Abstract
Nine 8 C57Bl6 mice (9 ± 0.5 months) were utilised for this dataset. Each animal was scanned twice on a 9.4T Bruker Magnetic Resonance Imaging (MRI) scanner using a cryogenically cooled coil with 0.1 mg/kg body weight/h (low) or 0.3 mg/kg body weight/h (high) medetomidine doses; 0.5% isoflurane was used in conjunction with both doses. The scans were one week apart, and the first session's dose was decided randomly. In each session, the animal had a pre-stimulation resting-state functional Magnetic Resonance Imaging (rs-fMRI) scan followed by 10 min where mild, constant electrical stimulation to the forepaw was applied, and a post-stimulation rs-fMRI scan. Each fMRI scan lasted 10 min, and there was 5 min break between fMRI scans.
The dataset included, for each animal, a pair of forward-phase and reverse-phase gradient echo Echo-Planar-Imaging (EPI) images for EPI distortion correction purpose and three (unprocessed) functional MRI images acquired using the same EPI sequence: prior, during, and post-stimulation. The MRI data was saved in compressed NIFTI format converted from Bruker DICOMs. The dataset also included the pre-processed functional MRI images, with the following pre-processing steps: slice-timing correction, temporal despiking, motion correction, distortion correction, band-pass filtration at 0.01–0.2 Hz, and spatial normalisation.
This dataset adds to the publicly available collection of resting-state functional MRI in the mice and facilitates reproducibility and validation of functional imaging and its analysis.
Keywords: Functional connectivity, Resting state, Magnetic Resonance Imaging, MRI, Medetomidine, Electrical stimulation
Specifications Table
| Subject | Neuroscience: General |
| Specific subject area | This dataset contains resting-state functional Magnetic Resonance Imaging data collected from the mice under different anaesthesia dosage and stimulation conditions. |
| Type of data | Compressed NIFTI Magnetic Resonance Imaging data. |
| How the data were acquired | The data was acquired a 9.4T MRI scanner (Bruker Biospin, Germany) using with a cryogenically cooled transmit and receive coil. The scanner was controlled by Paravision 6.0.1 software (Bruker Biospin, Germany). All MRI data were exported to DICOM format using Paravision 6.0.1 and then converted to NIFTI data format using MRIcron's dcm2nii tool. |
| Data format | Compressed NIFTI of: raw and pre-processed data. |
| Description of data collection | fMRI scans were acquired using a 2D gradient-echo echo-planar-imaging (GE-EPI) sequence: effective spatial resolution of 0.3 × 0.3 × 0.6 mm, TR/TE = 1000/14 ms, flip angle = 70°, and 10 min scan time, with fat suppression, FOV saturation (covering the head tissue inferior to the brain), and navigator pulse turned on. |
| Data source location |
|
| Data accessibility | Repository name: UQ eSpace https://doi.org/10.48610/3b35b94 Repository name: figshare https://doi.org/10.6084/m9.figshare.c.5728658.v1 |
| Related research article | [1] X.V. To, V. Vegh, F.A. Nasrallah, Towards data-driven group inferences of resting-state fMRI data in rodents: comparison of Group ICA, GIG-ICA, and IVA-GL, J. Neurosci. Methods. (2021) 109,411. https://doi.org/10.1016/j.jneumeth.2021.109411. |
Value of the Data
-
•
The data provided adds to the existing collections of rs-fMRI in the mice and can aid in data aggregation and reproducibility. The data represents rs-fMRI data collected from a high-field scanner and a cryogenically cooled coil (improved sensitivity and stability).
-
•
The data can benefit researchers seeking to establish consensus resting-state functional connectivity networks in the mice (such as the efforts in [2]) or testing different processing and analysis methods.
-
•
The data can be processed through a standardised processing pipeline to further add to the consensus baseline of mouse resting-state functional connectivity. They can also be processed and analysed through different procedures, programs, algorithms, etc. … for assessment of different methods’ stability and suitability for large-scale data.
1. Data Description
The data has been deposited in a publicly available data repository with the following Digital Object Identifier (DOI): https://doi.org/10.48610/3b35b94. The data can be downloaded as a compressed file. When extracted, there will be two folders with additionally a data_folder_structure.pdf and a data_folder_structure.xlsx files. The two data_folder_structure files (pdf and xlsx) explain the file and folder structure of the two folders.
The High-dose_constant folder contains data acquired in the session in which the animals were under higher medetomidine dose (0.3 mg/kg body weight (kg BW)/h) and the Low-dose_constant folder contains data acquired in the session in which the animals were under lower medetomidine dose (0.1 mg/kg BW/h); see Experimental design, materials, and methods for details. Inside the High-dose_constant or Low-dose_constant are folders containing data from each animal; folders with the same animal identification number (e.g. Animal1) indicated the data coming from the same animal. Inside each animal folder are the imaging files in compressed NIFTI (.nii.gz) formats. Note about image orientation, actual and displayed: while the experimenter positioned the animal in the “head first, prone” orientation, we registered the orientation in Paravision 6.0.1 as “head first, supine”. In our experience, the resulting orientation displayed on the console intuitive: the animal's left was on the radiographer's left and the images were “right side up”. All MRI data were exported to DICOM format using Paravision 6.0.1 and then converted to compressed NIFTI format (.nii.gz) using MRIcron's dcm2nii tool [3]. The exported NIFTI images, when displayed on FSL's FSLeyes (v.6.0.4 [4], https://fsl.fmrib.ox.ac.uk/fsl/fslwiki) will have the animal's left on the viewer's left, FSLeye's displayed anatomical orientation labelling will say “right”. The left-right orientation can be wrongly displayed, labelled, or both in many software packages, for different software interpret and display orientation information in the ANALYZE/NIFTI header file differently. Researchers acquiring new data may want to include a mean to independently and physically confirm the left-right orientation during scanning, displaying, or post-processing (e.g. a vitamin E capsule or a water phantom consistently placed on one side and visible on MRI scans). Researchers who use this data and want to maintain left-right orientation accuracy should cross-reference left-right display accuracy of the chosen software with the display and orientation described above. For the description of the experiments and scans for the specific file names, see Experimental design, materials, and methods.
Figs. 1 and 2, Supplementary data 1, and Supplementary data 2 are deposited in figshare, a publicly available data repository with the following Digital Object Identifier (DOI): https://doi.org/10.6084/m9.figshare.c.5728658.v1
Fig. 1.
Summarised spatial maps of the ``signal'' components from the Independent Vector Analysis of the functional Magnetic Resonance Imaging data, grouped into eight networks/anatomical groupings, namely the Anterior Cingulate – Retrosplenial areas axis (ACA – RSN), the Visual and Auditory areas (VA), the Hippocampal – Subcortical memory circuit (HP – MEM), the Primary Somatosensory areas (S1), the Striatum (STR), the Motor areas (MO), the Salience Network (SN), and the Supplementary Somatosensory areas (S2).
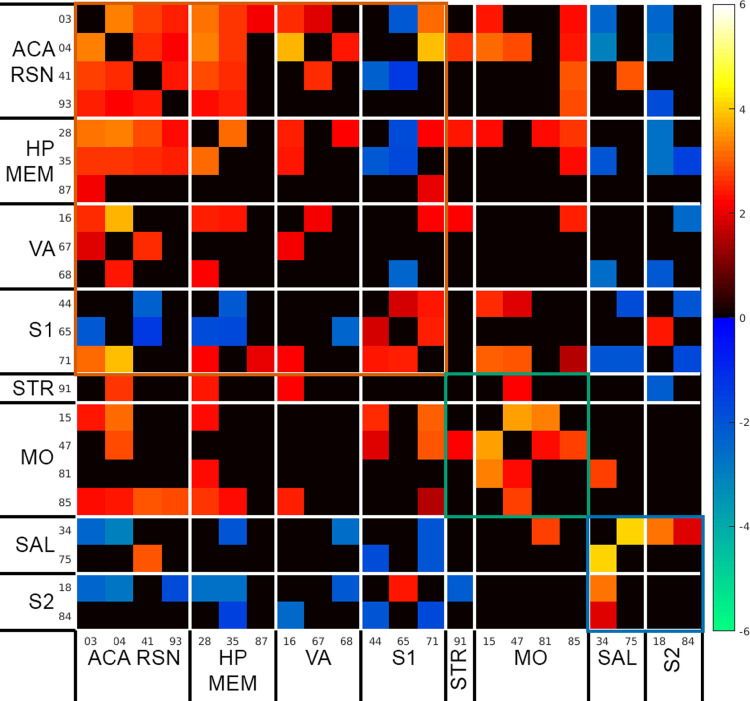
Fig. 2.
Calculated functional connectivity among the ``signal'' independent components shown in Fig. 1 under 0.3 mg/kg/h medetomidine dose after constant mild electrical stimulation to the forepaw (for how this was generated, see Experimental design, materials, and methods.). Functional connectivity matrix was generated using one sample t-test, corrected for multiple comparison correction using permutation-based test and False Discovery Rate, and thresholded at Q < 0.05. Matrix’s colours were scaled using the Z test statistics. Red–Yellow cells: positive correlation. Blue–Green cells: negative correlation.
Fig. 1 shows the summarised spatial maps of the “signal” components, grouped into eight networks/anatomical groupings, namely the Anterior Cingulate – Retrosplenial areas axis (ACA – RSN), the Visual and Auditory areas (VA), the Hippocampal – Subcortical memory circuit (HP – MEM), the Primary Somatosensory areas (S1), the Striatum (STR), the Motor areas (MO), the Salience Network (SN), and the Supplementary Somatosensory areas (S2). The components were overlaid on the generated study-specific EPI template.
Fig. 2 shows the calculated functional connectivity among the “signal” independent components shown in Fig. 1 under 0.3 mg/kg BW/h medetomidine dose after constant mild electrical stimulation to the forepaw (for how this was generated, see Experimental design, materials, and methods.). Functional connectivity matrices were generated using one sample t-test, corrected for multiple comparison correction using permutation-based test and False Discovery Rate, and thresholded at Q < 0.05. Matrix's colours were scaled using the Z test statistics. Red–Yellow cells: positive correlation. Blue–Green cells: negative correlation.
Supplementary data 1 shows the independent components’ spatial maps, frequency power spectra, and group-level time courses from an independent vector analysis of the processed functional MRI data (the data [resting_fMRI1.nii.gz, resting_fMRI2.nii.gz, and stim_fMRI.nii.gz] in the fMRIs/registered folder inside each animal's folder); for the exact processing steps to produce the processed functional MRI data, see Experimental design, materials, and methods. All 100 components were shown, including noise/artefacts and signal. The components were calculated from all functional MRI data (before, during, and after stimulation).
Supplementary data 2 is a comma-separated values file (.csv) containing a table showing the raw, unthresholded Z statistic of the functional connectivity matrix shown in Fig. 2, note: the first column and row contains the Independent Components’ labelling numbers.
2. Experimental Design, Materials and Methods
2.1. Experimental Design Overview
Nine female C57Bl6 mice (9 ± 0.5 months) were used for the generation of the data in this collection. The animals were kept in cages of 2–4 animals/cage in an animal holding facility under 12 h light-dark cycles with food and water available ad libitum.
Each animal underwent two separate scanning sessions: one at medetomidine doses of 0.1 mg/kg BW/h (low), the other at 0.3 mg/kg BW/h (high). The sessions were one week apart, and the dose used for the first session was determined randomly with a random number generator (https://www.random.org/). Within one session, each animal had a 10 min pre-stimulation resting-state scan (producing resting_fMRI1.nii.gz data), a 5 min break, a 10 min fMRI scan with constant mild electrical stimulation applied to the left forepaw (producing stim_fMRI.nii.gz data), another 5 min break, and a 10 min post-stimulation resting-state scan (producing resting_fMRI2.nii.gz data), in that order.
2.2. Animal Handling and Preparation for MRI Scans
Animals were acclimatised to the experimenter's handling and presence for two minutes per day, for five days prior to the first scanning session. The conditioning process was performed by holding the animal's tail and holding the animal on the back of the experimenter's gloved hands for 2 min. For the MRI scans, anaesthesia was initiated using 3% isoflurane in 60% air, 40% O2 mixture at 1 L/min in an animal anaesthesia induction box. Anaesthesia during the rest of the preparation (approximately 30 min) was delivered through a nose cone at 2–2.5% in the same air mixture at 0.5 L/min flow rate. Each mouse was placed on the manufacturer's (Bruker Biospin, Germany) MRI-compatible mouse cradle and the head was immobilised with ear bars and a bite bar. A polyethylene catheter was inserted into the peritoneal cavity and fixed to the mouse body with surgical tapes for the infusion of medetomidine (Domitor, Pfizer, USA). Subdermal electrodes were inserted near the 2nd and 4th digits of the left dorsal forepaw. Constant, mild stimulation was delivered with a current source (Isostim A320, World Precision Instrument USA) with the following parameters: 6 Hz pulse frequency, 0.3 ms pulse width, and 0.2 mA current. Respiration rate and pattern, and rectal temperature were monitored by an MRI-compatible animal physiological monitoring system (Model 1030, SA Instruments Inc, USA). The rectal temperature was kept at 36.5 ± 0.5 °C via a heated water circuit (SC100, Thermo Scientific, USA).
Medetomidine infusion solution was prepared from stock 1 mg/ml medetomidine solution (Domitor, Pfizer, USA) at 90 times dilution (0.0111 mg/ml medetomidine solution) for the 0.1 mg/kg BW/h low dose condition and 30 times dilution (0.0333 mg/ml medetomidine solution) for the 0.3 mg/kg BW/h high dose condition; the dilution strategy was to ensure that the infusion volume remained the same for either dosage. Once the animal was inside the scanner, medetomidine anaesthesia was initiated with an intraperitoneal bolus of 0.5 mg/kg BW and maintained with continuous infusion of 0.1 mg/kg BW/h for the low dose condition, or 0.15 mg/kg BW bolus and 0.3 mg/kg BW/h infusion for the high dose condition. Medetomidine infusion was started immediately after the bolus. After initiation of medetomidine, isoflurane was tapered off: reducing gradually at a rate of approximately 0.25% reduction in concentration every 5 min. Isoflurane tapering off took 30–40 min; during this period, isoflurane was reduced at a rate that maintain the animal's respiration rate at 80–120 breaths/minute. At the end of the isoflurane tapering process, isoflurane concentration was maintained at approximately 0.5% throughout the remaining of the experiment; respiration rate during the remaining of the experiment was not controlled by changing isoflurane concentration, unless there were obvious signs on the respiratory pattern monitor that the animal was waking up (with excessive motion) or having respiratory distress (respiratory rate < 60 breaths/minute). During this period, animals’ respiration rates were in the 90–180 breaths/minute range; despite this wide range, the most animals remained stable and had no excessive motions detrimental to the functional MRI scans. No obvious change to respiration rate was observed during stimulation. If the animal was waking up, isoflurane concentration was temporarily increased to 0.75–1% for approximately 5 min before reduced back to 0.5%. If the animal had respiratory distress, isoflurane was switched off and the animal monitored for the next 5 min. If the respiration rate did not improve, the scan would be stopped, and the animal removed from the scanner. No scan had to be stopped and cancelled for this data collection.
The total time under anaesthesia for each animal was approximately 2 h and 10 min. 1.25 mg/kg BW or 3.75 mg/kg BW atipamezole (Antisedan, Pfizer, USA) was given intraperitoneally as medetomidine reversal, for 0.1 mg/kg BW/h, and 0.3 mg/kg BW/h medetomidine doses, respectively. The animal was then allowed to recover in a heated box with food and water gel and returned to its home cage upon complete recovery.
Experimenter was not blinded to the specific medetomidine dose given to the animals during the scanning session.
2.3. MRI Experiments
MRI scans were performed on a 9.4T MRI scanner (Bruker Biospin, Germany) equipped with a cryogenically cooled transmit and receive coil, controlled by a console running Paravision 6.0.1 (Bruker Biospin, Germany).
For the fMRI scan, a 2D gradient-echo echo-planar-imaging (GE-EPI) sequence with the following parameters was used: matrix size = 64 × 64, FOV = 19.2 × 19.2 mm, 20 coronal slices of 0.5 mm thickness and 0.1 mm slice gap (effective spatial resolution = 0.3 × 0.3 × 0.6 mm), TR/TE = 1000/14 ms, flip angle = 70°, bandwidth = 200 kHz, and 600 vol (total 10 min scan time); fat suppression, FOV saturation (covering the head tissue inferior to the brain), and navigator pulse were turned on. The pre-stimulation rsfMRI scan was started approximately 55 min after the initiation of medetomidine. Each animal's pre-stimulation, during-stimulation, and post-stimulation functional MRI scans were converted and saved as compressed NIFTI files named resting_fMRI1.nii.gz, stim_fMRI.nii.gz, and resting_fMRI2.nii.gz, respectively, inside each animal's fMRIs folder.
A pair of EPI images with the exact EPI parameters and geometry but with the number of averages = 2 and opposite phase-encoding directions (Superior → Inferior then Inferior → Superior) was scanned for distortion correction processing. This pair of images was converted and saved as compressed NIFTI files named EPI_forward.nii.gz and EPI_reverse.nii.gz inside each animal's EPI_pair1 folder. “Forward” phase means acquisition using the same phase encoding directions as the stimulus-evoked fMRI and rs-fMRI.
2.4. MRI Data Processing
For the purpose of image processing, the images were given a header file with a voxel size 20 times larger than the original size (this was a necessary header modification to allow usage of software packages developed for human brain studies in animals) [5]. The personnel performing the data processing and analysis was not blinded to the identity of the animals though the procedure performed was applied uniformly and automatically across all subjects. The only part that required subjective judgement was the classification of Independent Vector Analysis components into “signal” or “noise” and this was done on an all-subjects-and-all-scans basis and the effect of this selection was applied uniformly across all scans and subjects.
The opposite phase-encoding direction EPI data were used to calculate the warping field required for distortion correction using FSL's TOPUP tool [6]. The distortion corrected EPI images can be corrected for signal inhomogeneity using N4ITK bias field correction [7] as implemented in Advanced Normalisation Tool (N4BiasFieldCorrection tool, ANTs v.2.3.4) [8]. The specification of the TOPUP and N4BiasFieldCorrection command can be found in our prior publication [9]. The distortion-corrected EPI data was linearly registered to the Australian Mouse Brain Mapping Consortium (AMBMC) MRI template resampled to 0.2 mm isotropic resolution using FSL's FLIRT [4]. This linearly registered EPI data were then used for an iterative image registration and EPI study-specific template creation procedure [10]. The iterative registration-template creation process was accomplished using ANTs’ antsMultivariateTemplateConstruction2.sh tool, with the following specification:
antsMultivariateTemplateConstruction2.sh -d 3 -b 1 -c 2 -g 0.2 -i 6 -j 12 -k 1 -q 1600×800×400×200×100 -f 4×2×2×1×1 -s 2×2×1×0×0 -n 1 -o <study_template> -m MI -v 8 input_images.txt
Each functional MRI dataset was processed through the following procedure: slice-timing correction with FSL's slicetimer tool, despiked using AFNI's 3dDespike (https://afni.nimh.nih.gov/) tool, and motion-corrected using FSL's MCFLIRT tool (FMRIB's Software Library) [4] with the forward phase EPI image as the reference image. Distortion correction warping field (from the opposite phase-encoding EPI pair) was applied on the processed functional images, then the distortion corrected functional data was band-pass filtered at 0.01–0.2 Hz using AFNI's 3dTproject (https://afni.nimh.nih.gov/), and spatially normalised to the study-specific EPI template.
The pre-processed and registered functional data (9 animals, 2 dosage levels sessions per animal, 3 functional scans per dosage level session: pre-, during-, and post-stimulation fMRI) were loaded into Independent Vector Analysis – Gaussian and Laplacian (IVA-GL) [11,12] as implemented in the Group ICA of fMRI Toolbox (GIFT v.3.0) [13] for data decomposition. The number of components were set to 100 Independent Components (ICs). Input data were pre-processed via default options by GIFT: automatic masking of non-brain voxels and de-meaning per time point. Individual dataset-level IC spatial maps and time courses were converted to Z statistics. Means of each component's spatial maps and frequency power spectra were calculated across all datasets and the mean component spatial maps were thresholded at Z > 1, for the purpose of display, manual classification, and sorting (in Supplementary data 1 and Fig. 1).
Manual classification of components into “signal” and “noise” components based on criteria outlined prior publications [14,15]. Components are considered “signal” if they meet all of the following criteria: (1) component's spatial map is clustered on spatially and functionally feasible grey matter, (2) component's frequency power spectrum shows power mostly in the 0.01–0.1 Hz range (or at least the power in this frequency range is larger than the power above 0.1 Hz), (3) component's spatial map has little or no overlap with white matter or cerebral-spinal fluid regions, (4) component is rejected as “functionally relevant” if its spatial map has a thickness in the rostral-caudal direction equivalent to one EPI slice in the acquisition resolution (single-EPI-slice artefacts). Components are rejected as “signal” and considered “noise” they meet one of the following criteria: (1) component's spatial map has a large overlap with white matter or cerebral-spinal fluid areas (2) component's spatial map has ring-like or crescent shapes around the edge of the brain or near regions with EPI distortion, (2) component's frequency power spectrum shows pan frequency distribution, (3) component's spatial map has alternating positive and negative correlation bands. All identified independent components’ time courses, and all-dataset-averaged complete spatial maps and frequency power spectra from 100 component IVA-GL decomposition are presented in Supplementary Fig. 1. “Signal” components’ spatial maps were identified, grouped into eight anatomical/network groupings and shown in Fig. 1.
One sample t-tests were performed through using GIFT's Mancovan toolbox [16] testing component-component functional network connectivity averages across all subjects’ post-stimulation scans during 0.3 mg/kg BW/h sessions; test was implemented as permutation-based test using FSL's randomise [17] corrected for multiple comparisons using False Discovery Rate at Q < 0.05. Thresholded functional connectivity matrix is shown in Fig. 2 and the unthresholded raw Z statistic of this test is shown in the comma-separated values file Supplementary Data 2.csv.
Ethics Statements
All experiments were approved by the Institutional Animal Ethics Committee at the University of Queensland (Animal Ethics Committee approval number QBI/SCMB/089/16/QBI). This manuscript was prepared in compliance to the ARRIVE guideline [18]. All experiments complied with the Australian code of practice for the care and use of animals for scientific purposes (2013, 8th edition) [19].
CRediT authorship contribution statement
Xuan Vinh To: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft. Fatima A. Nasrallah: Data curation, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This research was supported by the Motor Accident Insurance Commission (MAIC), Queensland Government, Australia (grant number: 2014000857). The MAIC played no role in study design, in the collection, analysis, interpretation, and publication of data; in the writing of this manuscript; and in the decision to submit the manuscript and the associated data for publication.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2022.108279.
Appendix. Supplementary materials
Data Availability
References
- 1.To X.V., Vegh V., Nasrallah F.A. Towards data-driven group inferences of resting-state fMRI data in rodents: comparison of group ICA, GIG-ICA, and IVA-GL. J. Neurosci. Methods. 2021 doi: 10.1016/j.jneumeth.2021.109411. [DOI] [PubMed] [Google Scholar]
- 2.Grandjean J., Canella C., Anckaerts C., Ayrancı G., Bougacha S., Bienert T., Buehlmann D., Coletta L., Gallino D., Gass N., Garin C.M., Nadkarni N.A., Hübner N.S., Karatas M., Komaki Y., Kreitz S., Mandino F., Mechling A.E., Sato C., Sauer K., Shah D., Strobelt S., Takata N., Wank I., Wu T., Yahata N., Yeow L.Y., Yee Y., Aoki I., Chakravarty M.M., Chang W.T., Dhenain M., von Elverfeldt D., Harsan L.A., Hess A., Jiang T., Keliris G.A., Lerch J.P., Meyer-Lindenberg A., Okano H., Rudin M., Sartorius A., Van der Linden A., Verhoye M., Weber-Fahr W., Wenderoth N., Zerbi V., Gozzi A. Common functional networks in the mouse brain revealed by multi-centre resting-state fMRI analysis. Neuroimage. 2020:205. doi: 10.1016/j.neuroimage.2019.116278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rorden C., Brett M. Stereotaxic display of brain lesions. Behav. Neurol. 2000;12:191–200. doi: 10.1155/2000/421719. [DOI] [PubMed] [Google Scholar]
- 4.Smith S.M., Jenkinson M., Woolrich M.W., Beckmann C.F., Behrens T.E.J., Johansen-Berg H., Bannister P.R., De Luca M., Drobnjak I., Flitney D.E., Niazy R.K., Saunders J., Vickers J., Zhang Y., De Stefano N., Brady J.M., Matthews P.M. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 5.Bajic D., Craig M.M., Mongerson C.R.L., Borsook D., Becerra L. Identifying rodent resting-state brain networks with independent component analysis. Front. Neurosci. 2017:11. doi: 10.3389/fnins.2017.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson J.L.R., Skare S., Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage. 2003;20:870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- 7.Tustison N.J., Avants B.B., Cook P.A., Zheng Yuanjie, Egan A., Yushkevich P.A., Gee J.C. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avants B.B., Tustison N.J., Stauffer M., Song G., Wu B., Gee J.C. The insight ToolKit image registration framework. Front. Neuroinform. 2014;8:1–13. doi: 10.3389/fninf.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.To X.V., Nasrallah F.A. Multi-modal magnetic resonance imaging in a mouse model of concussion. Sci. Data. 2021;8:1–13. doi: 10.1038/s41597-021-00985-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dohmatob E., Varoquaux G., Thirion B. Inter-subject registration of functional images: do we need anatomical images? Front. Neurosci. 2018;12:1–11. doi: 10.3389/fnins.2018.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J.H., Lee T.W., Jolesz F.A., Yoo S.S. Independent vector analysis (IVA): multivariate approach for fMRI group study. Neuroimage. 2008;40:86–109. doi: 10.1016/j.neuroimage.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Anderson M., Adali T., Li X.L. Joint blind source separation swith multivariate gaussian model: algorithms and performance analysis. IEEE Trans. Signal Process. 2012;60:1672–1683. doi: 10.1109/TSP.2011.2181836. [DOI] [Google Scholar]
- 13.Calhoun V.D., Adali T., Pearlson G.D., Pekar J.J. A method for making group inferences from functional MRI data using independent component analysis. Hum. Brain Mapp. 2001;14:140–151. doi: 10.1002/hbm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gri L., Douaud G., Bijsterbosch J., Evangelisti S., Alfaro-almagro F., Glasser M.F., Du E.P., Fitzgibbon S., Westphal R., Carone D., Beckmann C.F., Smith S.M. Hand classification of fMRI ICA noise components. Neuroimage. 2017;154:188–205. doi: 10.1016/j.neuroimage.2016.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerbi V., Grandjean J., Rudin M., Wenderoth N. Mapping the mouse brain with rs-fMRI: an optimized pipeline for functional network identification. Neuroimage. 2015;123:11–21. doi: 10.1016/j.neuroimage.2015.07.090. [DOI] [PubMed] [Google Scholar]
- 16.Allen E.A., Erhardt E.B., Damaraju E., Gruner W., Segall J.M., Silva R.F., Havlicek M., Rachakonda S., Fries J., Kalyanam R., Michael A.M., Caprihan A., Turner J.A., Eichele T., Adelsheim S., Bryan A.D., Bustillo J., Clark V.P., Feldstein Ewing S.W., Filbey F., Ford C.C., Hutchison K., Jung R.E., Kiehl K.A., Kodituwakku P., Komesu Y.M., Mayer A.R., Pearlson G.D., Phillips J.P., Sadek J.R., Stevens M., Teuscher U., Thoma R.J., Calhoun V.D. A baseline for the multivariate comparison of resting-state networks. Front. Syst. Neurosci. 2011;5:1–23. doi: 10.3389/fnsys.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winkler A.M., Ridgway G.R., Webster M.A., Smith S.M., Nichols T.E. Permutation inference for the general linear model. Neuroimage. 2014;92:381–397. doi: 10.1016/j.neuroimage.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.N.P. du Sert, A. Ahluwalia, S. Alam, M.T. Avey, M. Baker, W.J. Browne, A. Clark, I.C. Cuthill, U. Dirnagl, M. Emerson, P. Garner, S.T. Holgate, D.W. Howells, V. Hurst, N.A. Karp, S.E. Lazic, K. Lidster, C.J. MacCallum, M. Macleod, E.J. Pearl, O.H. Petersen, F. Rawle, P. Reynolds, K. Rooney, E.S. Sena, S.D. Silberberg, T. Steckler, H. Würbel, Reporting animal research: explanation and elaboration for the arrive guidelines 2.0, 2020. 10.1371/journal.pbio.3000411. [DOI] [PMC free article] [PubMed]
- 19.N. Health, Medical Research Council . 8th ed. National Health and Medical Research Council; Canberra: 2013. Australian Code of Practice For the Care and Use of Animals For Scientific Purposes. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.