also form UVR8‐COP1‐SPA complexes that repress UV‐B‐induced photomorphogenesis (Huang et al., 2014). SPA1 is required for several processes, including photoperiodic flowering (Ishikawa et al., 2006; Laubinger et al., 2006), thermosensory hypocotyl elongation (Martínez et al., 2018; Zhu et al., 2020), and jasmonate‐mediated inhibition of hypocotyl elongation (Zheng et al., 2017).
SPA2 was first demonstrated to function primarily in dark‐grown seedlings and to have limited effects on light‐grown seedlings and adult plants (Hoecker et al., 1998; Laubinger et al., 2004). Although the spa2‐100 single mutant characterized in the present study did not differ from the Col‐0 WT, the spa1‐100/spa2‐100 double mutant seedlings exhibited conspicuous de‐etiolation phenotypes with increased CAB3 expression under all light conditions (Figures 1c–e and S1b,c). Similar to transgenic Myc–SPA1 or Myc–SPA1–CT509 plants, plants overexpressing Myc–SPA2 or Myc–SPA2–CT472 displayed a hyper‐etiolation phenotype under FR, R, and B light conditions in the presence and absence of additional sugar (Figures 2, 3, and S6). The hyper‐etiolation phenotype caused by overexpression of full‐length or the central coiled‐coil and C‐terminal WD‐repeat domains of SPA2 indicates that SPA2 is involved in Arabidopsis light signaling (Figures 2 and 3). The suppressive effects of phyA, phyB, and cry1 on SPA2 activity suggest that SPA2 functions downstream of these photoreceptors in the FR, R, or B light pathways, respectively (Figure 6). In addition, phyA, phyB, and cry1 inhibited SPA2 protein activity through direct interactions in response to different light treatments (Figures 6, 7, 8). Taken together, these results suggest that SPA2 functions both in the dark and under multiple light conditions.
4.2. Compared with SPA1 activity, SPA2 activity is more strictly regulated by light
SPA1–SPA4 have overlapping but distinct functions in the inhibition of photomorphogenesis in dark‐ and light‐grown seedlings (Balcerowicz et al., 2011; Chen et al., 2016; Fittinghoff et al., 2006; Laubinger et al., 2004; Laubinger & Hoecker, 2003). SPA2 is more closely related to SPA1, while SPA3 is more similar to SPA4 (Figure S12a). SPA2 and SPA1 share 34.5%, 22.9%, and 66.3% identity at the amino acid level with the kinase‐like domain, coiled‐coil domain, and C‐terminal WD40‐repeat domain, respectively (Laubinger & Hoecker, 2003; Figure S12b). The SPA1 coiled‐coil domain and WD‐repeat domain are responsible for interacting with COP1 and the substrates of the COP1‐SPA1 complex (including HY5 and HFR1) (Saijo et al., 2003; Yang, Lin, Hoecker, et al., 2005). Both SPA1 domains mimic the function of full‐length SPA1 in repressing seedling de‐etiolation (Yang & Wang, 2006). Similar to SPA1, the SPA2 coiled‐coil and the WD40‐repeat domains are sufficient and necessary for promoting hypocotyl elongation, cotyledon folding, and hypocotyl negative gravitropism (Figures 3a,b and 4; Yang & Wang, 2006). Furthermore, the Myc–SPA2–CT472 (line 8) seedlings displayed 49%, 31%, 82%, and 61% longer hypocotyls than did Myc–SPA1–CT509 plants under FR, R, B, and W light conditions, respectively; their hyper‐etiolation phenotypes were comparable with the phenotypes of phyA‐211, phyB‐9, and cry1‐304 mutants (Figures 3a,b; Yang & Wang, 2006). These results indicate that the SPA2 coiled‐coil and WD40‐repeat domains are more effective than the domains of SPA1 in repressing seedling de‐etiolation.
In contrast to the hypocotyl hyper‐elongation caused by Myc–SPA1 or Myc–SPA2–CT472, the seedlings of transgenic Myc–SPA2 unexpectedly showed no effect on seedling etiolation in response to FR light (Figures 2a,b; Yang & Wang, 2006). Ectopic expression of a chimeric domain‐swap protein with the SPA1 kinase‐like domain and the SPA2 coiled‐coil and the WD40‐repeat domains causes long hypocotyls (similar to full‐length SPA1) under FR light (Chen et al., 2016). Thus, the SPA2 kinase‐like domain restrained the repressing activity on seedling photomorphogenesis from its coiled‐coil and WD40‐repeat domains. The SPA1 kinase‐like and coiled‐coil domains are required for interactions with the light‐activated photoreceptors cry2 and phyA (Sheerin et al., 2015; Zuo et al., 2011). In our experiments, phyA, phyB, and cry1 interacted with the SPA2 kinase‐like and coiled‐coil domains (Figure 7a–c) to repress SPA2 protein activity in response to different light treatments (Figure 8). SPA2 can interact with itself through its N‐terminus (Figure S9b). Thus, the SPA2 kinase‐like and coiled‐coil domains are necessary for COP1 and SPA2 to form a heterotetramer through direct interactions in plants (Figure S9; Zhu et al., 2008; Hoecker, 2017). The kinase‐like and coiled‐coil domains are also involved in COP1‐mediated degradation of SPA1 and SPA2 through the 26S proteasome pathway (Figures 5c,d, S7d,e, and S8; Laubinger et al., 2004; Yang & Wang, 2006; Chen et al., 2015). These findings suggest that the SPA2 kinase‐like domains may serve as a link to connect the light‐activation of photoreceptors and stability of the COP1‐SPA complexes. Compared with SPA1 activity, SPA2 activity is more strictly restrained through its N‐terminal kinase‐like domain in response to light.
4.3. Light‐activated phyA, phyB, and cry1 directly regulate SPA2 activity
Direct protein–protein interactions between photoreceptors and COP1‐SPA complexes are necessary for light signal transduction in plants. Physical interactions between photoreceptors (phyA, phyB, and cry2) and COP1‐SPA complexes are required for ubiquitination and degradation of photoreceptors through the 26S proteasome pathway to repress seedling de‐etiolation (Jang et al., 2010; Seo et al., 2004; Weidler et al., 2012). Conversely, photoreceptors modify the activities of COP1‐SPA complexes to activate light signaling through direct interactions with COP1 or SPAs (Lau & Deng, 2012; Podolec & Ulm, 2018). COP1 reportedly functions as an E3 ubiquitin ligase to mediate the degradation of SPA2 in light (Chen et al., 2015). In our experiments, FR, R, and B light induced rapid degradation of the SPA2 protein through the 26S proteasome pathway via direct interactions with COP1 (Figures 5 and S9a). In the cop1‐4 mutant background, Myc–SPA2 seedlings accumulated 5.2‐, 12.8‐, 18.7‐, 28.4‐, and 1.7‐fold higher levels of SPA2 protein in the dark or under FR, R, B, and W light conditions, respectively, compared with the WT background (Figures 5d, S7e, and S8). In addition, different light conditions lead to different SPA2 degradation efficiencies, among which B light is most effective, followed by R light. PhyA, phyB, and cry1 interact with the SPA2 protein; they also inhibit SPA2 activity in response to FR, R, or B light treatments, respectively (Figures 6 and 8). Both phyA and phyB significantly inhibited accumulation of the SPA2 protein in the cytoplasm in response to FR or R light treatments, while either phyB or cry1 greatly repressed the translocation of SPA2 to the nucleus under R or B light, respectively (Figures 8 and S11). In summary, light‐activated photoreceptors rapidly modulate repressing activity of SPA2 on plant de‐etiolation via direct interactions in response to different light treatments (Figures S13).
CONFLICT OF INTEREST
The authors declare no conflict of interest associated with the work described in this manuscript.
AUTHOR CONTRIBUTIONS
MS, LS, and JY designed the experiments. MS, LS, PZ, GL, and XJ carried out the experiments. MS, LS, PZ, WS, JG, HL, and BL collected and analyzed the data. MS, LS, and JY wrote the manuscript with input from all other authors.
Supporting information
Figure S1. Genotyping of the T‐DNA insertion of spa2–100 mutant and phenotypes of spa1–100/spa2–100 double mutant in the dark or under different light conditions; related to Figure 1.
Figure S2. Diagram of various domain deletions of SPA2; related to Figures 2, 3, 7, and S9.
Figure S3. qRT‐PCR analyses for comparing transcript abundances of transgenic Myc–SPA2, Myc–SPA2–NT703, and Myc–SPA2–CT473 with endogenous SPA2 in the wild type Col‐0; related to Figures 2 and 3.
Figure S4. Histograms comparing hypocotyl lengths relative to the dark control (hypocotyl length/hypocoty length in the dark) (average of 30 seedlings) of Col‐0, Myc–SPA1, Myc–SPA2 (#15, #33) seedlings under different intensities of FR light; related to Figure 2.
Figure S5. Anti‐Myc immunoblot analyses of transgenic lines of Myc–SPA2 and Myc–SPA1 in the dark (Dk) or under far‐red (FR), red (R), blue (B), and white (W) light‐conditions; related to Figure 2.
Figure S6. Evaluation of SPA2 functions on seedling etiolation in response to absence of additional sugars in the dark (Dk) or under far‐red (FR), red (R), blue (B), and white (W) light‐conditions; related to Figure 2 and Figure 3.
Figure S7. COP1 mediates SPA2 protein degradation through 26S proteasome‐dependent pathway; related to Figure 5.
Figure S8. Comparison of Myc–SPA2, Myc–SPA2–NT703, and Myc–SPA2–CT472 protein accumulations in the wild type and the cop1–4 mutant backgrounds under different light‐conditions; related to Figure 5.
Figure S9. The interactions between COP1 (a) with SPA2, and SPA2 with itself (b) in yeast.
Figure S10. The transcript abundance of transgenic Myc–SPA2 is comparable under different light conditions or in the wild type, phyA‐211, phyB‐9 and cry1–304 backgrounds; related to Figure 8.
Figure S11. Immunoblot analyses of Myc–SPA2 accumulation in total protein or in purified nuclear fraction of Myc–SPA2, phyA‐211/Myc–SPA2(a), phyB‐9/Myc–SPA2(b) and cry1–304/Myc–SPA2(c) seedlings in response to far‐red (FR), red (R), blue (B), and white (W) light‐treatments; related to Figure 8.
Figure S12. Phylogenetic analysis of four SPA proteins and architecture comparison of SPA2 with SPA1.
Figure S13. A model depicting the structure–function relationship of SPA2.
Table S1. List of primers used for plasmid constructions and RT‐PCR analysis in this study.
ACKNOWLEDGMENTS
We thank Drs. Qi Xie, Peter Quail, Ferenc Nagy, and Anthony Cashmore for providing the 35S:p19 vector and the PHYA–OE, PHYA–GFP, and CRY1–OE transgenic lines, respectively. We thank Drs. Daowen Wang, Hongjie Li, Pei Liu, Mingyue Gou, and Xu Zheng for their reading of, and comments on, the manuscript. This work was supported by the National Natural Science Foundation of China (Grants 31570268 to JY and 31400259 to MS), and the Natural Science Foundation of Beijing Municipality (Grant 5092019 to JY).
Su, L. , Zhou, P. , Guo, L. , Jia, X. , Wang, S. , Gao, J. , Li, H. , Liu, B. , Song, M. , & Yang, J. (2022). Arabidopsis SPA2 represses seedling de‐etiolation under multiple light conditions. Plant Direct, 6(5), e403. 10.1002/pld3.403
Su, Zhou, and Guo contributed equally to this article.
Contributor Information
Liang Su, Email: suliang_mp5@163.com.
Peng Zhou, Email: zhoupeng@caas.cn.
Lin Guo, Email: guolin@caas.cn.
Xiaolin Jia, Email: jiaxiaolin1129@163.com.
Shaoci Wang, Email: wwsscc@139.com.
Jianwei Gao, Email: jianweigao3@qq.com.
Hongyu Li, Email: lihongyu@caas.cn.
Bin Liu, Email: liubin05@caas.cn.
Meifang Song, Email: songmeifang@brc.ac.cn.
Jianping Yang, Email: jpyang@henau.edu.cn.
REFERENCES
- Ahmad, M. , Jarillo, J. A. , & Cashmore, A. R. (1998). Chimeric proteins between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. The Plant Cell, 10, 197–207. 10.1105/tpc.10.2.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, G. , & Choi, G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annual Review of Plant Biology, 59, 281–311. 10.1146/annurev.arplant.59.032607.092859 [DOI] [PubMed] [Google Scholar]
- Balcerowicz, M. , Fittinghoff, K. , Wirthmueller, L. , Maier, A. , Fackendahl, P. , Fiene, G. , Koncz, C. , & Hoecker, U. (2011). Light exposure of Arabidopsis seedlings causes rapid de‐stabilization as well as selective post‐translational inactivation of the repressor of photomorphogenesis SPA2. The Plant Journal, 65(5), 712–723. 10.1111/j.1365-313X.2010.04456.x [DOI] [PubMed] [Google Scholar]
- Baumgardt, R. L. , Oliverio, K. A. , Casal, J. J. , & Hoecker, U. (2002). SPA1, a component of phytochrome a signal transduction, regulates the light signaling current. Planta, 215(5), 745–753. 10.1007/s00425-002-0801-x [DOI] [PubMed] [Google Scholar]
- Boylan, M. T. , & Quail, P. H. (1991). Phytochrome a overexpression inhibits hypocotyl elongation in transgenic Arabidopsis . Proceedings of the National Academy of Sciences, 88(23), 10806–10810. 10.1073/pnas.88.23.10806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, W. R. , & Olney, M. A. (2001). Photoreceptors in plant photomorphogenesis to date, five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiology, 125, 85–88. 10.1104/pp.125.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Lory, N. , Stauber, J. , & Hoecker, U. (2015). Photoreceptor specificity in the light‐induced and COP1‐mediated rapid degradation of the repressor of Photomorphogenesis SPA2 in Arabidopsis. PLoS Genetics, 11(9), e1005516. 10.1371/journal.pgen.1005516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Wirthmueller, L. , Stauber, J. , Lory, N. , Holtkotte, X. , Leson, L. , Schenkel, C. , Ahmad, M. , & Hoecker, U. (2016). The functional divergence between SPA1 and SPA2 in Arabidopsis photomorphogenesis maps primarily to the respective N‐terminal kinase‐like domain. BMC Plant Biology, 16, 165. 10.1186/s12870-016-0854-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, J. M. (2007). Phototropin blue‐light receptors. Annual Review of Plant Biology, 58, 21–45. 10.1146/annurev.arplant.58.032806.103951 [DOI] [PubMed] [Google Scholar]
- Clough, S. J. , & Bent, A. F. (1998). Floral dip: A simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . The Plant Journal, 16(6), 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Deng, X. W. , & Quail, P. H. (1999). Signalling in light‐controlled development. Seminars in Cell & Developmental Biology, 10, 121–129. 10.1006/scdb.1999.0287 [DOI] [PubMed] [Google Scholar]
- Duek, P. D. , Elme, M. V. , van Oosten, V. R. , & Fankhauser, C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Current Biology, 14(24), 2296–2301. 10.1016/j.cub.2004.12.026 [DOI] [PubMed] [Google Scholar]
- Fankhauser, C. , & Casal, J. J. (2004). Phenotypic characterization of a photomorphogenic mutant. The Plant Journal, 39(5), 747–760. 10.1111/j.1365-313x.2004.02148.x [DOI] [PubMed] [Google Scholar]
- Feng, S. , & Deng, X. W. (2007). The role of ubiquitin/proteasome‐mediated proteolysis in photoreceptor action. Annual Plant Reviews, 30, 128–154. 10.1002/9780470988893.ch6 [DOI] [Google Scholar]
- Fittinghoff, K. , Laubinger, S. , Nixdorf, M. , Fackendahl, P. , Baumgardt, R. L. , Batschauer, A. , & Hoecker, U. (2006). Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. The Plant Journal, 47, 577–590. 10.1111/j.1365-313x.2006.02812.x [DOI] [PubMed] [Google Scholar]
- Gao, J. , Wang, X. , Zhang, M. , Bian, M. , Deng, W. , Zuo, Z. , Yang, Z. , Zhong, D. , & Lin, C. (2015). Trp triad‐dependent rapid photoreduction is not required for the function of Arabidopsis CRY1. Proceedings of the National Academy of Sciences, 112(29), 9135–9140. 10.1073/pnas.1504404112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker, U. (2017). The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Current Opinion in Plant Biology, 37, 63–69. 10.1016/j.pbi.2017.03.015 [DOI] [PubMed] [Google Scholar]
- Hoecker, U. , Tepperman, J. M. , & Quail, P. H. (1999). SPA1, a WD‐repeat protein specific to phytochrome a signal transduction. Science, 284, 496–499. 10.1126/science.284.5413.496 [DOI] [PubMed] [Google Scholar]
- Hoecker, U. , Xu, Y. , & Quail, P. H. (1998). SPA1: A new genetic locus involved in phytochrome A‐specific signal transduction. The Plant Cell, 10, 19–33. 10.1105/tpc.10.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, M. , & Deng, X. W. (1999). Structural organization and interactions of COP1, a light‐regulated developmental switch. Plant Molecular Biology, 41, 151–158. 10.1023/a:1006324115086 [DOI] [PubMed] [Google Scholar]
- Holtkotte, X. , Ponnu, J. , Ahmad, M. , & Hoecker, U. (2017). The blue light‐induced interaction of cryptochrome 1 with COP1 requires SPA proteins during Arabidopsis light signaling. PLoS Genetics, 13, e1007044. 10.1371/journal.pgen.1007044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Yang, P. , Ouyang, X. , Chen, L. , & Deng, X. W. (2014). Photoactivated UVR8‐COP1 module determines photomorphogenic UV‐B signaling output in Arabidopsis. PLoS Genetics, 10(3), e1004218. 10.1371/journal.pgen.1004218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, M. , Kiba, T. , & Chua, N. H. (2006). The Arabidopsis SPA1 gene is required for circadian clock function and photoperiodic flowering. The Plant Journal, 46(5), 736–746. 10.1111/j.1365-313x.2006.02737.x [DOI] [PubMed] [Google Scholar]
- Jang, I. C. , Henriques, R. , Seo, H. S. , Nagatani, A. , & Chua, N. H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. The Plant Cell, 22(7), 2370–2383. 10.1105/tpc.109.072520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, I. C. , Yang, J. Y. , Seo, H. S. , & Chua, N. H. (2005). HFR1 is targeted by COP1 E3 ligase for post‐translational proteolysis during phytochrome a signaling. Genes & Development, 19, 593–602. 10.1101/gad.1247205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, L. , Kirche, S. , Toth, R. , Adam, E. , Schäfer, E. , & Nagy, F. (2000). Light‐induced nuclear import of phytochrome‐a: GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis . The Plant Journal, 22, 125–133. 10.1046/j.1365-313x.2000.00729.x [DOI] [PubMed] [Google Scholar]
- Lau, O. S. , & Deng, X. W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends in Plant Science, 17, 584–593. 10.1016/j.tplants.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Laubinger, S. , Fittinghoff, K. , & Hoecker, U. (2004). The SPA quartet: A family of WD‐repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. The Plant Cell, 16(9), 2293–2306. 10.1105/tpc.104.024216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger, S. , & Hoecker, U. (2003). The SPA1‐like proteins SPA3 and SPA4 repress photomorphogenesis in the light. The Plant Journal, 35, 373–385. 10.1046/j.1365-313x.2003.01813.x [DOI] [PubMed] [Google Scholar]
- Laubinger, S. , Marchal, V. , Gentilhomme, J. , Wenkel, S. , Adrian, J. , Jang, S. , Kulajta, C. , Braun, H. , Coupland, G. , & Hoecker, U. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development, 133(16), 3213–3222. 10.1242/dev.02481 [DOI] [PubMed] [Google Scholar]
- Li, J. , Li, G. , Wang, H. , & Deng, X. W. (2011). Phytochrome signaling mechanisms. Arabidopsis Book, 9, e0123. 10.1199/tab.0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, H. L. , He, S. B. , Zhang, Y. C. , Zhu, D. M. , Zhang, J. Y. , Jia, K. P. , Sun, S. X. , Li, L. , & Yang, H. Q. (2011). Blue‐light‐dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes & Development, 25, 1023–1028. 10.1101/gad.2025111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. T. (2002). Blue light receptors and signal transduction. The Plant Cell, 14(suppl 1), S207–S225. 10.1105/tpc.000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. T. , & Shalitin, D. (2005). Cryptochrome structure and signal transduction. Annual Review of Plant Biology, 54, 469–496. 10.1146/annurev.arplant.54.110901.160901 [DOI] [PubMed] [Google Scholar]
- Liu, B. , Zuo, Z. C. , Liu, H. T. , Liu, X. , & Lin, C. T. (2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes & Development, 25, 1029–1034. 10.1101/gad.2025011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Zhang, Y. , Tang, S. , Zhao, Q. , Zhang, Z. , Zhang, H. , Dong, L. , Guo, H. , & Xie, Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana . The Plant Journal, 61(5), 893–903. 10.1111/j.1365-313x.2009.04109.x [DOI] [PubMed] [Google Scholar]
- Lu, X. D. , Zhou, C. M. , Xu, P. B. , Luo, Q. , Lian, H. L. , & Yang, H. Q. (2015). Red‐light‐dependent interaction of phyB with SPA1 promotes COP1‐SPA1 dissociation and photomorphogenic development in Arabidopsis. Molecular Plant, 8(3), 467–478. 10.1016/j.molp.2014.11.025 [DOI] [PubMed] [Google Scholar]
- Marine, J. C. (2012). Spotlight on the role of COP1 in tumorigenesis. Nature Reviews Cancer, 12(7), 455–464. 10.1038/nrc3271 [DOI] [PubMed] [Google Scholar]
- Martínez, C. , Nieto, C. , & Prat, S. (2018). Convergent regulation of PIFs and the E3 ligase COP1/SPA1 mediates thermosensory hypocotyl elongation by plant phytochromes. Current Opinion in Plant Biology, 45, 188–203. 10.1016/j.pbi.2018.09.006 [DOI] [PubMed] [Google Scholar]
- McNellis, T. W. , von Arnim, A. G. , Araki, T. , Komeda, Y. , Misera, S. , & Deng, X. W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. The Plant Cell, 6, 487–500. 10.1105/tpc.6.4.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler, T. C. , Guo, H. , Yang, H. , Duong, H. , & Lin, C. T. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development, 126, 2073–2082. 10.1242/dev.126.10.2073 [DOI] [PubMed] [Google Scholar]
- Podolec, R. , & Ulm, R. (2018). Photoreceptor‐mediated regulation of the COP1/SPA E3 ubiquitin ligase. Current Opinion in Plant Biology, 45, 18–25. 10.1016/j.pbi.2018.04.018 [DOI] [PubMed] [Google Scholar]
- Reed, J. W. , Nagatani, A. , Elich, T. D. , Fagan, M. , & Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiology, 104, 1139–1149. 10.1104/pp.104.4.1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J. W. , Nagpal, P. , Poole, D. S. , Furuya, M. , & Chory, J. (1993). Mutations in the gene for the red/far‐red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. The Plant Cell, 5, 147–157. 10.1105/tpc.5.2.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini, L. , Favory, J.‐J. , Cloix, C. , Faggionato, D. , O'Hara, A. , Kaiserli, E. , Baumeister, R. , Schafer, E. , Nagy, F. , Jenkins, G. I. , & Ulm, R. (2011). Perception of UV‐B by the Arabidopsis UVR8 protein. Science, 332(6025), 103–106. 10.1126/science.1200660 [DOI] [PubMed] [Google Scholar]
- Saijo, Y. , Sullivan, J. A. , Wang, H. , Yang, J. , Shen, Y. , Rubio, V. , Ma, L. , Hoecker, U. , & Deng, X. W. (2003). The COP1‐SPA1 interaction defines a critical step in phytochrome A‐mediated regulation of HY5 activity. Genes & Development, 17(21), 2642–2647. 10.1101/gad.1122903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H. S. , Watanabe, E. , Tokutomi, S. , Nagatani, A. , & Chua, N. H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome a signaling. Genes & Development, 18, 617–622. 10.1101/gad.1187804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H. S. , Yang, J. Y. , Ishikawa, M. , Bolle, C. , Ballesteros, M. , & Chua, N. H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature, 423(6943), 995–999. 10.1038/nature01696 [DOI] [PubMed] [Google Scholar]
- Serino, G. , Tsuge, T. , Kwok, S. , Matsui, M. , Wei, N. , & Deng, X. W. (1999). Arabidopsis cop8 and fuS5 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. The Plant Cell, 11, 1967–1980. 10.1105/tpc.11.10.1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin, D. J. , Menon, C. , zur Oven‐Krockhaus, S. , Enderle, B. , Zhu, L. , Johnen, P. , Schleifenbaum, F. , Stierhof, Y. D. , Huq, E. , & Hiltbrunner, A. (2015). Light‐activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. The Plant Cell, 27, 189–201. 10.1105/tpc.114.134775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short, T. W. (1999). Overexpression of Arabidopsis phytochrome B inhibits phytochrome a function in the presence of sucrose. Plant Physiology, 119, 1497–1506. 10.1104/pp.119.4.1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, M. F. , Zhang, S. , Hou, P. , Shang, H. Z. , Gu, H. K. , Li, J. J. , Xiao, Y. , Guo, L. , Su, L. , Gao, J. W. , & Yang, J. P. (2015). Ectopic expression of a phytochrome B gene from Chinese cabbage (Brassica rapa L. ssp. pekinensis) in Arabidopsis thaliana promotes seedling de‐etiolation, dwarfing in mature plants, and delayed flowering. Plant Molecular Biology, 87, 633–463. 10.1007/s11103-015-0302-5 [DOI] [PubMed] [Google Scholar]
- Tilbrook, K. , Arongaus, A. B. , Binkert, M. , Heijde, M. , Yin, R. , & Ulm, R. (2013). The UVR8 UV‐B photoreceptor: Perception, signaling and response. Arabidopsis Book, 11, e0164. 10.1199/tab.0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, M. , Chaban, C. , Schütze, K. , Batistic, O. , Weckermann, K. , Näke, C. , Blazevic, D. , Grefen, C. , Schumacher, K. , Oecking, C. , Harter, K. , & Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal, 40, 428–438. 10.1111/j.1365-313x.2004.02219.x [DOI] [PubMed] [Google Scholar]
- Wang, H. , Ma, L. G. , Li, J. M. , Zhao, H. Y. , & Deng, X. W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science, 294(5540), 154–158. 10.1126/science.1063630 [DOI] [PubMed] [Google Scholar]
- Weidler, G. , Zur Oven‐Krockhaus, S. , Heunemann, M. , Orth, C. , Schleifenbaum, F. , Harter, K. , Hoecker, U. , & Batschauer, A. (2012). Degradation of Arabidopsis CRY2 is regulated by SPA proteins and phytochrome a. The Plant Cell, 24(6), 2610–2623. 10.1105/tpc.112.098210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam, G. C. , & Halliday, K. J. (2007). Light and plant development. Blackwell Publishing. 10.1002/9780470988893 [DOI] [Google Scholar]
- Yang, J. P. , Lin, R. C. , Hoecker, U. , Liu, B. L. , Xu, L. , & Wang, H. Y. (2005). Repression of light signalling by Arabidopsis SPA1 involves post‐translational regulation of HFR1 protein accumulation. The Plant Journal, 43, 131–141. 10.1111/j.1365-313x.2005.02433.x [DOI] [PubMed] [Google Scholar]
- Yang, J. P. , Lin, R. C. , Sullivan, J. , Hoecker, U. , Liu, B. L. , Xu, L. , Deng, X. W. , & Wang, H. Y. (2005). Light regulates COP1‐mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis . The Plant Cell, 17(3), 804–821. 10.1105/tpc.104.030205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. P. , & Wang, H. Y. (2006). The central coiled‐coil domain and carboxyl‐terminal WD‐repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. The Plant Journal, 47, 564–576. 10.1111/j.1365-313x.2006.02811.x [DOI] [PubMed] [Google Scholar]
- Zheng, X. , Wu, S. W. , Zhai, H. Q. , Zhou, P. , Song, M. F. , Su, L. , Xi, Y. L. , Li, Z. Y. , Cai, Y. F. , Meng, F. H. , Yang, L. , Wang, H. Y. , & Yang, J. P. (2013). Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far‐red light. The Plant Cell, 25, 115–133. 10.1105/tpc.112.107086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y. , Cui, X. , Su, L. , Fang, S. , Chu, J. , Gong, Q. , Yang, J. , & Zhu, Z. (2017). Jasmonate inhibits COP1 activity to suppress hypocotyl elongation and promote cotyledon opening in etiolated Arabidopsis seedlings. The Plant Journal, 90(6), 1144–1155. 10.1111/tpj.13539 [DOI] [PubMed] [Google Scholar]
- Zhou, P. , Song, M. F. , Yang, Q. H. , Su, L. , Hou, P. , Guo, L. , Zheng, X. , Xi, Y. L. , Meng, F. H. , Xiao, Y. , Yang, L. , & Yang, J. P. (2014). Both PHYTOCHROME RAPIDLY REGULATED1 PAR1 and PAR2 promote seedling photomorphogenesis in multiple light signaling pathways. Plant Physiology, 164, 841–852. 10.1104/pp.113.227231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, D. M. , Maier, A. , Lee, J. H. , Laubinger, S. , Saijo, Y. , Wang, H. Y. , Qu, L. J. , Hoecker, U. , & Deng, X. W. (2008). Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. The Plant Cell, 20(9), 2307–2323. 10.1105/tpc.107.056580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W. , Zhou, H. , Lin, F. , Zhao, X. , Jiang, Y. , Xu, D. , & Deng, X. W. (2020). COLD‐REGULATED GENE 27 integrates signals from light and the circadian clock to promote hypocotyl growth in Arabidopsis. The Plant Cell, 32, 3155–3169. 10.1105/tpc.20.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, Z. , Liu, H. , Liu, B. , Liu, X. , & Lin, C. (2011). Blue light‐dependent interaction of Cry2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis . Current Biology, 21, 841–847. 10.1016/j.cub.2011.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Genotyping of the T‐DNA insertion of spa2–100 mutant and phenotypes of spa1–100/spa2–100 double mutant in the dark or under different light conditions; related to Figure 1.
Figure S2. Diagram of various domain deletions of SPA2; related to Figures 2, 3, 7, and S9.
Figure S3. qRT‐PCR analyses for comparing transcript abundances of transgenic Myc–SPA2, Myc–SPA2–NT703, and Myc–SPA2–CT473 with endogenous SPA2 in the wild type Col‐0; related to Figures 2 and 3.
Figure S4. Histograms comparing hypocotyl lengths relative to the dark control (hypocotyl length/hypocoty length in the dark) (average of 30 seedlings) of Col‐0, Myc–SPA1, Myc–SPA2 (#15, #33) seedlings under different intensities of FR light; related to Figure 2.
Figure S5. Anti‐Myc immunoblot analyses of transgenic lines of Myc–SPA2 and Myc–SPA1 in the dark (Dk) or under far‐red (FR), red (R), blue (B), and white (W) light‐conditions; related to Figure 2.
Figure S6. Evaluation of SPA2 functions on seedling etiolation in response to absence of additional sugars in the dark (Dk) or under far‐red (FR), red (R), blue (B), and white (W) light‐conditions; related to Figure 2 and Figure 3.
Figure S7. COP1 mediates SPA2 protein degradation through 26S proteasome‐dependent pathway; related to Figure 5.
Figure S8. Comparison of Myc–SPA2, Myc–SPA2–NT703, and Myc–SPA2–CT472 protein accumulations in the wild type and the cop1–4 mutant backgrounds under different light‐conditions; related to Figure 5.
Figure S9. The interactions between COP1 (a) with SPA2, and SPA2 with itself (b) in yeast.
Figure S10. The transcript abundance of transgenic Myc–SPA2 is comparable under different light conditions or in the wild type, phyA‐211, phyB‐9 and cry1–304 backgrounds; related to Figure 8.
Figure S11. Immunoblot analyses of Myc–SPA2 accumulation in total protein or in purified nuclear fraction of Myc–SPA2, phyA‐211/Myc–SPA2(a), phyB‐9/Myc–SPA2(b) and cry1–304/Myc–SPA2(c) seedlings in response to far‐red (FR), red (R), blue (B), and white (W) light‐treatments; related to Figure 8.
Figure S12. Phylogenetic analysis of four SPA proteins and architecture comparison of SPA2 with SPA1.
Figure S13. A model depicting the structure–function relationship of SPA2.
Table S1. List of primers used for plasmid constructions and RT‐PCR analysis in this study.


