Abstract
Vitiligo is characterized by chronic skin depigmentation arising from the autoimmune destruction of epidermal melanocytes. Systemic corticosteroid therapy is an effective immunosuppressive treatment for progressive generalized vitiligo. Circular RNAs (circRNAs) play various roles in diseases. In systemic corticosteroid therapy, however, how circRNAs function to counter vitiligo is still unclear. In this article, we identified the differentially expressed circRNAs (DEcircRNAs) in vitiligo patients before and after the administration of methylprednisolone. Total RNA was extracted from the peripheral blood of patients with vitiligo, and samples were hybridized into a circRNA array. A total of 375 (51 upregulated and 324 downregulated) circRNAs were differentially expressed. Box, scatter, volcano, and heatmap plots were generated to classify the samples. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed on DEcircRNAs. These DEcircRNAs were enriched in vitiligo-related biological processes, such as ferroptosis, organic substance transport, protein metabolic process, and cellular component organization or biogenesis. Two different databases, TargetScan and miRanda, were used to predict circRNA/miRNA interactions. Several circRNA/miRNA interactions were involved in ferroptosis. These circRNAs might serve as therapeutic targets in the treatment of vitiligo.
Keywords: vitiligo, circRNA, methylprednisolone, ferroptosis, treatment
Introduction
As a multifactorial disease, vitiligo brings not only white skin lesions, but also depression and self-abasement (1). Oxidative stress is involved in active vitiligo. Excessive oxygen radicals cause cellular oxidation, and even cell death (2, 3). Moreover, excessive generation of reactive oxygen species (ROS) is associated with autoimmune diseases (4), as evidenced by the destruction or apoptosis of vitiliginous melanocytes in vitiligo (5). Methylprednisolone exerts a moderate efficacy by suppressing inflammation and immune responses. However, the mechanism of systemic glucocorticoids in treating vitiligo remains a conundrum.
High-throughput sequencing techniques have discovered the roles of circular RNAs (circRNAs) in various diseases. circRNAs are generated from precursor mRNAs (pre-mRNAs), with a closed-loop without 5′-cap and 3′-end polyadenylated tails (6–8). circRNAs can function as miRNAs sponges to affect the stability or translation of target RNAs (9, 10). Moreover, circRNAs can also modify gene expression (11). Aberrant expression of circRNAs has been observed in cancer, neurodegenerative disorders, and metabolic disorders (12–14). For example, circAGO2 is upregulated to promote the proliferation, invasion, and metastasis of cancer in vitro and in vivo (15).
Increasing evidence suggests that dysregulation of circRNAs is implicated in the pathogenesis of vitiligo. Li et al. found circ_0087961-miR-27a-3p-PAXILLIN played a regulatory role in the pathology of vitiligo (16). Ouyang et al. demonstrated for the first time that UVB-induced circRNA ciRS-7 triggered melanogenesis in monocytes via the miR-7/STAT3 and AKT/FGF2 paracrine axis in both keratinocytes and fibroblasts (17). Jiang et al. verified that a ceRNA network of ENST00000606533, circ_0091223, and TYR mRNA was linked by miR-1291, suggesting that they may be novel biomarkers for skin pigmentation disorders (18). To date, treatment options for these disorders include topical agents, such as corticosteroids, calcineurin inhibitors, and vitamin D analogs (19). Repigmentation can be promoted with narrow-band ultraviolet B light (nb-UVB) (20, 21). Systemic corticosteroids are indicated for generalized and progressive vitiligo. Wada-Irimada et al. reported that methylprednisolone pulse therapy was safe and effective for progressive generalized vitiligo (22). The results of the initial screen of the microarray and the subsequent bioinformatics analysis provide an important basis for the in-depth study of the actions of circRNAs in the development of vitiligo (Table 1). However, we knew a little about the functions of circRNAs in the treatment of vitiligo with systemic glucocorticoid therapy.
TABLE 1.
Circular RNAs (circRNAs) expression profiles in vitiligo.
| Species | Sample | Circular RNA | Function of circRNAs | References |
| Human | Skin tissue | hsa_circ_0007716 | Upregulated in vitiligo | (16) |
| Human | Skin tissue | hsa_circ_0087961 | Downregulated in vitiligo | (16) |
| Human | Keratinocytes, Fibroblasts, and Melanocytes | ciRS-7 | Triggered melanogenesis in monocytes | (17) |
| Human | Melanocytes | circ_0091223 | Novel biomarker for skin pigmentation disorders | (18) |
| Human | Melanocytes | hsa_circ_0048910 | Oxidative stress injury of melanocytes | (43) |
| Human | Melanocytes | hsa_circ_0048909 | Oxidative stress injury of melanocytes | (43) |
| Mice | Skin tissue | circBub1b | Upregulated in black mice skin | (44) |
| Mice | Skin tissue | circTmem26 | Upregulated in black mice skin | (44) |
In this study, we explored the mechanism of circRNAs in the pathogenesis and treatment of vitiligo. We used Arraystar Circular RNA Microarray to explore the differentially expressed circRNAs (DEcircRNAs) in vitiligo patients before and after systemic glucocorticoids. Then, GO and KEGG analyses were performed to evaluate their enriched functions. Finally, we teased out functional circRNA-miRNA pairs. Our findings can provide novel insights into the pathogenesis and treatment of vitiligo.
Materials and Methods
Patients and Samples
Peripheral blood specimens were obtained from the Department of Dermatology, The Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University, with written consent from all patients. All experiments were conducted in full compliance with governmental policies and guidelines. All four patients (two men and two women, aged 50–60 years) enrolled in this study were diagnosed with non-segmental vitiligo, without a history of systemic therapy or topical steroids before blood sampling. Under sterile conditions, whole blood (3 ml) was sampled by venipuncture into heparinized vacutainers from the patients before and after systemic glucocorticoid therapy (oral methylprednisolone tablets, 12 mg daily for 8 weeks). All patients showed improvement in symptoms at week 8 after glucocorticoid therapy. These patients were hence considered glucocorticoid therapy responders and evaluated by genomic and bioinformatic analysis. The experimental protocols described in this study were approved by the ethics review committee of The Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University and were conducted in compliance with the guidelines. The certificate number is [2020]KY205-1.
RNA Isolation
Total RNA of 4 pairs of vitiligo samples was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. After the addition of chloroform, the samples were centrifuged at 12,000 rpm for 10 min. The aqueous phase was mixed with isopropanol and RNA pellets were collected by centrifugation (12,000 rpm, 15 min, 4°C). RNA pellets were washed with 70% ethanol and dissolved in RNase-free, DEPC (diethylpyrocarbonate)-treated water (Waltham, MA, United States).
RNA Sample QC, Labeling, and Array Hybridization
Total RNA from each sample was quantified using the NanoDrop ND-1000, and the concentration of RNA was determined by OD260. First, total RNA was digested with Rnase R (Epicenter, Inc.) to remove linear RNA and enrich circular RNA. Then, the enriched circular RNA was amplified and transcribed into fluorescent cRNA utilizing a random priming method (Arraystar Super RNA Labeling Kit; Arraystar). The labeled cRNA was hybridized onto the Arraystar Human circRNA Array V2 (8 × 15K, Arraystar). Finally, the slides were washed and scanned by the Agilent Scanner G2505C. The sample preparation and microarray hybridization were performed based on Arraystar’s standard protocols.
Microarray Differential Expression Analysis
Array image analysis was based on Agilent Feature Extraction software (version 11.0.1.1). The Limma R package was used to conduct differential expression analysis. circRNAs having a fold change > 1.5 and a P-value < 0.05 were considered significantly differentially expressed. The volcano plot and hierarchical clustering heat map showed the DEcircRNAs between the two groups. The statistical significance of the difference was estimated by a t-test.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analyses
The overlapped DEcircRNAs were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analyses performed on the R software clusterProfiler package. In GO analysis, the biological processes (BPs), cellular compositions (CCs), and molecular functions (MFs) involved in circRNAs were analyzed. The pathway diagram was derived from the KEGG database. A P-value < 0.05 was deemed to be significant functional terms and pathways.
Annotation With Circular RNAs-miRNA Interaction
Arraystar’s homemade miRNA target prediction software based on TargetScan (23) and miRanda (24) was used to predict circRNA/miRNA interaction. Upregulation and downregulation of circRNA and miRNA were identified.
Statistical Analysis
The data from this study were presented as means ± standard deviation (SD). The difference between groups was assessed using a two-sided student t-test. P < 0.05 was considered statistically significant.
Results
Differentially Expressed CircRNAs
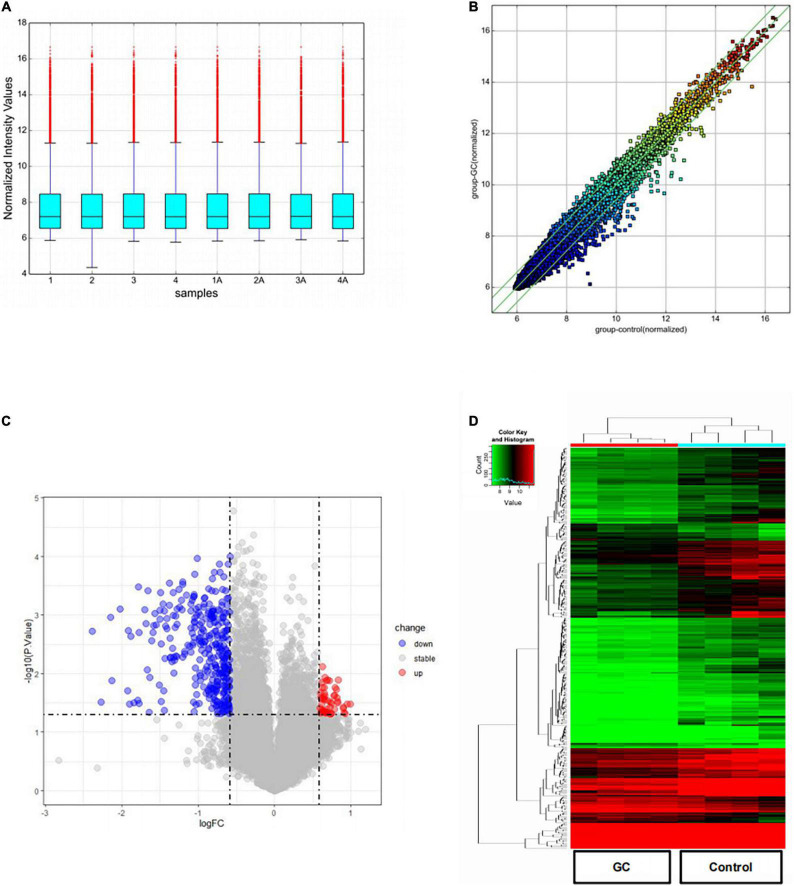
The box plot shows the distribution of expression data from all the samples. After normalization, the total gene expression was basically the same, indicating insignificant batch effects and system deviations (Figure 1A). The scatter plot shows circRNA expression variation between the two groups of samples (Figure 1B). The circRNAs above the top green line and below the bottom green line showed fold changes of more than 1.5.
FIGURE 1.
DEcircRNAs in patients with vitiligo before and after systemic glucocorticoid therapy. (A) A box plot shows the distribution of expression data in each sample after quantile normalization. (B) Scatter plot was performed to evaluate the centralized tendency of chip data in two different groups. (C) Volcano plot of the P-value as a function of weighted fold change for DEcircRNAs (DEcircRNAs) (fold change > 1.5, P < 0.05). (D) Heat map for potential DEcircRNAs (n = 375) shows 51 upregulated and 324 downregulated.
The volcano plot visualizes the circRNA microarray results after a 2-month treatment of systemic glucocorticoids (Figure 1C). The vertical lines correspond to 1.5-fold up or down, respectively, and the horizontal line represents a P-value of 0.05. The red and blue points in the plot represent the up-regulated and down-regulated circRNAs with statistical significance, separately. Compared to the control group, 51 up-regulated and 324 down-regulated circRNAs were detected in the group treated with systemic glucocorticoid.
The number of down-regulated circRNAs was largely more than that of up-regulated circRNAs. Tables 2, 3 demonstrate the top 10 upregulated and downregulated circRNAs. The heatmap of hierarchical clustering analysis shows that these DEcircRNAs could distinguish samples collected before and after treatment (Figure 1D).
TABLE 2.
Top 10 upregulated circRNAs.
| CircRNA | P-value | FC | Regulation |
| hsa_circRNA_105055 | 0.007582547 | 1.5388999 | Up |
| hsa_circRNA_103796 | 0.011590286 | 1.5802079 | Up |
| hsa_circRNA_009550 | 0.012917238 | 1.779194 | Up |
| hsa_circRNA_104747 | 0.012932469 | 1.6126129 | Up |
| hsa_circRNA_103948 | 0.013086105 | 1.6376076 | Up |
| hsa_circRNA_100982 | 0.013325807 | 1.5911831 | Up |
| hsa_circRNA_403865 | 0.014330979 | 1.5287481 | Up |
| hsa_circRNA_044065 | 0.017245821 | 1.5710328 | Up |
| hsa_circRNA_100412 | 0.017501176 | 1.5438505 | Up |
| hsa_circRNA_025984 | 0.017539967 | 1.7253039 | Up |
TABLE 3.
Top 10 downregulated circRNAs.
| circRNA | P-value | FC | Regulation |
| hsa_circRNA_011228 | 0.000100102 | 1.5007234 | Down |
| hsa_circRNA_403694 | 0.00010878 | 2.0222611 | Down |
| hsa_circRNA_101738 | 0.000133793 | 1.652627 | Down |
| hsa_circRNA_100966 | 0.00013521 | 1.5250118 | Down |
| hsa_circRNA_002122 | 0.000184899 | 1.644309 | Down |
| hsa_circRNA_061482 | 0.000188585 | 1.7258389 | Down |
| hsa_circRNA_102949 | 0.000201268 | 2.0667206 | Down |
| hsa_circRNA_102479 | 0.000221586 | 1.5848103 | Down |
| hsa_circRNA_102849 | 0.000240042 | 1.7620554 | Down |
| hsa_circRNA_402533 | 0.000272795 | 2.3031702 | Down |
Gene Ontology Enrichment Analysis
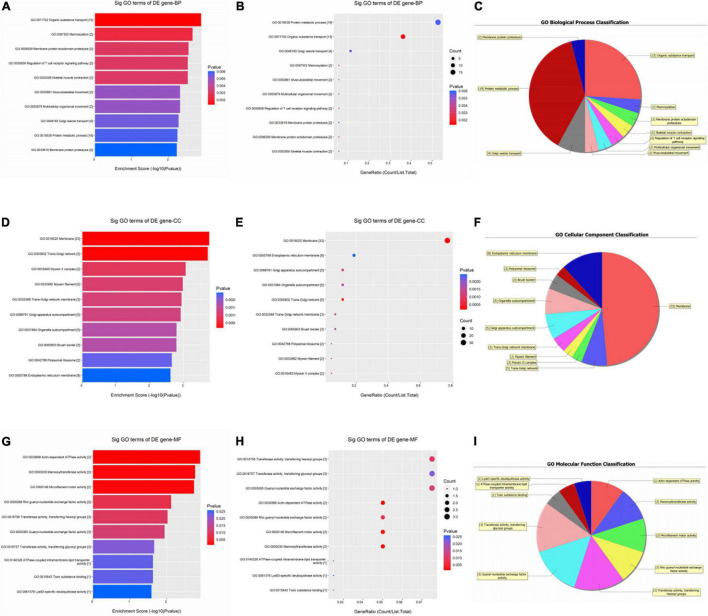
As shown in Figure 2, up-regulated circRNAs were significantly enriched in skin epidermis-related biological processes, such as organic substance transport, mannosylation, membrane protein ectodomain proteolysis, protein metabolic process, and Golgi vesicle transport (Figures 2A–C). As for CCs, these DEcircRNAs were significantly enriched in membrane, trans-Golgi network, myosin II complex, endoplasmic reticulum membrane, and Golgi apparatus subcompartment (Figures 2D–F). For MFs, they were significantly enriched in actin-dependent ATPase activity, mannosyltransferase activity, microfilament motor activity, transferase activity, transferring hexosyl, and glycosyl groups (Figures 2G–I).
FIGURE 2.
Gene ontology (GO) enrichment analysis of upregulated circRNAs. For bubble plot, the y-axis shows the enriched terms in three GO categories and the x-axis shows the Enrichment Score [−log10(P-value)] or GeneRatio (Count/List. Total): (A,B) biological process, (D,E) cellular component, (G,H) molecular function. Pie chart shows the distribution of significantly GO terms: (C) biological process, (F) cellular component, (I) molecular function.
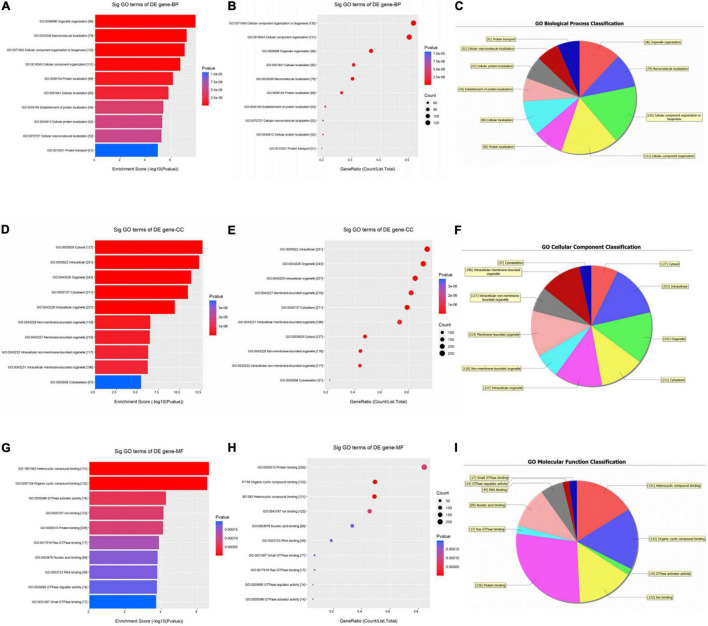
As shown in Figure 3, the down-regulated circRNAs were enriched in biological processes such as organelle organization, macromolecule localization, and cellular component organization (Figures 3A–C). For CCs, these genes were significantly enriched in the cytosol and intracellular organelle (Figures 3D–F). As for MFs, they were significantly enriched in heterocyclic compound binding, organic cyclic compound binding, GTPase activator activity, and protein binding (Figures 3G–I).
FIGURE 3.
Gene ontology (GO) enrichment analysis of downregulated circRNAs. For bubble plot, the y-axis shows the enriched terms in three GO categories and the x-axis shows the Enrichment Score [−log10(P-value)] or GeneRatio (Count/List. Total): (A,B) biological process, (D,E) cellular component, (G,H) molecular function. Pie chart shows the distribution of significantly GO terms: (C) biological process, (F) cellular component, (I) molecular function.
Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analysis
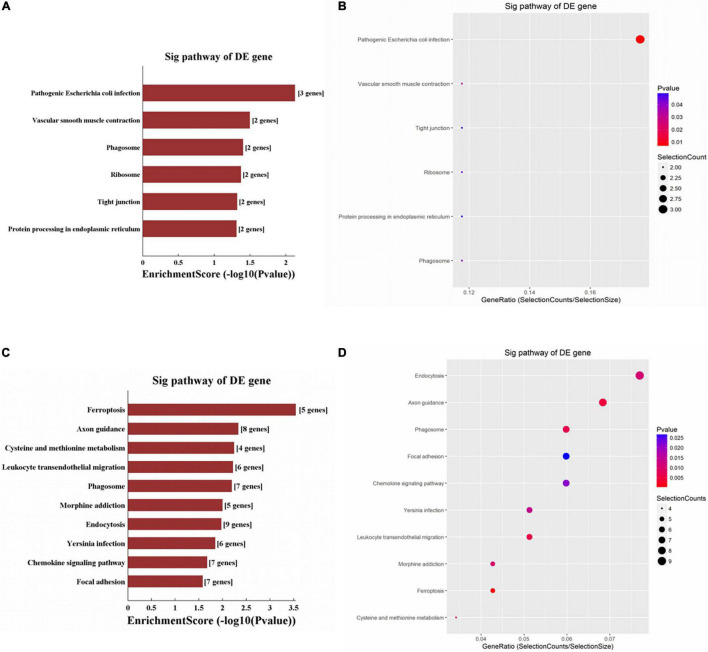
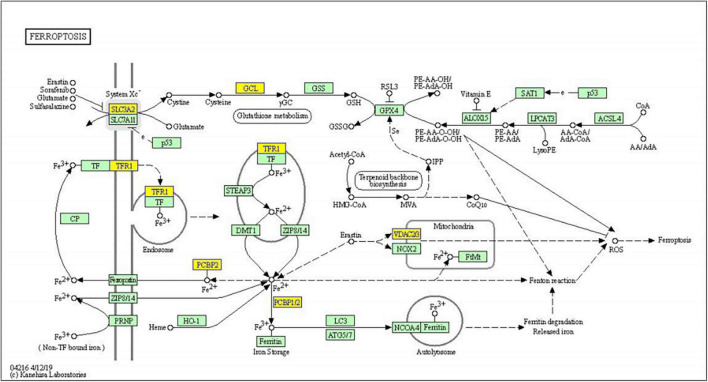
Significantly enriched pathways are shown in Figure 4. The up-regulated circRNAs were significantly involved in the processes of pathogenic Escherichia coli infection, vascular smooth muscle contraction, phagosome, and ribosome (Figures 4A,B). The down-regulated circRNAs were significantly enriched in ferroptosis, axon guidance, cysteine and methionine metabolism, endocytosis, and phagosome (Figures 4C,D). The genes involved in the ferroptosis pathway included SLC3A2, GCLM, PCBP2, TFRC, and VDAC3 (Figure 5), respectively, corresponding to hsa_circRNA_100842, hsa_circRNA_100283, hsa_circRNA_101076, hsa_circRNA_103556, and hsa_circRNA_104600.
FIGURE 4.
KEGG enrichment analysis of circRNAs. For bar plot and bubble plot, the y-axis shows the enriched pathways of genes and the x-axis shows the Enrichment Score [−log10(P-value)] or GeneRatio (Count/List. Total): (A,B) upregulated circRNAs, (C,D) downregulated circRNAs.
FIGURE 5.
KEGG pathway diagram of ferroptosis involved in differentially expressed genes. Yellow nodes are associated with down-regulated genes, and green nodes have no significance.
Annotation With Circular RNAs/miRNA Interaction
It is reported that the circRNA ciRS-7 can act as an endogenous “sponge” of miRNA-7 (25). As shown in Figure 6, the circRNA/miRNA interaction was predicted, and the DEcircRNAs involved in ferroptosis were annotated in detail with circRNA/miRNA interaction. The circRNAs and corresponding miRNAs involved in ferroptosis pathway were demonstrated, including hsa-miR-23b-5p and hsa-miR-429 containing conserved binding sites with mmu_hsa_circRNA_100842 (circSLC3A2); hsa-miR-576-5p and hsa-miR-383-5p containing conserved binding sites with mmu_hsa_circRNA_100283 (circGCLM); hsa-miR-608 and hsa-miR-873-3p containing conserved binding sites with mmu_hsa_circRNA_101076 (circPCBP2); hsa-miR-149-5p and hsa-miR-653-3p containing conserved binding sites with mmu_hsa_circRNA_103556 (circTFRC); hsa-miR-609 and hsa-miR-758-5p containing conserved binding sites with mmu_hsa_circRNA_104600 (circVDAC3).
FIGURE 6.

Complementary miRNAs of DEcircRNAs. (A) The 2D structure of the binding sites of hsa_circRNA_100842 (circSLC3A2) with hsa-miR-23b-5p and hsa-miR-429. (B) The 2D structure of the binding sites of hsa_circRNA_100283 (circGCLM) with hsa-miR-576-5p and hsa-miR-383-5p. (C) The 2D structure of the binding sites of hsa_circRNA_101076 (circPCBP2) with hsa-miR-608 and hsa-miR-873-3p. (D) The 2D structure of the binding sites of hsa_circRNA_103556 (circTFRC) with hsa-miR-149-5p and hsa-miR-653-3p. (E) The 2D structure of the binding sites of hsa_circRNA_104600 (circVDAC3) with hsa-miR-609 and hsa-miR-758-5p.
Discussion
In this study, we explored the function of DEcircRNAs in patients with vitiligo treated with systemic glucocorticoid therapy. A total of 51 up-regulated and 324 down-regulated circRNAs were detected in the group of patients with vitiligo treated with systemic glucocorticoids. These DEcircRNAs provide clues to design new targeted therapies for vitiligo.
According to KEGG enrichment analysis, the down-regulated DEcircRNAs were significantly enriched in ferroptosis. Ferroptosis regulates cell death in an iron and lipid peroxidation−dependent manner (26, 27). The ferroptosis is repressed after systemic glucocorticoid therapy in patients with vitiligo. It is reported that ferroptosis is implicated in IFN−γ−associated apoptosis, and that IFN−γ can lead to melanocyte death in the case of vitiligo (28–30). Increased ROS and lip-ROS caused by oxidative stress play a vital role in the process of ferroptosis (26, 31, 32). Previous studies have provided that circRNAs can regulate ferroptosis. circKIF4A can suppress ferroptosis by sponging miR-1231 and upregulating GPX4 in the malignant progression of papillary thyroid cancer (33). GPX4 has been known as a biomarker of ferroptosis (34, 35). circEPSTI1 modulates the proliferation of cervical cancer via the miR-375/409-3P/515-5p-SLC7A11 axis relative to ferroptosis (36).
After systemic glucocorticoid therapy, we detected more down-regulated circRNAs in patients with vitiligo. They were also significantly enriched in the pathway of ferroptosis. Moreover, we elucidated that several miRNAs interacted with the DEcircRNAs involved in the ferroptosis pathway. circRNAs interact with miRNAs to regulate disease development (37). ciRS-7 functions in neurological disorders and tumor. miR-7 directly targeting a-synuclein protein in Parkinson’s disease and ubiquitin protein ligaseA (UBE2A) in Alzheimer’s disease (38).
As we all know, circRNAs play critical roles in autoimmune diseases. circPTPN22, synthesized from the protein tyrosine phosphatase non-receptor type 22 (PTPN22) gene, shows an expression negatively correlated with the activity of systemic lupus erythematosus (39). Lodde et al. have described several circRNAs that function mainly as biomarkers for autoimmune diseases, including ciRS-7 for rheumatoid arthritis (40) and the circ_0000479 for systemic lupus erythematosus (41, 42). However, we found that circSLC3A2 and circTFRC were down-regulated after treatment with systemic glucocorticoid therapy. Furthermore, downregulation of circGCLM, circPCBP2, and circVDAC3, three circRNAs involved in the ferroptosis, was first reported in the treatment of vitiligo.
Our findings demonstrated the DEcircRNAs in the treatment of vitiligo with systemic glucocorticoid therapy for the first time. Additional experiments will be required to address the significance of these findings, the binding characteristics of the identified circRNAs, and their functional activity after exposure to glucocorticoid. Moreover, it would be interesting to compare differential circRNA expression in cases where remission is induced by a different therapy, to control for confounding effects independent from glucocorticoid. A limitation of this study mainly lies in that most of the patients with vitiligo in this study were between 50 and 60 years old, while no younger patients were included. We will explore the circRNA data of younger patients and compare the difference in expression profile among patients of different age groups in a further study. Moreover, the data were obtained from a circRNA microarray; although enrichment analyses provided several related pathways, there is still a lack of verification with wet-lab biochemical experiments. In addition, molecular biology experiments should be carried out to verify our results.
Data Availability Statement
The data presented in the study are deposited in the Gene Expression Omnibus repository, accession number GSE197415.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Board (IRB) of The Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JZ and YC conceived the main idea and analyzed the data. JZ drafted the manuscript. YC reviewed drafts of the article and improved the manuscript. NL collected the data and performed the statistical analysis. ML provided statistical advice. YC and ML supervised the study and provided funding. All authors read and commented on the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
This study was supported by the Youth Program of Changzhou Second People’s Hospital (2020K002). The funder had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.839066/full#supplementary-material
References
- 1.Ezzedine K, Lim HW, Suzuki T, Katayama I, Hamzavi I, Lan CC, et al. Revised classification/nomenclature of vitiligo and related issues: the vitiligo global issues consensus conference. Pigment Cell Melanoma Res. (2012) 25:E1–13. 10.1111/j.1755-148X.2012.00997.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bickers DR, Athar M. Oxidative stress in the pathogenesis of skin disease. J Invest Dermatol. (2006) 126:2565–75. 10.1038/sj.jid.5700340 [DOI] [PubMed] [Google Scholar]
- 3.Plaza-Rojas L, Guevara-Patino JA. The role of the NKG2D in vitiligo. Front Immuno. (2021) 12:624131. 10.3389/fimmu.2021.624131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Dalmazi G, Hirshberg J, Lyle D, Freij JB, Caturegli P. Reactive oxygen species in organ-specific autoimmunity. Auto Immun Highlights. (2016) 7:11. 10.1007/s13317-016-0083-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. (2009) 417:1–13. 10.1042/BJ20081386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. (2014) 32:453–61. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 8.Xie H, Wang J, Wang B. Circular RNA Circ_0003221 promotes cervical cancer progression by regulating miR-758-3p/CPEB4 axis. Cancer Manag Res. (2021) 13:5337–50. 10.2147/CMAR.S311242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun H, Sun Y, Yu X, Gao X, Wang H, Zhang L, et al. Analysis of age-related circular RNA expression profiles in mesenchymal stem cells of rat bone marrow. Front Genet. (2021) 12:600632. 10.3389/fgene.2021.600632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aird KM, Zhang R. Metabolic alterations accompanying oncogene-induced senescence. Mol Cell Oncol. (2014) 1:e963481. 10.4161/23723548.2014.963481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. (2015) 365:141–8. 10.1016/j.canlet.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 12.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. (2018) 187:31–44. 10.1016/j.pharmthera.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Li C, Tan C, Mai G, Liu X. [Circular RNA in human disease and their potential clinic significance]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. (2017) 34:133–7. 10.3760/cma.j.issn.1003-9406.2017.01.031 [DOI] [PubMed] [Google Scholar]
- 14.Qian P, Linbo L, Xiaomei Z, Hui P. Circ_0002770, acting as a competitive endogenous RNA, promotes proliferation and invasion by targeting Mir-331-3p in melanoma. Cell Death Dis. (2020) 11:264. 10.1038/s41419-020-2444-x [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. (2019) 26:1346–64. 10.1038/s41418-018-0220-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Xie Z, Qian X, Wang T, Jiang M, Qin J, et al. Identification of a potentially functional circRNA-MiRNA-mRNA regulatory network in melanocytes for investigating pathogenesis of vitiligo. Front Genet. (2021) 12:663091. 10.3389/fgene.2021.663091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouyang Y, Chen J, Jiang L, Li Y, Hu Y, Li S, et al. UVB-induced ciRS-7 activates melanogenesis by paracrine effects. DNA Cell Biol. (2021) 40:523–31. 10.1089/dna.2020.5489 [DOI] [PubMed] [Google Scholar]
- 18.Jiang L, Huang J, Hu Y, Lei L, Ouyang Y, Long Y, et al. Identification of the ceRNA networks in α-MSH-induced melanogenesis of melanocytes. Aging (Albany N Y). (2020) 13:2700–26. 10.18632/aging.202320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel NS, Paghdal KV, Cohen GF. Advanced treatment modalities for vitiligo. Dermatol Surg. (2012) 38:381–91. 10.1111/j.1524-4725.2011.02234.x [DOI] [PubMed] [Google Scholar]
- 20.Ocampo-Candiani J, Salinas-Santander M, Trevino V, Ortiz-Lopez R, Ocampo-Garza J, Sanchez-Dominguez CN. Evaluation of skin expression profiles of patients with vitiligo treated with narrow-band UVB therapy by targeted RNA-Seq. An Bras Dermatol. (2018) 93:843–51. 10.1590/abd1806-4841.20187589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan J, Chen H, Yan R, Cui S, Li YH, Wu Y, et al. Fractional Co2 lasers contribute to the treatment of stable non-segmental vitiligo. Eur J Dermatol. (2016) 26:592–8. 10.1684/ejd.2016.2875 [DOI] [PubMed] [Google Scholar]
- 22.Wada-Irimada M, Tsuchiyama K, Sasaki R, Hatchome N, Watabe A, Kimura Y, et al. Efficacy and safety of i.v. methylprednisolone pulse therapy for vitiligo: a retrospective study of 58 therapy experiences for 33 vitiligo patients. J Dermatol. (2021) 48:1090–3. 10.1111/1346-8138.15858 [DOI] [PubMed] [Google Scholar]
- 23.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. Microrna targets in drosophila. Genome Biol. (2003) 5:R1. 10.1186/gb-2003-5-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasquinelli AE. Micrornas and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. (2012) 13:271–82. 10.1038/nrg3162 [DOI] [PubMed] [Google Scholar]
- 25.Lukiw WJ. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet. (2013) 4:307. 10.3389/fgene.2013.00307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. (2012) 149:1060–72. 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. (2017) 171:273–85. 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W, Green M, Choi JE, Gijon M, Kennedy PD, Johnson JK, et al. Cd8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. (2019) 569:270–4. 10.1038/s41586-019-1170-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rashighi M, Agarwal P, Richmond JM, Harris TH, Dresser K, Su MW, et al. CXCL10 is critical for the progression and maintenance of depigmentation in a mouse model of vitiligo. Sci Transl Med. (2014) 6:223ra23. 10.1126/scitranslmed.3007811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Li S, Li C. Mechanisms of melanocyte death in vitiligo. Med Res Rev. (2021) 41:1138–66. 10.1002/med.21754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu T, Shi L, Yu C, Dong Y, Qiu F, Shen L, et al. Ferroptosis promotes photodynamic therapy: supramolecular photosensitizer-inducer nanodrug for enhanced cancer treatment. Theranostics. (2019) 9:3293–307. 10.7150/thno.32867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Yang Y, Xiang L, Zhang C. The fate of melanocyte: mechanisms of cell death in vitiligo. Pigment Cell Melanoma Res. (2021) 34:256–67. 10.1111/pcmr.12955 [DOI] [PubMed] [Google Scholar]
- 33.Chen W, Fu J, Chen Y, Li Y, Ning L, Huang D, et al. Circular RNA circkIF4A facilitates the malignant progression and suppresses ferroptosis by sponging miR-1231 and upregulating GPX4 in papillary thyroid cancer. Aging (Albany N Y). (2021) 13:16500–12. 10.18632/aging.203172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou Y, Palte MJ, Deik AA, Li H, Eaton JK, Wang W, et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun. (2019) 10:1617. 10.1038/s41467-019-09277-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. (2014) 16:1180–91. 10.1038/ncb3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu P, Li C, Ye DM, Yu K, Li Y, Tang H, et al. Circular RNA circEPSTI1 accelerates cervical cancer progression Via miR-375/409-3P/515-5p-SLC7A11 axis. Aging (Albany N Y). (2021) 13:4663–73. 10.18632/aging.202518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular rna potentially associated with disease and traits. Front Genet. (2013) 4:283. 10.3389/fgene.2013.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microrna sponges. Nature. (2013) 495:384–8. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 39.Miao Q, Zhong Z, Jiang Z, Lin Y, Ni B, Yang W, et al. RNA-seq of circular RNAs identified circPTPN22 as a potential new activity indicator in systemic lupus erythematosus. Lupus. (2019) 28:520–8. 10.1177/0961203319830493 [DOI] [PubMed] [Google Scholar]
- 40.Tang X, Wang J, Xia X, Tian J, Rui K, Xu H, et al. Elevated expression of ciRS-7 in peripheral blood mononuclear cells from rheumatoid arthritis patients. Diagn Pathol. (2019) 14:11. 10.1186/s13000-019-0783-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo Q, Zhang L, Fang L, Fu B, Guo Y, Huang Z, et al. Circular RNAS hsa_circ_0000479 in peripheral blood mononuclear cells as novel biomarkers for systemic lupus erythematosus. Autoimmunity. (2020) 53:167–76. 10.1080/08916934.2020.1728529 [DOI] [PubMed] [Google Scholar]
- 42.Lodde V, Murgia G, Simula ER, Steri M, Floris M, Idda ML. Long noncoding RNAs and circular RNAs in autoimmune diseases. Biomolecules. (2020) 10:1044. 10.3390/biom10071044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Zeng H, Huang J, Lu J, Chen J, Zhou Y, et al. Identification of the competing endogenous RNA networks in oxidative stress injury of melanocytes. DNA Cell Biol. (2021) 40:192–208. 10.1089/dna.2020.5455 [DOI] [PubMed] [Google Scholar]
- 44.Zhu Z, Ma Y, Li Y, Li P, Cheng Z, Li H, et al. The comprehensive detection of mirna, lncrna, and circrna in regulation of mouse melanocyte and skin development. Biol Res. (2020) 53:4. 10.1186/s40659-020-0272-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in the study are deposited in the Gene Expression Omnibus repository, accession number GSE197415.