Abstract
In order to scale up medium-chain-length polyhydroxyalkanoate (mcl-PHA) production in recombinant microorganisms, we generated and investigated different recombinant bacteria containing a stable regulated expression system for phaC1, which encodes one of the mcl-PHA polymerases of Pseudomonas oleovorans. We used the mini-Tn5 system as a tool to construct Escherichia coli 193MC1 and P. oleovorans POMC1, which had stable antibiotic resistance and PHA production phenotypes when they were cultured in a bioreactor in the absence of antibiotic selection. The molecular weight and the polydispersity index of the polymer varied, depending on the inducer level. E. coli 193MC1 produced considerably shorter polyesters than P. oleovorans produced; the weight average molecular weight ranged from 67,000 to 70,000, and the polydispersity index was 2.7. Lower amounts of inducer added to the media shifted the molecular weight to a higher value and resulted in a broader molecular mass distribution. In addition, we found that E. coli 193MC1 incorporated exclusively the R configuration of the 3-hydroxyoctanoate monomer into the polymer, which corroborated the enantioselectivity of the PhaC1 polymerase enzyme.
The poly-(R)-3-hydroxyalkanoic acids (PHA) constitute a growing family of polyesters which are accumulated as storage products and can account for significant fractions of the cell matter in many microorganisms (1). A polymer is normally accumulated as an internal reserve of carbon and energy when cells are cultured in the presence of an excess carbon source and when growth is limited by the lack of an essential nutrient. If the conditions for growth are restored, the PHA is used as a carbon and energy source (39). Recently, PHA have attracted considerable attention due to their potential use as biodegradable thermoplastics and as sources of chiral monomers (1, 5, 26, 27). Thus, research is currently being performed to improve productivity, to reduce production costs, and, more importantly, to produce specific functionalized PHA. One of the most attractive approaches is to use heterologous microorganisms, such as Escherichia coli, for PHA production (10, 18, 22, 29, 30, 32, 38). This is because using recombinant E. coli strains for production of biopolymers has several potential advantages, including fast growth, a wide range of possible carbon substrates, well-understood genetics and metabolic pathways, the availability of well-established high-cell-density culture techniques, and possibly easier and less costly downstream processing techniques (32).
PHA with medium-chain-length monomers (mcl-PHA), which are composed of 3-hydroxyalkanoic acids that have 6 to 14 carbon atoms, occur naturally as storage products of fluorescent pseudomonads (8, 14, 16, 39, 41) and are suitable for applications in which flexibility and elasticity are required (28). One of the best-studied mcl-PHA producers is Pseudomonas oleovorans GPo1 (8). This bacterium relies on the β-oxidation pathway to convert fatty acid intermediates into (R)-3-hydroxyacyl-coenzyme A [(R)-3-hydroxyacyl-CoA] thioesters, which are the substrates of the PHA polymerases that catalyze the committed step of mcl-PHA biosynthesis (15, 17, 19, 21). P. oleovorans contains two PHA polymerases, which are encoded by phaC1 and phaC2 of the pha gene cluster (17) (Fig. 1); the substrate specificities of these enzymes differ slightly (18). Moreover, it has been demonstrated that both of these polymerases are functional proteins which are able to catalyze PHA formation independent of each other; i.e., one of the polymerase-encoding genes is enough to produce mcl-PHA in heterologous hosts (18).
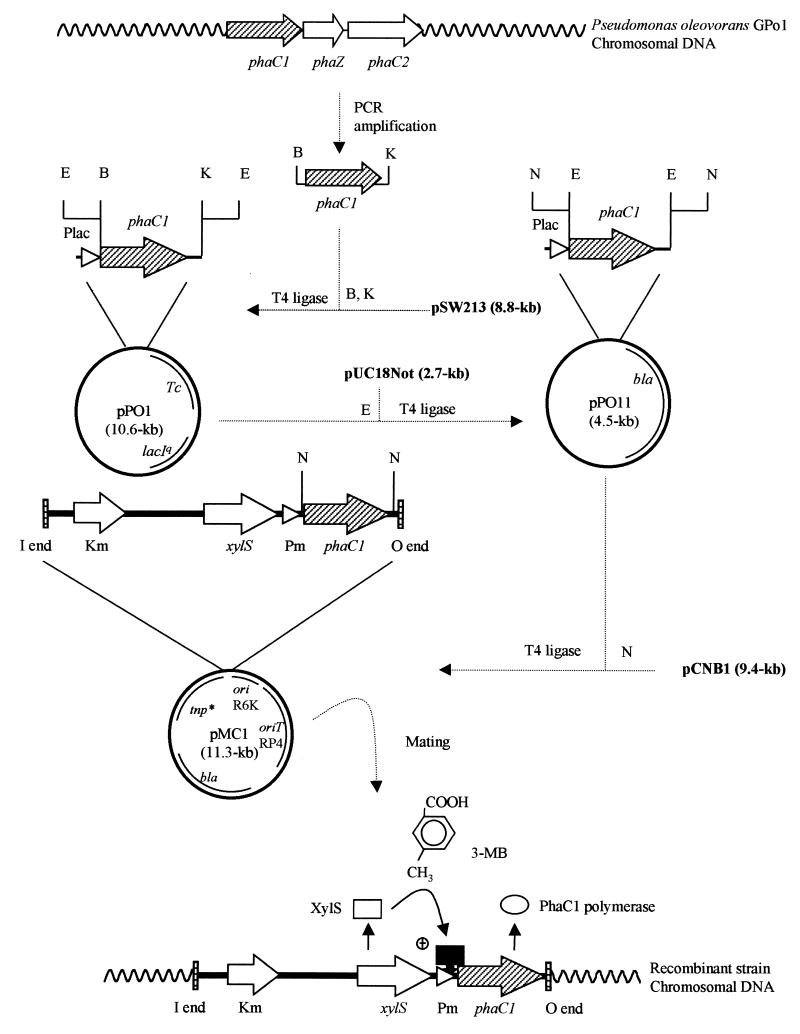
FIG. 1.
Construction of a phaC1 expression system and chromosomal integration. The molecular organization of the phaZ, phaC1, and phaC2 genes, which code for the PHA depolymerase and two PHA polymerases, respectively, on the chromosome of P. oleovorans GPo1, is indicated at the top. The final minitransposon construct is shown at the bottom. Addition of 3-MB to a bacterial culture activates the XylS regulatory protein, which induces expression of the phaC1 gene, resulting in production of PhaC1 polymerase. Abbreviations: Tc, tetracycline resistance; bla, ampicillin resistance; Km, kanamycin resistance; tnp*, Tn5 transposase. The positions of the 19-bp Tn5 I and O ends, oriT, RP4, and oriR6K are indicated. B, E, K, and N represent restriction enzymes BamHI, EcoRI, KpnI, and NotI, respectively.
Recently, it was reported that E. coli strains blocked in the 3-ketoacyl thiolase (FadA) or 3-hydroxyacyl-CoA dehydratase (FadB) enzyme activity of the β-oxidation pathway were able to accumulate mcl-PHA when only the phaC1 or phaC2 gene of P. oleovorans GPo1 was expressed (32). Similar results were observed for the PHA polymerases of Pseudomonas aeruginosa (22, 29). The use of heterologous expression systems and/or high-copy-number plasmids has shown that the amounts of PHA in these recombinant organisms depend on the polymerase dosage (32). Thus, it is possible to produce significant amounts of PHA in E. coli, but this production does require stable and constant expression of phaC genes. A major problem in using such expression systems in large-scale fermentation is plasmid maintenance and stability. The classical approach is to add antibiotics to the culture medium to maintain the phenotype of the recombinant strain. This can have a considerable effect on the reproducibility of the results and the final cost of the product. An attractive alternative is to develop a stable, regulated, inexpensive expression system for phaC1 gene expression as a first step toward establishing PHA production in recombinant strains. In this paper we describe the design and use of minitransposons to create recombinant strains that carry a single copy of the desired heterologous phaC gene in the chromosome, based on a set of tools developed by de Lorenzo and coworkers (6, 7). Furthermore, we exploited the stability of the system to culture mcl-PHA-producing recombinant E. coli in a bioreactor operated in the batch or continuous cultivation mode in the absence of a selection marker. We isolated the PHA produced and determined its monomer composition and molecular weight and the chirality of the 3-hydroxyoctanoic acid monomers.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Table 1 shows the bacterial strains and plasmids used in this study.
TABLE 1.
Bacterial strains and plasmids and their relevant genotypes and phenotypes
| Strain or plasmid | Relevant genotype or phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| P. oleovorans | ||
| GPo1 | OCT, PHA+ | 36 |
| POMC1 | GPo1 derivative, xylS/Pm::phaC1, Kmr | This study |
| P. putida | ||
| GPp104 | NTG mutant of P. putida KT2442 (2), PHA− | 17 |
| GPMC1 | GPp104 derivative, xylS/Pm::phaC1, Kmr | This study |
| E. coli | ||
| DH10B | Host for E. coli plasmids | 11 |
| CC1118λpir | Host for pUT-derived plasmids | 13 |
| JMU193 | fadR::Tn10 fadB64, Tcr | 33 |
| 193MC1 | JMU193 derivative, xylS/Pm::phaC1, Kmr Tcr | This study |
| Plasmids | ||
| pPSW213 | RK2 derivative, Tcr, lacIq/Plac | 4 |
| pUC18Not | pUC18 derivative, Apr, NotI-flanking MCS | 13 |
| pCNB1 | mini-Tn5 delivery plasmid, Kmr Apr, xylS/Pm in mobile segment | 6 |
| pPO1 | pSW213 derivative, Tcr, lacIq/Plac::phaC1 | This study |
| pPO11 | pUC18Not derivative, Apr, 1.8-kb NotI cassette containing phaC1 | This study |
| pMC1 | pCNB1 derivative, Apr Kmr, xylS/Pm::phaC1 | This study |
OCT, natural plasmid enabling growth on C6 to C12 alkanes; NTG, N′-methyl-N′-nitro-N′-nitrosoguanidine; MCS, multiple cloning site.
DNA manipulation.
DNA manipulation and other molecular biology techniques were performed essentially as described previously (34). E. coli cells were transformed by using the RbCl method or by electroporation (Gene Pulser; Bio-Rad) (9). Minitransposon elements were inserted into the chromosomes of the target strains by using the filter-mating technique (13).
Isolation of the phaC1 gene.
The DNA fragment containing the phaC1 gene of P. oleovorans GPo1 was amplified by PCR by using 1 μg of P. oleovorans GPo1 chromosomal DNA as the template and the following primers: NC1 (5′-GATC GATCGGATCCCGGTACTCGTCTCAGGACAACGGAGCGTCGTAGAT G-3′) and CC1 (5′-GATCGATCGGTACCTGAAATGAACACCGTGGCGTCCCGCAGGTGGCC-3′) (the engineered BamHI and KpnI sites are underlined).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis.
The proteins in whole-cell samples were separated by 10% polyacrylamide sodium dodecyl sulfate–polyacrylamide gel electrophoresis as described previously (20). Polymerase C1 antibodies were prepared and a Western blot analysis was performed as reported previously (19).
Cultivation conditions and media.
Unless otherwise stated, bacteria were grown in 500-ml Erlenmeyer flasks containing 100 ml of Luria-Bertani (LB) medium (34) at 30°C (P. oleovorans) or 37°C (E. coli) with vigorous shaking. Appropriate selection markers (50 μg of kanamycin per ml, 12.5 μg of tetracycline per ml, 100 μg of ampicillin per ml) and the inducer isopropyl-β-d-1-thiogalactopyranoside (IPTG) were added if necessary. E2 minimal medium supplemented with 0.1% (vol/vol) MT microelement solution (21), 20 mM glycerol, and 1 mM 3-methylbenzoic acid (3-MB) was used for PhaC1 polymerase production by E. coli in shaking flasks. For PHA production by Pseudomonas strains in shaking flasks, cells were cultured overnight in 0.1 N E2 minimal medium containing 15 mM octanoic acid (18).
Chemostat cultures of P. oleovorans POMC1 (Table 1) were grown in a 3-liter reactor that had a working volume of 1 liter and was equipped as previously described (42). The cells were precultured overnight at 30°C in 500-ml Erlenmeyer flasks containing 100 ml of E2 medium supplemented with 10 mM citric acid and kanamycin. The precultures were used to inoculate 1 liter of continuous culture medium containing 8.35 mM (NH4)2SO4, 7.4 mM KH2PO4, 1 mM MgSO4, 10 M FeSO4, and 0.1% (vol/vol) MT microelement solution (12). Octanoic acid was used as the carbon source. The carbon/nitrogen (C/N) ratio was 15, and the 3-MB concentration was varied as part of the experiment. After inoculation, the culture was grown in batch mode to a density of 1.0 g · liter−1 and then was switched to the continuous operation mode at a dilution rate of 0.2 h−1. We assumed that a steady state was present when the optical density of the culture at 450 nm and the dissolved-oxygen tension were constant for at least three mean residence times. The PHA content was analyzed by using cells from the outflow.
E. coli 193MC1 (Table 1) was cultivated in a 3-liter reactor with a 1.5-liter working volume containing E2 minimal medium supplemented with 100 mM glycerol as the carbon source. Palmitic acid was added constantly at a rate of 65 mg · h−1 · liter for 40 h from the onset of the exponential phase. 3-MB was added as indicated below in order to induce production of PhaC1 polymerase. The cells were precultured overnight at 37°C in 500-ml Erlenmeyer flasks containing 100 ml of E2 minimal medium supplemented with 20 mM glycerol and kanamycin.
The standard cultivation conditions in chemostat cultures were pH 7 and 30°C for P. oleovorans POMC1 and 37°C for E. coli 193MC1. The cultures were agitated at 1,500 rpm constantly, and air was supplied at a rate of 1.4 liter · min−1. The pH was automatically controlled by using 4 N sodium hydroxide. The dissolved-oxygen tension was monitored with an in situ amperometric polarographic Ingold oxygen sensor (Mettler Toledo) with a type S membrane (Silicon) and was always more than 30% of saturation. Cell densities, expressed as milligrams of cell dry weight (CDW) per milliliter, were determined gravimetrically by using tared 0.2-μm-pore-size filters (Costar).
Polymer isolation and analysis.
For qualitative detection of PHA inclusion bodies, cells were observed by phase-contrast light microscopy after they were stained with Sudan Black (35). The amounts of total cellular 3-hydroxyalkanoates (free and PHA-incorporated monomers) were determined with a gas chromatograph (model GC8000; Fisons) equipped with a 25-m type CP-Sil5CB capillary column (Chrompack) as described previously (21). Polymers were extracted by lyophilization (1 mbar, 48 to 144 h) and subsequent Soxhlet extraction (10% [wt/vol] CH2Cl2, 50°C, 8 h) of dried cells. The resulting solution was filtered through a porous disperger, and mcl-PHA were then precipitated in ice-cold methanol (10-fold excess) with vigorous stirring. After the methanol-CH2Cl2 solvent mixture was decanted, the polymers were air dried overnight and stored at 4°C. After the polymers were hydrolyzed and the monomers were methylated, the PHA-derived methyl-3-hydroxyalkanoates were identified by gas chromatography and by gas chromatography-mass spectrometry. Mass spectra were obtained by performing an electron impact analysis with a mass spectrometer (model MD800; Fisons) at 70 eV after trimethylsilyl derivatization of the methyl-3-hydroxyalkanoates as described by Lee and Choi (23).
The molecular weights of purified mcl-PHA were determined by gel permeation chromatography (GPC). Samples were dissolved in tetrahydrofuran and injected onto a PL-Gel mixed-C, 5-m column (7.5 by 600 mm; Polymer Laboratories). The GPC system (Knauer) was equipped with a low-angle laser light-scattering detector (model KMX-6 LALLS; Chromatix), a viscosity detector (model H502; Viskotek), and a differential refractive index detector (Knauer). Universal calibration with narrowly dispersed polystyrene standards (Polymer Laboratories) was used to calculate average molecular weights.
The absolute configurations of the 3-hydroxyoctanoic methyl ester monomers obtained after methanolysis of the polymer were determined by gas chromatography performed with a type Beta-DEX 120 column (fused silica capillary column; length, 60 m; inside diameter, 0.25 mm; film thickness, 0.25 μm; Supelco). The temperature profile started with an isothermal oven temperature of 115°C for 15 min; then the temperature was increased from 115 to 122°C at a rate of 0.5°C · min−1, and this was followed by an additional isothermal period consisting of 15 min at 122°C. Compounds were detected with a flame ionization detector. R and S enantiomers were identified on the basis of retention times by using commercially available standards (Larodan Lipids) and R-3-hydroxyoctanoic methyl ester monomers obtained from PHA of a P. oleovorans wild-type strain.
RESULTS
Engineering of a monocopy phaC1 expression system for mcl-PHA production in recombinant strains.
Integration of the mcl-PHA polymerase-encoding genes into the chromosomes of recombinant microorganisms might be a particularly well-suited method for generating genetically stable strains in order to scale up mcl-PHA production. Therefore, a 1.8-kb DNA fragment containing the phaC1 gene of P. oleovorans GPo1 was amplified by PCR by using the chromosomal DNA of the wild-type strain as the template (Fig. 1). In order to simplify the subsequent cloning, the amplification primers were designed to create a BamHI site upstream of the start codon and a KpnI site downstream of the stop codon of the phaC1 gene (Fig. 1). The PCR product was ligated into the RK2-derived shuttle vector pSW213 cut with the endonucleases indicated above. This cloning step was performed to test the functionality of the phaC1 gene product by complementation of the PHA-negative strain Pseudomonas putida GPp104 (Table 1) in order to avoid a nonfunctional phaC1 gene derived from a mutation generated during PCR amplification. Several clones containing the plasmid with the 1.8-kb fragment were selected and transferred by triparental mating to P. putida GPp104. PHA accumulation by the exconjugants was assessed qualitatively by phase-contrast microscopy after Sudan Black staining (data not shown). This method allowed us to isolate plasmid pPO1 (Fig. 1 and Table 1), which complemented mcl-PHA production in P. putida GP104 in the presence of the inducer IPTG. Based on an active PHA polymerase encoded on pPO1, we constructed the mini-Tn5 delivery plasmid pMC1 (Table 1), as shown in Fig. 1. Plasmid pMC1 is a pCNB1 (Table 1) derivative containing a phaC1 expression system (xylS/Pm::phaC1) which is based on the regulatory system of the well-characterized meta-cleavage catabolic operon of the TOL plasmid of P. putida as part of its mobile element (6); XylS is the regulatory protein that turns on the Pm promoter when it is activated by benzoates or toluates (31) (Fig. 1). The mini-Tn5 transposon encoded on plasmid pMC1 was transferred into the chromosomes of P. putida GPp104, P. oleovorans GPo1, and E. coli JMU193 by mating and subsequent transposition, which generated the transconjugants P. putida GPMC1, P. oleovorans POMC1, and E. coli 193MC1, respectively (Table 1). These recombinant strains carry in their chromosomes a single copy of the expression system xylS/Pm::phaC1 (Fig. 1). mcl-PHA production was determined qualitatively and quantitatively in P. putida GPMC1, in which PHA accounted for 20% of the CDW in the presence of 1 mM 3-MB (data not shown). However, this recombinant strain was not analyzed further due to the low yield of biomass and the longer generation time observed in batch fermentations compared to P. oleovorans POMC1 (data not shown).
mcl-PHA production in a P. oleovorans recombinant strain.
Compared to the parental strain P. oleovorans GPo1, P. oleovorans POMC1 contains an additional copy of the phaC1 gene in its chromosome. First, we cultured POMC1 in shaking flasks in the presence and absence of 3-MB to ascertain whether the new phaC1 expression system (i.e., an increased number of molecules of PhaC1 polymerase) might influence mcl-PHA production in P. oleovorans. The results showed that strain POMC1 produced 1.8-fold more mcl-PHA when it was grown in the presence of inducer (0.4 mM) or when it was compared to parental strain GPo1, which suggested that xylS/Pm::phaC1 does functionally enhance PhaC1 production (Table 2). However, it is well-known that limitation of an essential nutrient like oxygen or nitrogen during growth of P. oleovorans results in an increase in mcl-PHA production (8). Thus, the higher level of polymer production in POMC1 in the presence of the inducer could have been due to an unknown growth limitation or another factor which imposes stress conditions on the cells. To gain better insight into the contributions of these factors to overall PHA production, we cultured P. oleovorans POMC1 in a chemostat and monitored all growth parameters (pH, aeration, volume, nutrient content, etc.) so that they could be maintained at desired levels (see above). The stability of insertion of mobile element xylS/Pm::phaC1 into the chromosome of strain POMC1 was confirmed by plating onto LB medium and LB medium containing kanamycin, which revealed that all colonies were kanamycin resistant in every steady state. The results obtained during this fermentation experiment showed that addition of 3-MB to the POMC1 culture resulted in only a slight increase in mcl-PHA synthesis under the conditions used (Table 2).
TABLE 2.
PHA production by P. oleovorans POMC1 in batch and continuous culturesa
| Culture | 3-MB concn (mM) | CDW (g · liter−1) | Amt of PHA (% of CDW) |
|---|---|---|---|
| Batch fermentation | 0 | 0.36 | 20.6 ± 1.9 |
| 0.20 | 0.40 | 34.6 ± 3.4 | |
| 0.40 | 0.42 | 37.9 ± 3.7 | |
| 0.60 | 0.42 | 35.5 ± 3.5 | |
| Continuous fermentation | 0 | 1.50 | 42.7 ± 1.7 |
| 0.10 | 1.64 | 53.2 ± 1.3 | |
| 0.15 | 1.56 | 47.0 ± 3.0 | |
| 0.20 | 1.78 | 49.7 ± 1.6 | |
| 0.30 | 1.66 | 47.3 ± 1.8 |
Each value is the average of values from three different experiments.
mcl-PHA production in an E. coli recombinant strain.
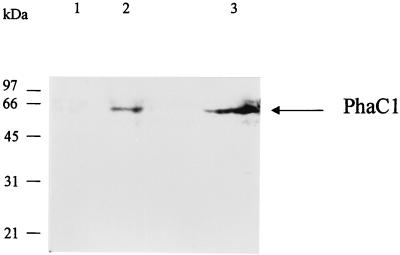
We have shown previously that strain E. coli JMU193 (fadR− fadB−) produces mcl-PHA when it is transformed with a high-copy-number plasmid containing the phaC1 gene and that palmitic acid is one of the best substrates for mcl-PHA production (10% of the CDW) (32). Therefore, E. coli 193MC1 was cultivated under similar conditions. PhaC1 production was analyzed by Western blotting by using cultures grown in the absence or presence of the inducer 3-MB, which revealed that PhaC1 polymerase was detected only when 3-MB was added to the culture medium (Fig. 2). This result confirmed that the xylS/Pm::phaC1 expression system is functional in this strain.
FIG. 2.
Detection of PhaC1 polymerase by Western blot analysis of E. coli 193MC1. Western blotting was performed with polyclonal antibodies raised against PhaC1 of P. oleovorans. Cells of E. coli 193MC1 were cultured in E2 minimal medium in the absence (lane 1) and in the presence (lane 2) of 1 mM inducer (3-MB). Lane 3 contained purified PhaC1 polymerase. All lanes contained 25 μg of protein. The molecular masses (in kilodaltons) of the marker proteins and the position of the PhaC1 polymerase are indicated on the left and right, respectively.
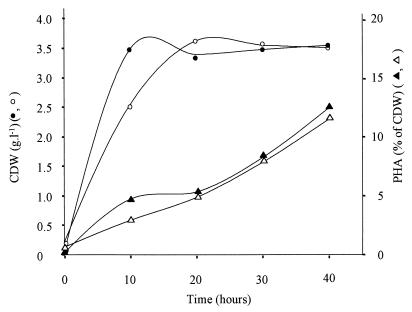
To demonstrate that a single copy of the phaC1 gene gives E. coli 193MC1 the ability to produce mcl-PHA in the absence of a selection marker, we cultured 193MC1 in fed-batch mode in a bioreactor in the absence or presence of 0.25 or 1 mM 3-MB by using glycerol as the growth substrate and palmitic acid as the mcl-PHA precursor (Fig. 3). The final amounts of mcl-PHA produced under these growth conditions were similar (11 to 12% of the CDW) independent of the concentration of 3-MB in the medium, suggesting that PhaC1 polymerase was not limiting in these experiments. This is in agreement with previous findings that PhaC1 polymerase is needed only in very small amounts for mcl-PHA production in P. oleovorans GPo1 (19). The phenotype stability of strain 193MC1 was determined as described above for strain POMC1, and 193MC1 turned out to be 100% phenotypically stable throughout the fermentation process.
FIG. 3.
mcl-PHA production by E. coli 193MC1 in a fed-batch culture. Symbols: ○ and ▵, 0.25 mM 3-MB used during fermentation; ● and ▴, cultivation in the presence of 1 mM 3-MB. The inducer 3-MB was supplied during the exponential phase after 5 h of growth. Palmitic acid was added at a constant feeding rate of 65 mg · h−1 · liter−1 for 40 h.
Relative monomer compositions of isolated mcl-PHA.
The polymers produced by 193MC1 and POMC1 were isolated and used for additional characterization experiments. Table 3 shows that the polyester formed by E. coli 193MC1 contained 1.3-fold less 3-hydroxyoctanoate (C8) than Pseudomonas strains contain (86 to 89 mol%). The relative amount of the 3-hydroxydecanoate monomer (C10) was 20-fold higher in the polymer produced by E. coli than in the mcl-PHA produced by the Pseudomonas strain. However, this is not surprising since E. coli 193MC1 was grown on fatty acids with longer chain lengths than the fatty acids on which Pseudomonas strain POMC1 was grown. Cultivation of the Pseudomonas strain on longer-chain-length fatty acids could also have shifted the molar ratio of the monomers towards C10 (16). In all of the cases tested, increased amounts of inducer that presumably resulted in increased amounts of PhaC1 polymerase which had a higher level of substrate specificity towards shorter 3-hydroxyalkanoates (18, 19) led to a shift in monomer composition towards higher relative contents of 3-hydroxyhexanoate monomers (C6). Similar effects were observed with a recombinant P. oleovorans strain that overproduced PhaC1 polymerase (19).
TABLE 3.
Relative monomer compositions of isolated PHA produced by recombinant strains
| Strain | Culture | 3-MB concn (mM) | CDW (g · liter−1) | Amt of PHA (% of CDW) | Monomer composition (mol%)a
|
||
|---|---|---|---|---|---|---|---|
| C6 | C8 | C10 | |||||
| E. coli 193MC1b | Fed batch fermentation | 0.25 | 3.48 | 11 | 13 | 66 | 21 |
| 1.00 | 3.55 | 12 | 14 | 66 | 20 | ||
| P. oleovorans POMC1c | Continuous fermentation | 0.00 | 1.50 | 43 | 10 | 89 | <1 |
| 0.10 | 1.67 | 53 | 13 | 86 | <1 | ||
| P. oleovorans GPo1d | 10 | 89 | <1 | ||||
Monomer compositions are expressed as molar percentages. C6, 3-hydroxyhexanoate; C8, 3-hydroxyoctanoate; C10, 3-hydroxydecanoate.
Polymers produced by E. coli 193MC1 were isolated from the culture described in the legend to Fig. 3.
Polymers produced by P. oleovorans POMC1 were isolated from the continuous culture (Table 2).
Relative monomer composition of PHA lab stock produced by wild-type P. oleovorans GPo1. Each value is the average of values from three different assays.
Molecular weights of the polymers.
It has been found previously that in a recombinant polyhydroxybutyrate (PHB)-producing E. coli strain the molecular weight of the PHB is controlled by PHB synthase activity (37). The more synthase molecules, the shorter the chain length of the polymer, suggesting that the enzyme is part of a system which controls the polymer chain length. In order to study this effect in E. coli 193MC1 and P. oleovorans POMC1, we determined the molecular mass distribution of the isolated mcl-PHA by GPC. The mcl-PHA produced by P. oleovorans POMC1 had molecular weights that varied between 180,000 and 230,000 depending on the inducer concentration added; these values are consistent with the values obtained for the P. oleovorans parental strain (28).
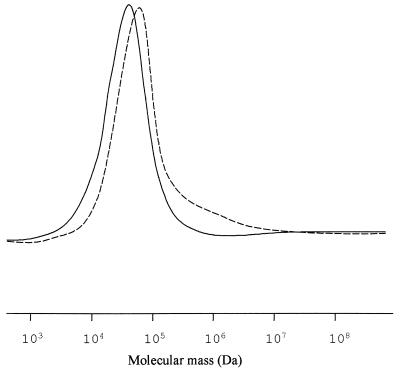
The behavior of E. coli 193MC1 was different. When induced with 1 mM 3-MB, strain 193MC1 produced considerably shorter polyesters, whose molecular weights varied between 67,000 and 70,000 and whose polydispersity was 2.7. Like the E. coli PHB producer, induction with only 0.25 mM 3-MB shifted the median molecular weight of the polymer to a higher value compared to the molecular weight of the polymer induced with 1 mM 3-MB (Fig. 4), and the molecular weight distribution was considerably broader. This effect was not observed in the case of P. oleovorans POMC1, in which the mcl-PHA produced had a polydispersity of 4.3 independent of the amount of inducer added to the medium.
FIG. 4.
Molecular mass distribution of mcl-PHA produced by E. coli 193MC1. The PHA isolated from the cultures described in the legend to Fig. 3 were analyzed by GPC. Solid line, polymer produced in the presence of 1 mM 3-MB; dashed line, PHA isolated from cells induced with 0.25 mM 3-MB.
Chirality of the monomers produced by E. coli 193MC1.
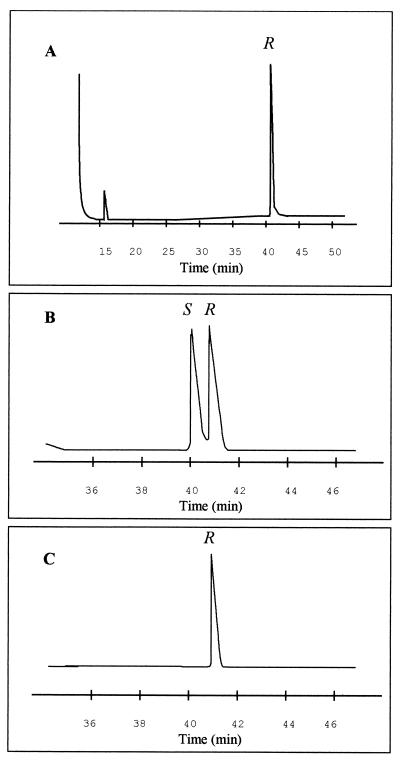
It has been demonstrated that the monomers produced by parental strain P. oleovorans GPo1 have only the R configuration (21). We used chiral gas chromatography to analyze the chirality of the C8 monomers obtained from the mcl-PHA produced by E. coli 193MC1 cultured in the presence of 1 mM 3-MB. We determined the retention time of the R-methyl-C8 monomer produced by the wild-type strain in order to identify the corresponding R and S forms in the commercially available methyl-C8 standard racemic mixture (Fig. 5A and B). A retention time of 41.5 min was obtained for the R-methyl-C8 enantiomer. This result was confirmed by coinjecting a sample containing the standard mixture and the methyl monomers of P. oleovorans GPo1 (data not shown). The chromatogram in Fig. 5C shows that the PHA polymer produced by E. coli 193MC1 contained only monomers having the R configuration.
FIG. 5.
Gas chromatography of the C8-monomer methyl esters produced by E. coli 193MC1. (A) Methanolysis products of standard PHA from our lab stock isolated from parental strain P. oleovorans GPo1. (B) Racemic reference material. (C) Identification of the R enantiomer of the C8-monomer methyl esters of mcl-PHA produced by E. coli 193MC1.
DISCUSSION
Previously, it has been shown that biosynthesis of short-chain-length PHA (PHB) in heterologous microorganisms that do not naturally produce polyester granules allows manipulation of the biosynthetic enzyme levels and hence modification and control of the molecular weight and the polydispersity of the polymer (37). However, the lack of stability of recombinant microorganisms is often a major drawback for the production of sufficient amounts of PHA (32). In order to generate stable mcl-PHA-producing recombinants, we used an E. coli host strain blocked in the 3-hydroxyacyl-coenzyme A (CoA) dehydratase (FadB) enzyme activity of the β-oxidation pathway, which provides enough 3-hydroxyacyl-CoA precursors for PHA synthesis (32), and introduced an mcl-PHA polymerase-encoding gene of P. oleovorans into the chromosome by using a minitransposon technique.
Minitransposons developed by de Lorenzo and coworkers are recombinant transposons in which only the elements essential for transposition have been retained and arranged in such a manner that the transposase-encoding gene is adjacent to but outside the mobile DNA element (6, 7). The advantages of using these tools include the fact that once inserted in a target strain, the minitransposons are inherited in a stable fashion and, unlike natural transposons, do not provoke DNA rearrangements or other forms of genetic instability since they lack the cognate transposase-encoding gene and the major part of the insertion elements present in wild-type transposons. In this study, we found that minitransposons can be used not only to clone and express genes in heterologous microorganisms but also to generate stable recombinant strains for production purposes, which allowed us to culture bacteria in a bioreactor in the absence of selection markers.
Using our system, we found that the chain length of the polymer produced by an E. coli 193MC1 recombinant expressing the mcl-PHA polymerase-encoding gene varies depending on the amount of inducer added to the medium. A reduction in the inducer concentration resulted in an increase in the number of polymer molecules with longer chains (Fig. 4), which can only be explained by fewer molecules of PhaC1 polymerase. This is in agreement with the hypothesis that higher enzyme levels could lead to an increased number of chain initiation events, resulting in shorter polymer chains (18). If we assume that the xylS/Pm expression system is working properly in Pseudomonas, as shown previously (6), and that the recombinant phaC1 gene is expressed to a similar extent, as seen in E. coli, our results also imply (based on the molecular weight of the PHA formed by the recombinant P. oleovorans POMC1, which did not differ significantly from the molecular weight of the PHA produced by the wild-type strain) that such a strategy cannot be used in a native PHA-producing organism. The additional copy of the phaC1 gene affected only the polydispersity of the polymer, which was much higher than the polydispersity of polymers produced by wild-type Pseudomonas strains. Whether this effect is caused by the additional polymerase molecules in a presumably unchanged precursor concentration environment remains to be determined.
Our results confirmed the enantioselectivity of the PhaC1 polymerase of P. oleovorans GPo1. Although it is known that mcl-PHA precursors are formed via β-oxidation, not much is known about the β-oxidation pathway of Pseudomonas. For E. coli it has been shown that the 3-hydroxyacyl-CoA intermediates of the β-oxidation pathway have the S configuration (3, 24). However, the monomers of mcl-PHA produced by the wild-type strain of P. oleovorans occur exclusively in the R configuration (21). In this study, we found that the main monomer (3-hydroxyoctanoate) of the polymer produced by E. coli 193MC1 also has the R configuration exclusively (Fig. 5). Therefore, it would still be interesting to determine how S-β-oxidation intermediates are converted to R monomers during incorporation into PHA.
The enantioselectivity of the PhaC1 polymerase could be very useful in using recombinant microorganisms to produce optically active intermediates which are difficult to produce by synthetic chemistry methods. In fact, utilization of R-hydroxycarboxylic acids as precursors for the synthesis of captopril and β-lactams has been proposed by Ohashi and Hasegawa (25). Based on the versatile metabolism of E. coli, alcohols, aldehydes, and carboxylic acids can be taken up by cells, and depending on the metabolic pathways used, the corresponding hydroxycarboxylic acid intermediates can be formed. Since mcl-PHA polymerases have a broad substrate range (40), we expect that many of these as-yet-untested hydroxycarboxylic acid intermediates can be incorporated into mcl-PHA. Thus, this work opens a new scenario in the use of recombinant E. coli strains not only for production of various plastics but also for production of chiral R-3-hydroxycarboxylic acids.
ACKNOWLEDGMENTS
We thank Q. Ren for helpful comments. We are indebted to M. Colussi of the Institute of Polymer Sciences, ETHZ, for help with the GPC analysis and to A. Prieto for help with the chirality analysis. We appreciate the contribution of G. de Roo to the Western blot analysis. We also thank M. Röthlisberger and H.-J. Feiten for excellent technical assistance.
M. A. Prieto was the recipient of an EMBO long-term fellowship, and D. Radnovic received support from the Fund for an Open Society, Belgrade.
REFERENCES
- 1.Anderson A J, Dawes E A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagdasarian M, Lurz R, Ruckert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 3.Black P N, DiRusso C C. Molecular and biochemical analyses of fatty acid transport, metabolism, and gene regulation in Escherichia coli. Biochim Biophys Acta. 1994;1210:123–145. doi: 10.1016/0005-2760(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 4.Chen C-Y, Stephen C, Winass C. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J Bacteriol. 1991;173:1139–1144. doi: 10.1128/jb.173.3.1139-1144.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Koning G J M, Kellerhals M B, van Meurs C, Witholt B. Poly(hydroxyalkanoates) from fluorescent pseudomonads in retrospect and prospect. J Environ Polymer Deg. 1996;4:243–252. [Google Scholar]
- 6.de Lorenzo V, Fernandez S, Herrero M, Jakubzik U, Timmis K N. Engineering of alkyl- and haloaromatic-responsive gene expression with mini-transposons containing regulated promoters of biodegradative pathways of Pseudomonas. Gene. 1993;130:41–46. doi: 10.1016/0378-1119(93)90344-3. [DOI] [PubMed] [Google Scholar]
- 7.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 8.de Smet M J, Eggink G, Witholt B, Kingma J, Wynberg H. Characterization of intracellular inclusions formed by Pseudomonas oleovorans during growth on octane. J Bacteriol. 1983;154:870–878. doi: 10.1128/jb.154.2.870-878.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fidler S, Dennis D. Polyhydroxyalkanoate production in recombinant Escherichia coli. FEMS Microbiol Rev. 1992;103:231–236. doi: 10.1016/0378-1097(92)90314-e. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D. Studies on transformation of Escherichia coli with plasmid. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 12.Hazenberg W M. Production of poly(3-hydroxyalkanoates) by Pseudomonas oleovorans in two-liquid-phase media. Ph.D. thesis. Zürich, Switzerland: ETH; 1997. [Google Scholar]
- 13.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic selection markers for cloning and stable chromosomal insertion of foreign DNA in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huijberts G N M, Eggink G, de Waard P, Huisman G W, Witholt B. Pseudomonas putida KT2442 cultivated on glucose accumulates poly(3-hydroxyalkanoates) consisting of saturated and unsaturated monomers. Appl Environ Microbiol. 1992;58:536–544. doi: 10.1128/aem.58.2.536-544.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huijberts G N M, de Rijk T C, de Waard P, Eggink G. 13C nuclear magnetic resonance studies of Pseudomonas putida fatty acid metabolic routes involved in poly(3-hydroxyalkanoate) synthesis. J Bacteriol. 1994;176:1661–1666. doi: 10.1128/jb.176.6.1661-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huisman G W, de Leeuw O, Eggink G, Witholt B. Synthesis of poly-3-hydroxyalkanoates is a common feature of fluorescent pseudomonads. Appl Environ Microbiol. 1989;55:1949–1954. doi: 10.1128/aem.55.8.1949-1954.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huisman G W, Wonink E, Meima R, Kazemier B, Terpstra P, Witholt B. Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. J Biol Chem. 1991;266:2191–2198. [PubMed] [Google Scholar]
- 18.Huisman G W, Wonink E, de Koning G J M, Preusting H, Witholt B. Synthesis of poly(3-hydroxyalkanoates) by mutant and recombinant Pseudomonas strains. Appl Microbiol Biotechnol. 1992;38:1–5. [Google Scholar]
- 19.Kraak M N, Smits T H M, Kessler B, Witholt B. Polymerase C1 levels and poly(R-3-hydroxyalkanoate) synthesis in wild-type and recombinant Pseudomonas strains. J Bacteriol. 1997;179:4985–4991. doi: 10.1128/jb.179.16.4985-4991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lageveen R G, Huisman G W, Preusting H, Ketelaar P, Eggink G, Witholt B. Formation of polyesters by Pseudomonas oleovorans: effect of substrates on formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl Environ Microbiol. 1988;54:2924–2932. doi: 10.1128/aem.54.12.2924-2932.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langenbach S, Rehm B H A, Steinbüchel A. Functional expression of the PHA synthase gene phaC1 from Pseudomonas aeruginosa in Escherichia coli results in poly(3-hydroxyalkanoate) synthesis. FEMS Microbiol Lett. 1997;150:303–309. doi: 10.1016/s0378-1097(97)00142-0. [DOI] [PubMed] [Google Scholar]
- 23.Lee E Y, Choi C Y. Gas chromatography-mass spectrometric analysis and its application to a screening procedure for novel bacterial polyhydroxyalkanoic acids containing long chain saturated and unsaturated monomers. J Ferment Bioeng. 1995;80:408–414. [Google Scholar]
- 24.Magnuson K, Jackowski S, Rock C O, Cronan J E. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohashi T, Hasegawa J. d(−)-β-Hydroxycarboxylic acids as raw materials for captopril and β-lactams. In: Collins A N, Sheldrake G N, Crosby J, editors. Chirality in industry. Manchester, United Kingdom: ZENECA Specialities; 1992. pp. 269–278. [Google Scholar]
- 26.Page W J. Bacterial polyhydroxyalkanoates, natural biodegradable plastics with a great future. Can J Microbiol. 1995;41:1–3. [Google Scholar]
- 27.Poirier Y, Dennis D E, Klomparens K, Sommerville C. Polyhydroxybutyrate, a biodegradable thermoplastic, produced in transgenic plants. Science. 1992;256:520–523. doi: 10.1126/science.256.5056.520. [DOI] [PubMed] [Google Scholar]
- 28.Preusting H, Nijenhuis A, Witholt B. Physical characteristics of poly(3-hydroxyalkanoates) and poly(3-hydroxyalkenoates) produced by Pseudomonas oleovorans grown on aliphatic hydrocarbons. Macromolecules. 1990;23:4220–4224. [Google Scholar]
- 29.Qi Q, Rehm B H A, Steinbüchel A. Synthesis of poly(3-hydroxyalkanoate) in Escherichia coli expressing the PHA synthase gene phaC2 from Pseudomonas aeruginosa: comparison of PhaC1 and PhaC2. FEMS Microbiol Lett. 1997;157:155–162. doi: 10.1111/j.1574-6968.1997.tb12767.x. [DOI] [PubMed] [Google Scholar]
- 30.Qi Q, Steinbüchel A, Rehm B H A. Metabolic routing towards polyhydroxyalkanoic acid synthesis in recombinant Escherichia coli (fadR): inhibition of fatty acid β-oxidation by acrylic acid. FEMS Microbiol Lett. 1998;167:89–94. doi: 10.1111/j.1574-6968.1998.tb13212.x. [DOI] [PubMed] [Google Scholar]
- 31.Ramos J L, Mermod N, Timmis K N. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta-cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol Microbiol. 1987;1:293–300. doi: 10.1111/j.1365-2958.1987.tb01935.x. [DOI] [PubMed] [Google Scholar]
- 32.Ren Q. Biosynthesis of medium chain length poly-3-hydroxyalkanoates: from Pseudomonas to Escherichia coli. Ph.D. thesis. Zürich, Switzerland: ETH; 1997. [Google Scholar]
- 33.Rhie H G, Dennis D. Role of fadR and atoC (Con) mutations in poly(3-hydroxybutyrate-Co-3-hydroxyvalerate) synthesis in recombinant pha+ Escherichia coli. Appl Environ Microbiol. 1995;61:2487–2492. doi: 10.1128/aem.61.7.2487-2492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Schaad N W. Laboratory guide for identification of plant pathogenic bacteria. St. Paul, Minn: American Phytopathological Society; 1988. [Google Scholar]
- 36.Schwartz R D, McCoy C J. Pseudomonas oleovorans hydroxylation-epoxidation system: additional strain improvements. Appl Microbiol. 1973;26:217–218. doi: 10.1128/am.26.2.217-218.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sim S J, Snell K D, Hogan S A, Stubbe J, Rha C, Sinskey A J. PHA synthase activity controls the molecular weight and polydispersity of polyhydroxybutyrate in vivo. Nat Biotechnol. 1997;15:63–67. doi: 10.1038/nbt0197-63. [DOI] [PubMed] [Google Scholar]
- 38.Slater S, Gallaher T, Dennis D. Production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in a recombinant Escherichia coli strain. Appl Environ Microbiol. 1992;58:1089–1094. doi: 10.1128/aem.58.4.1089-1094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinbüchel A, Hustede E, Liebergesell M, Pieper U, Timm A, Valentin H. Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoic acids in bacteria. FEMS Microbiol Rev. 1992;103:217–230. doi: 10.1111/j.1574-6968.1992.tb05841.x. [DOI] [PubMed] [Google Scholar]
- 40.Steinbüchel A, Valentin H E. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol Lett. 1995;128:219–228. [Google Scholar]
- 41.van der Leij F R, Witholt B. Strategies for the sustainable production of new biodegradable polyesters in plants: a review. Can J Microbiol. 1995;41:222–238. [Google Scholar]
- 42.Wubbolts M G, Favre-Bulle O, Witholt B. Biosynthesis of synthons in two-liquid-phase media. Biotechnol Bioeng. 1996;52:301–308. doi: 10.1002/(SICI)1097-0290(19961020)52:2<301::AID-BIT10>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]