Abstract
Metabolite biosynthesis is regulated by gene expression, which is altered by DNA methylation in the promoter region. Chalcone isomerase (CHI) gene encodes a key enzyme in the Lithocarpus polystachyus Rehd flavonoid pathway, and the expression of L. polystachyus CHI (LpCHI) is closely related to the synthesis of flavonoid metabolites. In this study, we analyzed the DNA methylation site of the LpCHI promoter and its effect on gene expression and metabolite accumulation. The proportions of three types of LpCHI promoter DNA methylation are 7.5%, 68.75%, 18.75%, determined by bisulfite sequencing. Transcriptome sequencing shows that LpCHI is strongly up-regulated in LpCHI promoter methylation Type A but down-regulated in LpCHI promoter methylation Type B and Type C. The expression of LpCHI shows no significant difference between Type B and Type C. Moreover, nine kinds of differentially expressed transcription factors (DETFs) bind to seven CpG-sites of the LpCHI promoter region to regulate LpCHI expression. The results of metabolomics show that differentially accumulated flavonoids are higher in LpCHI promoter methylation Type A than in LpCHI promoter methylation Type B and Type C. Additionally, a positive correlation was found between the LpCHI expression and flavonoids accumulation. These results show that the effect of CpG site-specificity on gene transcription is great than that of overall promoter DNA methylation on gene transcription. The mechanisms of flavonoid genes regulating metabolite accumulation are further revealed.
Keywords: Chalcone isomerase, DNA methylation, Transcription, Flavonoid
1. Introduction
As a traditional Chinese herbal medicine, Lithocarpus polystachyus Rehd belongs to Fagaceae and is evergreen. Due to its young leaves can be used as tea leaves, L. polystachyus is called “sweet tea” in folk [1,2]. In addition, L. polystachyus is rich in flavonoid metabolites, and trilobatin and phlorizin of dihydrochalcone are its main active components [3,4]. Leaf aqueous extract of L. polystachyus can inhibit proliferation, migration, and invasion of cancer cells and has anticancer activity [5]. The flavonoid fraction of L. polystachyus could reduce fasting blood glucose, improve serum lipid levels, and exerts therapeutic and preventive effects on hyperlipidemia and hyperglycemia in diabetes rats [6]. Flavonoids are common secondary metabolites in plants and are synthesized by phenylalanine derivatives through the phenylpropane synthesis pathway [7,8]. Chalcone synthase (CHS), the first rate-limiting enzyme in the biosynthesis pathway of flavonoids, can catalyze three malonyl CoA and one coumarin CoA to produce naringin chalcone (6′-hydroxychalcone). Chalcone isomerase (CHI) is the second key enzyme in the biosynthesis pathway of flavonoids. Naringin chalcone (6′-hydroxychalcone) isomerizes to 5,7,4′-trihydroxyflavanone (naringenin) via CHI. The CHI superfamily is divided into four families: Type Ⅰ CHI widely exists in vascular plants and has activity; Type Ⅱ CHI with activity is mainly present in legumes; Type Ⅲ CHI (FAP) has fatty acid catalytic activity; Type Ⅳ CHI (CHIL) has no catalytic activity [9,10]. Since the cDNA of CHI was first cloned from pea in 1987, scholars have successively analyzed the CHI gene sequence of Mirabilis himalaica [11], Carthamus tinctorius [12], Dracaena cambodiana [13], and other species. In addition, the CHI gene families of ferns [14], cotton [15], soybeans [16], and other plants are analyzed. Three types of CHIs are identified in the genomes of Quercus robur L, Castanea mollissima, and other Fagaceae [17].
DNA methylation is an important epigenetic mechanism. DNA methylation of plant genes is involved in X-chromosome inactivation, transposable element silencing, genomic imprinting, and genome stability. Additionally, it regulates embryogenesis, flowering time, and leaf morphology [18]. DNA methylation within the promoter region of plant functional genes prevents transcription factors (TFs) from binding to methylated promoters, thereby inhibiting gene expression [19]. The promoter methylation level of the osmotic stress Wdreb2 TF is negatively correlated with its gene expression level in wheat [20]. The different methylation sites and levels of anthocyanidin synthase (ANS) gene promoter in red and white cultivars may result in different expression levels of ANS, leading to different flower colors of two lout cultivars [21]. At the developmental stages of Malus halliana, different methylation levels of R2 and R8 in the MtMYB10 promoter could cause differentially expressed MtMYB10, thus leading to different anthocyanin contents in flowers [22]. The evaluation of the interconnection of the DNA methylome and transcriptome shows melatonin effects on promoter methylation of flavonoid biosynthesis genes and gene expression and controls flavonoid accumulation [23]. Additionally, whole-genome DNA methylation patterns of apple fruit skin show that hypomethylation of mCG and mCHG contexts in flavonoid biosynthesis pathway genes leads to transcriptional activation and promotes anthocyanin accumulation [24]. There have been reports about the effect of DNA methylation of L. polystachyus CHI (LpCHI) gene promoter on its gene expression. However, the molecular mechanism of LpCHI promoter DNA methylation regulation has not been thoroughly studied. In this study, we analyzed the DNA methylation in the LpCHI promoter region by bisulfite sequencing. Further, we revealed the influence of the CHI gene on flavonoid synthesis based on transcriptomics and metabolomics of L. polystachyus. Technical and theoretical supports for promoting the industrial synthesis of L. polystachyus flavonoids can be provided by artificially regulating the DNA methylation state of the key enzyme gene promoter in engineered bacteria.
2. Materials and methods
2.1. Plant materials
In July 2018, L. polystachyus leaves were sampled in Bama Yao Autonomous County of Guangxi Zhuang Autonomous Region, China. The identification of the plant materials was performed by Prof. Zhaobin Xing. These samples were immediately frozen in liquid nitrogen and stored at −80 °C for subsequent DNA bisulfite, transcriptome, and metabolite analysis. Three replicates of all tests were performed for each sample.
Plant DNA Isolation Kit (FOREGENE, Chengdu, China); DNA Bisulfite Conversion Kit, Methylation-specific kit, RNAprep Pure Plant Plus Kit (Polysaccharides & Polyphenolics-rich), Talent qPCR PreMix (SYBR Green) (TIANGEN, Beijing, China); phloretin, phlorizin, trilobatin, naringenin and apigenin (MREDA, Beijing, China); kaempferol (Shanghai yuanye Bio-Technology Co., Ltd, Shanghai, China); Plant PAL ELISA KIT, Plant C4H ELISA KIT, Plant 4CL ELISA KIT, Plant CHS ELISA KIT, and Plant CHI ELISA KIT (Jing Kang Bio, Shanghai, China); the kit for the determination of total flavonoids in plants (Solarbio, Beijing, China).
2.2. Bisulfite sequencing and analysis of DNA methylation of the LpCHI promoter
According to Lin et al. [24], total DNA was extracted from the mature leaves of L. polystachyus. Afterward, the PCR amplification was performed using the methylation-specific kit according to the manufacturer's instructions. A 25 μL reaction mixture containing 1 μL of methylation DNA, 1 μL of CHIjhS1 (CHIjhS3), 1 μL of CHIjhX1 (CHIjhX3), 0.4 μL of MSP DNA polymerase, 2 μL of 10 × Msp PCR buffer, and 13 μL of ddH2O was prepared. The following cycling conditions were used for amplification: 94 °C for 5 min, (94 °C for 20 s, 55 °C for 30 s, 72 °C for 20 s (10 s)) × 35 cycles, 72 °C for 5 min. The sequences of the primers are shown in Supplementary Table S6.
PCR products of bisulfite-treated DNA sequences were analyzed by ethidium bromide-stained 1% agarose gel, and the corresponding band was sequenced at the Nuosai Genome Research Center (Beijing, China). Sequencing results of bisulfite-treated DNA sequences were analyzed using BiQ Analyzer 2.0, and DNA sequencing [17] was performed to screen DNA methylation sites.
2.3. RNA-seq analysis
Total RNA was extracted from frozen mature L. polystachyus leaves, and the mRNA libraries for each sample were constructed and sequenced in the Illumina Nova Seq 6000 system. After filtering and quality control of the original data obtained by sequencing, high-quality clean reads were obtained. Sequences from each library were spliced using the Trinity (http://trinityrnaseq.github.io/) software. In addition, all isoforms with dad FPKM (Fragments Per Kilobase of transcript per Million mapped reads) < 1, TPM (Transcripts Per Million) < 1, or IsoPct (Isoform PerCen Tage) < 5.0% were removed. The TGICL program (version 2.0.6) (https://sourceforge.net/projects/tgicl/) was used to cluster transcripts into unigenes. The BLAST algorithm was used to compare the unigene sequences with sequences from the NCBI nonredundant (NR), Swiss-Prot, Gene Ontology (GO), Clusters of Orthologous Groups (COG) of proteins, and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Moreover, KOBAS v2.0 (http://kobas.cbi.pku.edu.cn) was applied to identify enriched KEGG pathways associated with unigenes. After predicting the amino acid sequences encoded by the unigenes, the sequences were compared with those in the Pfam database using the HMMER (http://www.hmmer.org/) program for functional annotation of the unigenes.
2.4. Sample extraction and UPLC-MS/MS analysis
The freeze-dried mature leaves were pulverized using a mixer mill (MM 400, Retsch, Haan, German) at 30 Hz for 1.5 min with zirconia beads. A total of 100 mg powder was weighed and extracted with 1.0 mL 70% aqueous methanol at 4 °C overnight. After centrifugation at 10,000 g for 10 min, the supernatant was absorbed and filtered by 0.22 μm Pro size microporous membrane for UPLC-MS/MS analysis.
The extracted samples were analyzed using a UPLC-EIS-MS/MS system (UPLC, shim-pack UFLC SHIMADZU CBM30A system, https://www.shimadzu.com.cn/; MS/MS, Applied Biosystems 4500 QTRAP, http://www.appliedbiosystems.com.cn/). The analytical conditions were as follows. UPLC: column, Waters ACQUITY UPLC HSS T3 C18 (1.8 μm, 2.1 mm × 100 mm); solvent system, water (0.04% acetic acid): acetonitrile (0.04% acetic acid); gradient program, 0.00–10.00 min, 95:5 v/v to 5:95 v/v; 10.00–11.00 min, 5:95 v/v; 11.00–11.10 min, 5:95 v/v to 95: 5 v/v; 11.10–14.00 min, 95:5 v/v; flow rate, 0.35 mL/min; temperature, 40 °C; injection volume: 4 μL. MS/MS: electrospray ionization (ESI) was maintained at 40 °C; mass spectrometry voltage 5500 V; curtain gas (CUR) was 30 psi; the collision-activated ionization (CAD) parameter was set to high. Triple quadrupole (QQQ) scans of the ion pair were obtained while further optimizing the declustering potential (DP) and collision energy (CE) [25]. The software Analyst 1.6.3 was used to process mass spectrum data.
Based on MWDB (Metware Database), a qualitative analysis was conducted according to secondary spectrum information. Metabolites were quantitatively analyzed by multiple reaction monitoring (MRM) with QQQ mass spectrometry. The above analysis was completed by Metware Biotechnology Co., Ltd. (Wuhan, China).
2.5. RT-qPCR (real-time quantitative PCR)
Total RNA was extracted and purified from L. polystachyus leaves of three types of DNA methylation at different stages using the RNAprep Pure Plant kit (Tiangen, Beijing, China). Afterward, the purity, concentration, and integrity of the RNA were determined with the NanoDrop spectrophotometer (Thermo Fisher, Waltham, MA, USA) and a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). According to the manufacturer's instructions, 11 μL total RNA was reverse transcribed using the RevertAid Frist Strand cDNA synthesis kit (Beijing Bayerdi Biotechnology). Primer 5.0 (Premier Biosoft Interpairs, Palo Alto, CA, USA) was used to design primers specific for the selected genes (Supplementary Tables S6) and a dissolution curve analysis was used to determine specificity. The L. polystachyus GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene was used as the internal reference gene. The Applied Biosystems 7900HT Fast Real-Time PCR System (Life Technologies, Waltham, MA, USA) with a 96-well plate was used for amplification reactions consisting of 3 min at 95 °C, followed by 40 cycles of 5 s at 95 °C, and 15 s at 60 °C in a volume of 10 μL. The qRT-PCR assay was performed in three biological replicates, and relative expression levels were calculated based on the 2−ΔΔCt method.
2.6. Metabolite extraction and determination
The leaves were oven-dried to a constant weight and ground. A total of 10 mL extracting solution (60% ethanol) was added to 0.100 g of leaf powder for ultrasonic extraction. After adding 1 mL of extracting solution (70% ethanol), mixture A was extracted by ultrasonic method (ultrasonic power: 300 W, crushing time: 5 s, interval time: 8 s) at 60 °C for 30 min. The supernatant was collected after centrifugation at 12,000 rpm for 10 min. The supernatant was then processed according to the instructions of the kit for the determination of total flavonoids in plants. Total flavonoid content was calculated using the standard rutin curve.
The supernatant was collected and filtered by a 0.22 μm filter membrane to determine phloretin, phlorizin, trilobatin, naringenin, apigenin, and kaempferol. The stock solution of the four standards was prepared by dissolving the accurately weighed standards in 70% ethanol and fixing the volume to 50 mL. All chromatographic separations were performed using an ultra-performance liquid chromatography (UPLC) system (Waters, UK). Reversed-phase separation was performed using an ACQUITY UPLC BEN C18 column (2.1 mm × 50 mm, 1.7 μm, Waters, UK). The column oven was maintained at 30 °C with a 0.2 mL/min flow rate. The mobile phases were solvent A (acetonitrile) and solvent B (0.1% formic acid of water). Gradient elution conditions were set as follows: 0∼25 min, 95%–70% B; 25∼26min, 70%–95% B; 26–27 min, 95% B. The kaempferol (apigenin) standard was accurately weighed and dissolved in methanol as the stock solution at a concentration of 28.3 (58.7) μg/mL. Gradient elution conditions were set as follows: 0∼8 min, 95%–40% B; 8∼9min, 40%–95% B; 9–10 min, 95% B (0∼7 min, 90%–50% B; 7∼8min, 50%–90% B; 8∼9 min, 90% B).
2.7. Determination of enzyme activity
A 0.1 g of L. polystachyus leaves were ground in liquid nitrogen. The sample was transferred into 15 ml centrifuge tubes containing 9 mL of PBS buffer with pH = 7.4. After centrifugation at 8000 g for 30 min at 4 °C, the supernatant was isolated. The supernatants were processed separately using Plant PAL ELISA KIT, Plant C4H ELISA KIT, Plant 4CL ELISA KIT, Plant CHS ELISA KIT and Plant CHI ELISA KIT (Shanghai Crystal Antibody, Shanghai, China). The OD values of standards and samples were measured using the enzyme-labeled instrument Infinite M200 (TECAN, Switzerland). The linear regression equation of the standard curve was calculated from the concentration of the standards and OD values, and the sample enzyme activity was calculated.
3. Results
3.1. DNA methylation analysis of LpCHI gene promoter
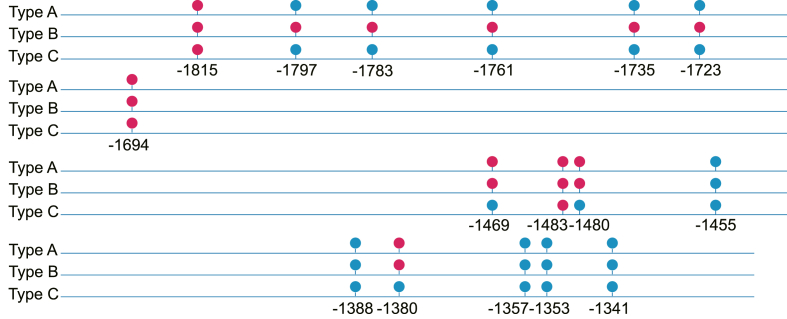
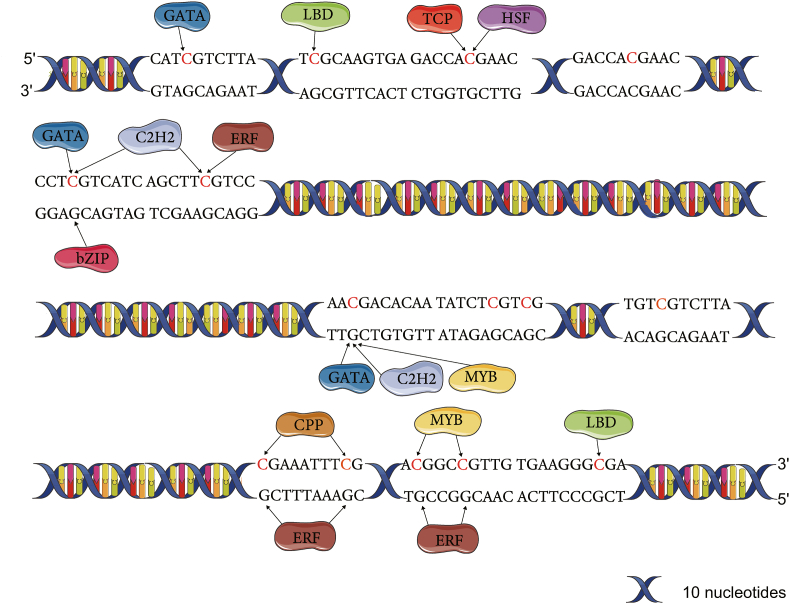
Three contexts of methylation in plants and fungi are CpG, CpHpG, and CpHpH (where H represents A, T, or C), with CpG being the major context [26]. The LpCHI promoter has 16 CpG-type cytopyrimidine DNA methylation sites, and three DNA methylation types are detected, named LpCHI promoter methylation Type A, LpCHI promoter methylation Type B, and LpCHI promoter methylation Type C. The three types of LpCHI promoter DNA methylation have 6, 11, and 3 methylation sites, respectively. Among the 16 sites, cytosines at -1815, -1694, and -1483 sites are methylated in all types of DNA methylation; cytosines at - 1469, 1480, and 1380 sites are methylated in Type A and Type B; cytosines at -1797, -1783, - 1761, -1735, and -1723 sites only have DNA methylation in Type B (Fig. 1). By combining analysis with the cis-acting elements predicted by PlantCARE, we found that 6 of the 16 DNA methylated cytosines are in cis-regulatory elements. They are -1353 (MYB recognition site/CCAAT-box), -1388 (LTR), -1496 (TGA-element), -1735 (CGTCA-motif/TGACG-motif/as-1), -1783 (Unnamed_1:CGTGG), and -1797 (Unnamed_4:CTCC).
Fig. 1.
DNA methylation sites of LpCHI promoter. Blue circle: corresponding to methylated Cs; red circle: unmethylated Cs. Number: site distance to ATG translation initiation codon (bp).
A1-A3, B1-B3, and C1-C3 denote the three biological replicates of Type A, Type B, and Type C, respectively. Q20: percentage of bases with a Phred quality score greater than 20. Q30: percentage of bases with a Phred quality score greater than 30.
3.2. Transcriptome sequencing and unigene annotation
We collected samples of LpCHI promoter methylation Type A, LpCHI promoter methylation Type B, and LpCHI promoter methylation Type C for RNA-seq analysis. On average, 25.0 million clean reads were obtained for each sample. The GC content of sequences for the nine samples is about 44%, and more than 92% of the bases have a Phred quality score greater than 30 (Table 1). Transcripts were spliced by Trinity, and 254,492 transcripts and 216,116 unigenes were obtained. The total length of unigenes is 107,119,884 bp, with the longest being 15,721 bp and the shortest being 200 bp, and the average length is 497 bp. N50 is 641 bp, and N90 is 240 bp.
Table 1.
Summary of sequencing reads after filtering.
| SampleID | ReadSum | BaseSum | GC (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| Type A1 | 24,188,556 | 7,256,566,800 | 43.90% | 97.32% | 92.65% |
| Type A2 | 24,628,061 | 7,388,418,300 | 44.10% | 97.38% | 92.77% |
| Type A3 | 26,965,807 | 8,089,742,100 | 44.03% | 97.51% | 93.05% |
| Type B1 | 24,168,850 | 7,250,655,000 | 44.28% | 97.70% | 93.44% |
| Type B2 | 23,120,916 | 6,936,274,800 | 44.34% | 97.48% | 92.96% |
| Type B3 | 23,850,671 | 7,155,201,300 | 44.37% | 97.78% | 93.62% |
| Type C1 | 28,963,964 | 8,689,189,200 | 44.56% | 97.56% | 93.11% |
| Type C2 | 26,283,334 | 7,885,000,200 | 44.53% | 97.43% | 92.87% |
| Type C3 | 23,086,670 | 6,926,001,000 | 44.44% | 97.53% | 93.12% |
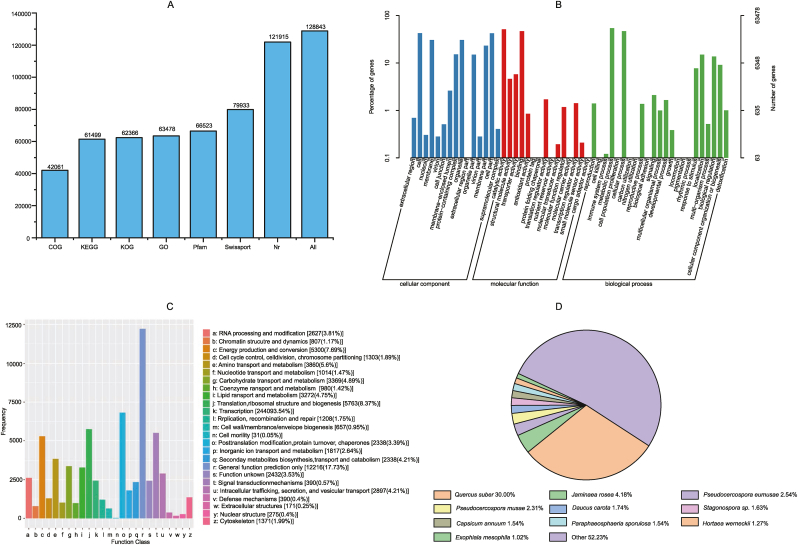
A total of 128,843 unigenes functions were predicted in seven public databases, including NR, Swiss-Prot, GO, COG, KOG, KEGG, Pfam (Fig. 2A). Among them, 37,562 (29.15%) genes are 300 bp to 1000 bp in length, and 19,374 (15.04%) genes were greater than 1000 bp in average length. The unigene number of the COG database is the lowest (42,061 unigenes), while the number of unigene annotations in the NR database is the highest (63,478 unigenes) (Fig. 2A). A total of 53 GO terms are significantly enriched in 3 GO ontologies. The main terms of the cell component are “membrane” (19,422 unigenes), “membrane part” (14,740 unigenes), “cell” (26,900 unigenes), and “cell part” (26,827 unigenes). The main terms of the molecular function are “binding” (29,798 unigenes) and “catalytic activity” (32,600 unigenes). The main terms of the biological process are “cellular process” (29,367 unigenes) and “metabolic process” (34,301unigenes) (Fig. 2B). In addition, 17.73% unigene annotations of the KOG database are “General function prediction only”, and 0.50% unigenes annotations are “Cell motility” (Fig. 2C). Sequence match analysis of the NR database reveals that L. polystachyus unigenes are most similar to genes from Quercus suber (30%) (Fig. 2D).
Fig. 2.
Unigenes annotation. A: Unigenes annotations of seven public databases. B: Unigene functional annotations based on the gene ontology database. C: Classifications based on the euKaryotic Orthology Groups database. D: gene annotations of NR database.
3.3. Expression of flavonoid synthesis-related genes
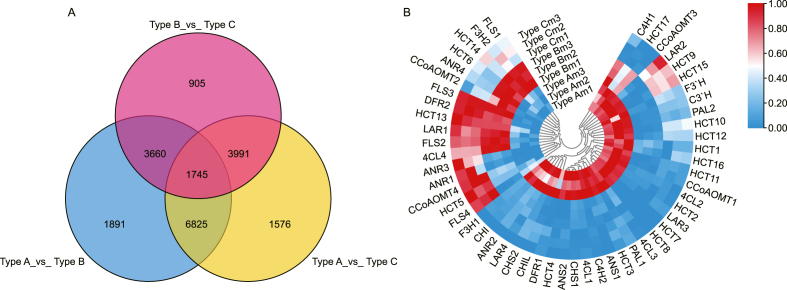
Based on significance level (FDR ≤0.01) and log2 fold change (FC), the screening conditions for differentially expressed genes (DEGs) serve as the threshold for up-regulation (FC≥ + 0.5) and down-regulation (FC ≤ - 0.5). A total of 20,593 DEGs were obtained in the transcriptome sequencing (Fig. 3A). There are 14,121 DEGs from Type A_vs_Type B, including 6852 up-regulated genes and 7269 down-regulated genes. Additionally, 14,137 DEGs, including 7547 up-regulated genes and 6590 down-regulated genes, were identified between LpCHI promoter methylation Type A and Type C. We identified 10,301 DEGs, including 7547 up-regulated and 5757 down-regulated genes in LpCHI promoter methylation Type B compared with LpCHI promoter methylation Type C. The Venn diagram shows the overlap between DEGs in the three comparisons (Type A_vs_Type B, Type A_vs_Type C, and Type B_vs Type_C). A total of 1745 DEGs are different in all three comparisons, with 1891 DEGs only present in Type A_vs_Type B and 905 DEGs only present in Type B_vs_Type C (Fig. 3A). In addition, there are 8570 DEGs between the group of Type A_vs_Type B and Type A_vs_Type C, suggesting that the transcript expressions of LpCHI promoter methylation Type A, Type B, and Type C differ significantly.
Fig. 3.
Analysis of DEGs in three types of LpCHI promoter DNA methylation L. polystachyus. A: Venn diagram of DEGs among the three groups. B: Heatmap Clustering is based on normalized FPKM of DEGs in the flavonoid pathway.
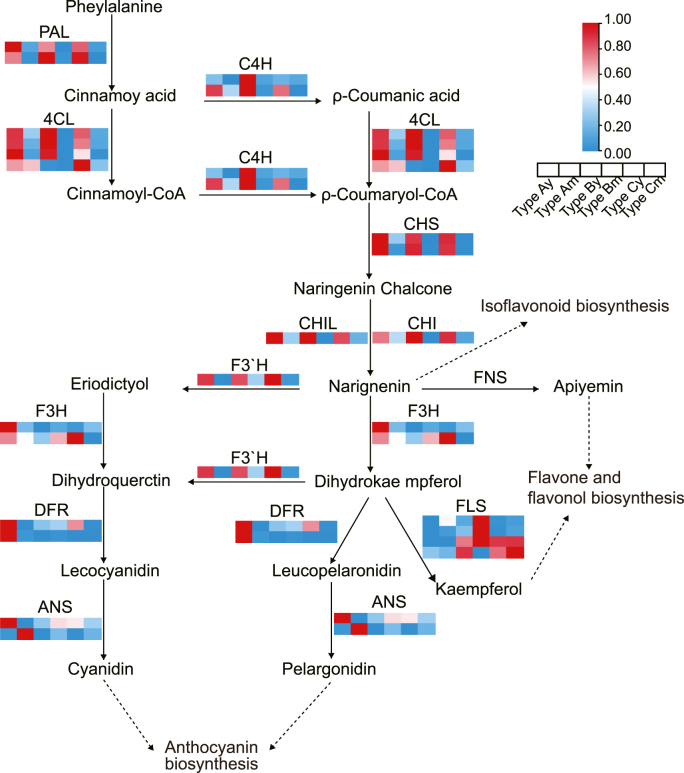
In plants, phenylalanine can be converted by phenylalanine ammonia-lyase (PAL) to produce cinnamic acid, catalyzed by cinnamate 4-hydroxylase (C4H) and 4-coumarate CoA ligase (4CL) to produce ρ-coumaroyl CoA, and then enters the flavonoid biosynthesis pathway [27]. According to the KO numbers of the three genes and the flavonoid biosynthesis pathway (ko00941), 53 DEGs were identified in all comparisons (Fig. 3B, Supplementary Table 1). Specifically, TRINITY_DN22537_c0_g2 and TRINITY_DN256813_c0_g1 annotate to CHI in KEGG, Pfam, and Nr; TRINITY_DN22537_c0_g2 is identical to the LpCHI (MZ733721) [28] sequence cloned in the previous study; TRINITY_DN256813_c0_g1 is the gene encoding Type Ⅳ CHI (CHIL). Their expressions are high in LpCHI promoter methylation Type A but are not significantly different in LpCHI promoter methylation Type B and Type C (Fig. 3B). Two unigenes encoding LpPAL exhibit high expression in Type A but do not differ significantly in Type B and Type C. In addition, LpC4H1 transcription is not significantly different in Type A and Type B, while LpC4H2 is up-regulated in Type A but not significantly different in Type B and Type C. Four transcriptions encoding 4CL show the same expression trend as LpPAL, except that Lp4CL4 has high expression in Type A but low expression in Type B (Fig. 3B). CHS catalyzes three malonyl CoA and one coumaroyl CoA to naringenin. The two LpCHS identified from transcriptome sequencing data are differentially expressed in the three types of LpCHI promoter methylation and show the same aggregation trend as LpCHI (Fig. 3B). Downstream genes of LpCHI in the flavonoid biosynthetic pathway, including LpDFR1 (dihydroflavonol 4-reductase), LpF3′H (flavonoid 3′-monooxygenase), LpANS1 (anthocyanidin synthase), LpANS2, LpANR2 (anthocyanidin reductase), and three LpLARs (leucoanthocyanidin reductase), are down-regulated in Type B and Type C. However, LpFLSs (flavonol synthase) are principally expressed in Type B (Fig. 3B).
In order to validate the reliability of the transcriptome data, real-time PCR (qRT-PCR) was conducted to DEGs of the flavonoid biosynthetic pathway. The results show that the relative expression trend of DGEs is consistent with transcriptome sequencing data, proving the reliability and accuracy of transcriptome sequencing data (Fig. 4).
Fig. 4.
Heatmap Clustering is based on normalized flavonoids pathway genes relative expressions (qRT-PCR) of three types of LpCHI promoter DNA methylation L. polystachyus at different stages. Solid arrows indicate flavonoids pathway, and dotted arrows indicate downstream branches of flavonoids. The “y” indicates young leaves, and “m” indicates mature leaves.
The transcript expression of DEGs in the flavonoid biosynthetic pathway in young and mature leaves was assessed by qRT-PCR, and the changing transcript expression at different developmental stages was analyzed. Expressions of LpFLSs involved in the flavone and flavonol biosynthesis pathways and LpANS2 involved in the anthocyanin biosynthesis pathway are greater at the young stage compared with the mature stage. Expressions of LpPALs, LpCHSs, LpCHI, LpCHIL are lower at the mature stage than at the young stage (Fig. 4).
3.4. Analysis of TFs associated with LpCHI
Transcription factor binding sites in the LpCHI promoter region were predicted using Transcriptional Regulatory Map with Castanea mollissima as a reference. The results suggest that 33 kinds, 148 TFs of C. mollissima can bind to the LpCHI promoter region with 284 binding sites. There are nine kinds TFs that can bind DNA methylation sites in LpCHI promoter CpG-island. Each TF can bind 1 to 3 CpG-sites (Fig. 5, Supplementary Table 2). Additionally, TF binding sites are absent in -1455, -480, -1483, and -1761 sites, and the remaining 12 CpG-sites have TF binding sites (Fig. 5). The -1469 and -380 CpG-sites occur in LpCHI promoter DNA methylation Type A and Type B; the -1469 site has C2H2, ERF, NAC, and MYB TF binding sites; the −1380 site has ERF binding site. CpG-sites are only present in Type C, where LBD, TCP, HSF, GATA, bZIP, C2H2, and ERF binding sites are found.
Fig. 5.
Predictions of binding sites of CpG-sites and TFs in LpCHI promoter (1848 bp to 1339 bp upstream of the ATG start codon).
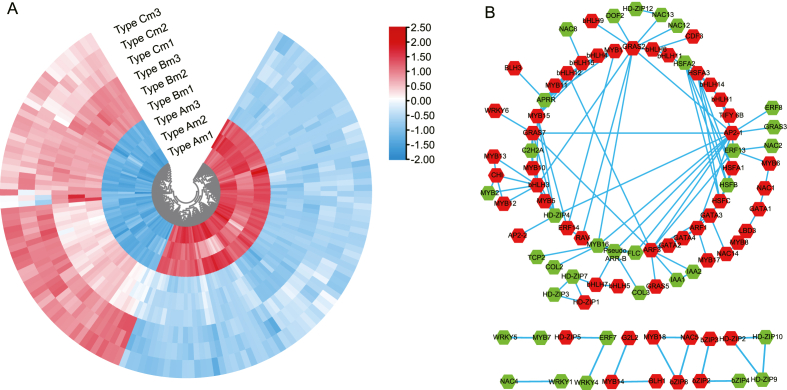
Multiples sequences of C. mollissima 148 TF amino acid sequences and L. polystachyus amino acid sequences in the transcriptome database were aligned by BLAST, and 427 DETFs were identified. Correlation analysis was performed between 427 DETFs and LpCHI expressions by the R package. PCC (Pearson correlation coefficient) > 0.9 and P < 0.01 were set as the cutoff criterion. A total of 148 DETFs were filtered, which strongly correlated with LpCHI expression, including bHLH, bZIP, ERF, MYB, and NAC. Among them, 85 DETFs were positively correlated, and 63 DETFs were negatively correlated (Fig. 6A; Supplementary Table 3). The protein-protein interaction network of 148 DETFs and LpCHI was reconstructed with STRING 11, and 91 proteins had protein-protein interaction (Fig. 6B). In addition, LpMYB2, LpMYB12, LpMYB13, and LpbHLH3 directly interact with LpCHI; LpMYB2 is down-regulated in LpCHI promoter DNA methylation Type A, while LpbHLH3, LpMYB12, and LpMYB13 are up-regulated. LpbHLH3 interacts with the proteins of LpMYB2, LpMYB12, LpMYB13, LpMYB5, LpMYB10, LpMYB15, LpGRAS2, and LpHD-ZIP4. However, LpMYB2, LpMYB12, and LpMYB13 do not interact with other DETFs. Furthermore, there is an interaction among 5 LpHSFs. Specifically, LpHSFA3 interacts with LpbHLH14, and LpHSFC interacts with LpGATA3. LpGATA3, LpGATA2, and LpGATA4 interact with LpARF1 and LpARF3; LpGATA1 has interactions with the proteins of LpLBD3 and LpNAC1; LpC2H2A, LpMYB10, and LpGRAS7 have interaction. For protein-protein interactions, four LpERFs interact with other TFs; LpAP2-1 has interplays of proteins with LpERF8 and LpERF13; LpERF14 interacts with three IFs, and one of them is LpRAV of the AP2/ERF family.
Fig. 6.
Analysis of DETFs of 3 types LpCHI promoter DNA methylation in L. polystachyus. A: Cluster analysis for DETFs. Red boxes indicate high expression levels, and blue boxes indicate low expression levels; m1-m3: 3 biological repeats of L. polystachyus mature leaves. B: protein-protein interaction of DETFs and LpCHI. Green hexagon indicates low-expression in LpCHI promoter DNA methylation Type A, and red hexagon indicates high-expression in LpCHI promoter DNA methylation Type A.
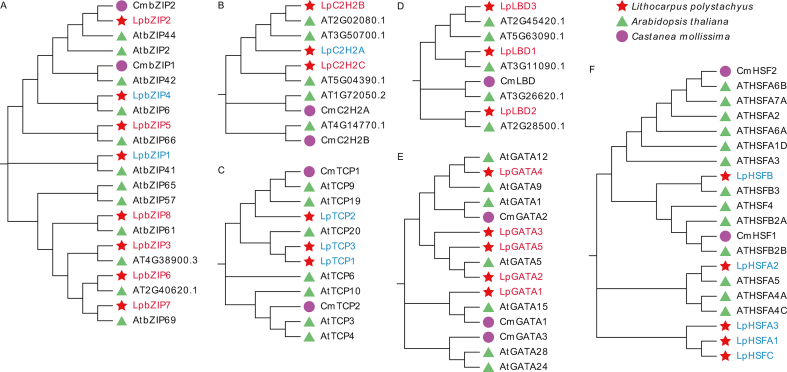
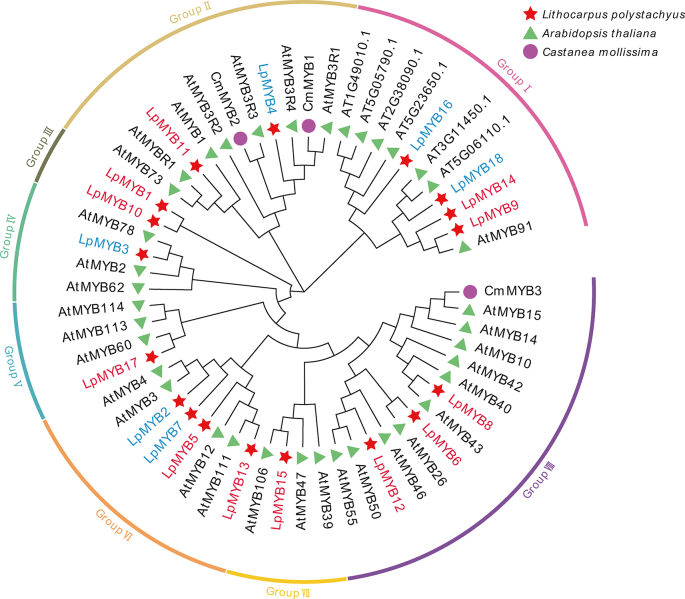
Nine kinds of DETFs that can bind to the CpG-site of LpCHI promoter and predict TFs of C. mollissima, and Arabidopsis thaliana TFs were aligned with Muscle W and NJ trees constructed by MEGA X. Additionally, three out of seven LpbZIPs are clustered with CmbZIP1 and CmbZIP2; LpbZIP2 has a positive correlation with LpCHI gene expression and the highest sequence similarity with CmbZIP2 (78.08%); they are clustered with AtbZIP44, a TF affecting seed germination of A. thaliana [29] (Supplementary Fig. 1A). LqTCPs are grouped with CmTCP1, LqGATA1 displays closer relationships with CmGATA1, and LqGATA4 is closely related to CmGATA2. Five LpHSFs expressions are negatively correlated with LpCHI. Only LpHSFB clusters with CmHSF1. LpC2H2s, LpLBDs, and predicted TFs of C. mollissima are not clustered (Supplementary Figs. 1B–F). Eighteen LpMYBs were classified into eight groups. Among them, LpMYB4, LpMYB11, CmMBY1, and CmMBY2 are clustered in Group Ⅱ; LpMYB8, LpMYB6, LpMYB12, and CmMBY3 are in Group Ⅷ. However, LpMYB1 and LpMYB10 in Group Ⅲ do not cluster with A. thaliana TFs (Supplementary Fig. 2). Sixteen LpERFs and ten CmERFs are classified into nine branches. Seven branches cluster with LpERFs and CmERFs apart from Group Ⅰ and Group Ⅱ. LpERF15 shows 97.07% sequence similarity to CmERF9 in Group Ⅶ. The sequence similarity between LpERF6 and CmERF8 in group VIII is 90.64%. LpERF15 and LpERF6 positively correlate with LpCHI expression (Supplementary Fig. 3). Three in the identified six branches of LpNACs have negative relations to LpCHI and are clustered with CmNAC. LpNAC13 exhibits relatively high sequence similarity with CmNAC at 95.95%, and it clusters with ANAC042, a TF involved in the regulation of camalexin biosynthesis [30] (Supplementary Fig. 2).
3.5. Identified metabolites involved in flavonoid biosynthesis
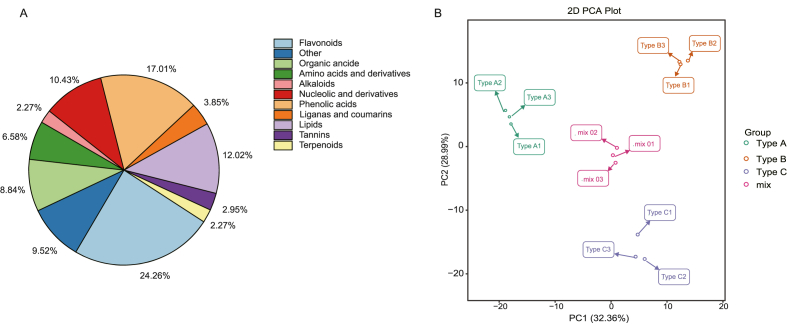
To further evaluate the effects of different LpCHI promoter DNA methylation on flavonoid biosynthesis, metabolites in L. polystachyus leaves of three types of LpCHI promoter DNA methylation were analyzed using UPLC–MS/MS. A total of 441 metabolites containing 11 classes were detected and quantified. Among them, 107 (24.26%) are flavonoids, followed by 75 (17.01%) phenolic acids (Fig. 7A).
Fig. 7.
Analysis of metabolome in L. polystachyus three types of LpCHI promoter DNA methylation. A: Distribution of metabolites. B: PCA of three types of LpCHI promoter DNA methylation L. polystachyus.
The results of principal component analysis (PCA) show that metabolites from different types of LpCHI promoter DNA methylation are clearly separated in the score plots, where the first principal component (PC1) is plotted against the second principal component (PC2). PC1 and PC2 represent 32.36% and 28.99% of the total variations, respectively (Fig. 7B). These results suggest that the samples between groups are scattered and clustered within groups, indicating good duplication within groups and significant differences between groups (Fig. 7B).
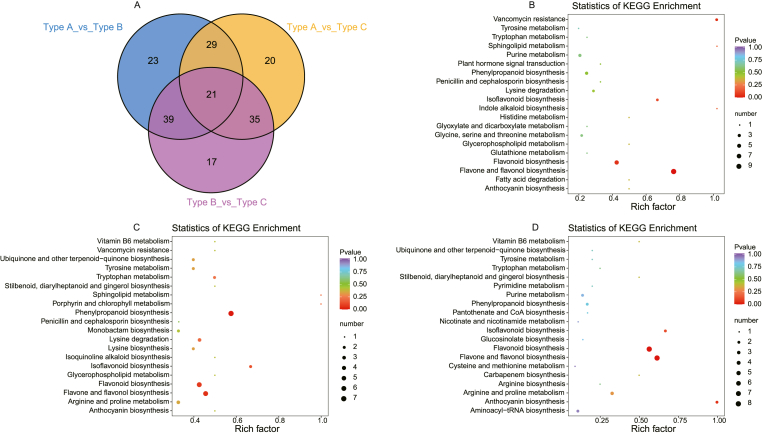
The screening criterion for differentially accumulated metabolites (DAMs) is VIP≥1, FC ≥ 2 or ≤0.5. All 184 kinds of DAMs are filtered out (Fig. 8A). In the comparison between Type A and Type B, 66 and 46 metabolites present up-regulation and down-regulation, respectively. Additionally, 105 kinds of DAMs are filtered out in Type A_vs_Type C, of which 46 are up-regulated, and 59 are down-regulated; 111 kinds of metabolites are differentially expressed among Type B and Type C, of which 59 are up-regulated, and 52 are down-regulated. We detected 21 kinds of DAMs that are differentially expressed in the three comparisons. Among them, 13 belong to flavonoids: apigenin, ladanein, tilianin (acacetin-7-O-β-D-glucoside), tricin 7-O-glucoside, luteolin-7-O-rutinoside, chrysoeriol-O-sinapoylhexoside, myricetin, 3,7-Di-O-methylquercetin, quercetin-3-O-α-l-arabinopyranoside (guaijaverin), quercetin-3-O-(6″-trans-p-coumaroyl)-β-d-galactopyranoside, malonylglucoside, sissotrin, and malvidin 3-O-galactoside (primulin). In addition, DAMs were mapped to the KEGG database and tested for enrichment. The results show that four pathways (“Flavonoid biosynthesis”, “Anthocyanin biosynthesis”, “Isoflavonoid biosynthesis”, and “Flavone and flavonol biosynthesis”) related to flavonoid metabolism are enriched in the three comparisons. The “Anthocyanin biosynthesis” pathway is significantly enriched in Type A_vs_Type B (Fig. 8B–D).
Fig. 8.
Identification of candidate DAMs. A: Venn diagram showing the distribution of metabolites altered in different types LpCHI promoter DNA methylation. B: KEGG enrichment of Type A_vs Type_B; C Venn Diagram of DAMs. B: KEGG enrichment of Type A_vs_Type C. D: Venn Diagram of DAMs. B KEGG enrichment of Type B_vs_Type C.
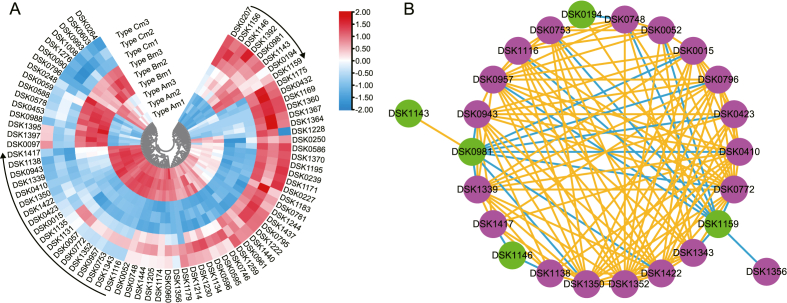
Cluster analysis for flavonoids of DAMs shows that 26 of 79 differentially accumulated flavonoids (DAFs) are significantly different among LpCHI promoter DNA methylation Typer A, Type B, and Type C; the accumulation of 8 flavonoids (DSK0207-DSK1158) is lower in Type A than in Type B and Type C; 8 flavonoids (DSK1116-DSK1417) have a higher accumulation in Type A than in Type B and Type C (Fig. 9A, Supplementary Table 4).
Fig. 9.
The DAFs of L. polystachyus three types of LpCHI promoter DNA methylation. A: Heatmap shows the relative distribution of flavonoids. B: Network diagram demonstrates the correlation between DAFs. Green circles indicate low accumulation in LpCHI promoter DNA methylation Type A, and magenta circles indicate high accumulation in LpCHI promoter DNA methylation Type A. Blue lines indicate negative, and orange lines indicate positive.
To further study the relationship among metabolites, a total of 26 DAFs were subjected to a correlation assay. Correlations were found among 24 flavonoids except for malonylglucoside and quercetin-3-O-(6″-trans-p-Coumaroyl)-β-d-galactopyranoside (Fig. 9B). Additionally, 6-Hydroxykaempferol-7-O-glucoside and Kaempferol-7-O-β-D-(6″-O-(E)-p-coumaroyl) glucoside present positive relativity, but nine flavonoids present negative relativity. Trilobatin and acetylphlorizin present positive relativity, but eight flavonoids present negative relativity. There is only a positive association between 3′-Methoxydaidzin and other flavonoids.
3.6. Correlation of metabolites and DEG of the flavonoid biosynthesis pathway
To investigate the effect of flavonoid synthesis gene expression on flavonoid content accumulation in L. polystachyus, we screened transcriptome data and qRT-PCR data for flavonoid synthesis genes consistent with LpCHI gene expression trends. The PCC (PCC>0.8, P < 0.01) between DEG and DAFs was calculated, and the correlation network diagram between genes and metabolites was constructed.
Among the 26 differential flavonoid metabolites screened, 22 metabolites are correlated with the DEG of the flavonoid biosynthesis pathway, except for quercetin-3-O-α-l-arabinopyranoside (guaijaverin) (DSK0239), myricetin (DSK0781), luteolin-7-O-rutinoside (DSK0796), and chrysoeriol-O-sinapoylhexoside (DSK0961). The LpCHI is positively correlated with 15 metabolites, negatively correlated with hydroxykaempferol-7-O-glucoside (DSK0981), acetylphlorizin (DSK0194), quercetin-3-O-(6″-trans-p-Coumaroyl)-β-d-galactopyranoside (DSK1156). Compared with LpCHI, LpCHIL is correlated with Tilianin (Acacetin-7-O-β -d-glucoside) (DSK1356), but not with Apigenin (DSK1138) and Malvidin 3-O-Galactoside (Primulin) (DSK1417). The LpC4H2 is positively correlated with 16 flavonoids, negatively correlated with 3 metabolites: 6-hydroxykaempferol-7-O-glucoside (DSK0981), acetylphlorizin (DSK0194), quercetin-3-O-(6″-trans-p-Coumaroyl)-β-d-galactopyranoside (DSK1156). The LpC4H2 is positively correlated with 16 flavonoids and negatively correlated with 3 metabolites. The LpF3′H is positively correlated with 16 flavonoids and negatively correlated with 6-hydroxykaempferol-7-O-glucoside (DSK0981) and aetylphlorizin (DSK0194). A total of 20 metabolites are correlated with Lp4CLs, but tilianin (acacetin-7-O-β-D-glucoside) (DSK1356) is positively correlated with Lp4CL1. There was no correlation between malonylglucoside (DSK0207) and Lp4CL2. All 16 metabolites are correlated with LpCHS1 and LpCHS2, while malonylglucoside (DSK0207), apigenin (DSK1138), and malvidin 3-O-galactoside (Primulin) (DSK1417) are only correlated with LpCHS1, and tilianin (Acacetin-7-O-β-D-glucoside) (DSK1356) is only correlated with LpCHS2 (Fig. 10A–G).
Fig. 10.
Correlation network in DEG and DAFs. A: correlation network of LpCHI and DAFs; B: correlation network of LpCHL and DAFs; C: correlation network of LpC4H2 and DAFs; D: correlation network of LpF3′H and DAFs; E: correlation network of LpPALs and DAFs; F: correlation network of Lp4CLs and DAFs; G: correlation network of LpCHSs and DAFs. Green circles indicate low accumulation in LpCHI promoter DNA methylation Type A; magenta circles indicate high accumulation in LpCHI promoter DNA methylation Type A; red hexagonals indicate high-expression in LpCHI promoter DNA methylation Type A; blue lines indicate negativity and orange lines indicate positive.
3.7. Flavonoids content, expression of key genes, and enzyme activity
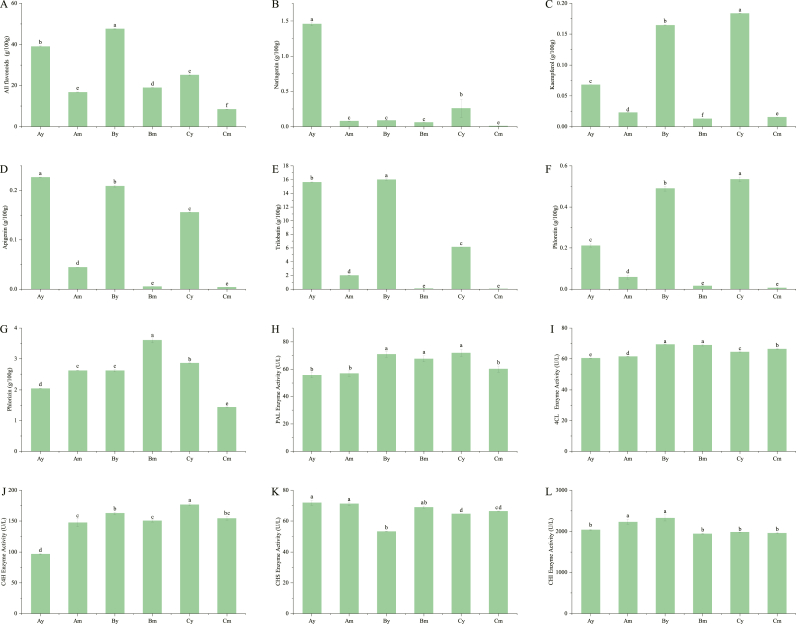
The total flavonoid content is 47.5713 (±0.1653) g/100 g to 8.3803 (±0.1210) g/100 g for the leaves of L. polystachyus. The total flavonoid content of young leave is more than twice that of mature leaves. At different developing stages, the total flavonoids content of the three types of LpCHI promoter DNA methylation leave is the highest in Type B, followed by Type A and Type C (Fig. 11A). Naringenin is a CHI catalyzed product with 1.4568 (±0.0246) g/100 g for Type Ay and 0.2592 (±0.1273) g/100 g for Type Cy. The content of other samples is less than 0.1000 g/100 g. The naringenin content was significantly higher in the early stage than in the mature stage (Fig. 11B). In an integrative analysis of LpCHI expression and metabolome, kaempferol and apigenin are significantly correlated with LpCHI. These results are consistent with our metabolome results, indicating that Type A has higher kaempferol and apigenin content than Type B and C at the mature stage. The kaempferol content of Type A is significantly lower than that of Type B and Type C at early stages (Fig. 11C and D). Trilobatin, phlorizin, and phloretin are the major chalcones and medicinal constituents in L. polystachyus. The content of trilobatin range from 0.0380 (±0.0015) g/100 g to 16.0065 (±0.1115) g/100 g, and the content of phloretin range from 0.0065 (±0.0023) g/100 g to 0.5345 (±0.0091) g/100 g. Both substances decrease during leaf maturation. Phlorizin content range from 1.4331 (±0.01113) g/100 g to 3.6023 (±0.0816) g/100 g and increase during leaf maturation (Fig. 11E–G).
Fig. 11.
Flavonoid content and enzymes activities of three types of LpCHI promoter DNA methylation at different growth stages. The “y” indicates young leaves, and “m” indicates mature leaves. Different lowercase letters represent significant differences (P < 0.05).
SPSS 25.0 was used to analyze the correlation between the relative expression of synthetic genes of flavonoid biosynthesis pathway and the content of total flavonoids and 6 key flavonoids in two growth and development stages (PCC>0.7, P < 0.05) (Supplementary Table 5). The relative expression of LpCHI is significantly correlated with the content of apigenin and kaempferol, which is consistent with our integrative analysis of transcriptome and metabolite (P < 0.01). The correlation between the relative expression of LpCHI and total flavonoid content is highly significant (P < 0.01). The relative expression of LpCHI is highly correlated with the content of trilobatin and phloretin, as well as the total content of flavonoid, trilobatin, and apigenin (P < 0.01). In addition, the PCC of LpCHIL is greater than that of LpCHI. The relative expression of Lp4CL1-3, LpCHS2, LpC4H2, and LpF3′H are also significantly correlated with the total content of flavonoids, trilobatin, and apigenin (P < 0.01).
The activities of PAL, C4H, 4CL, CHS, and CHI enzymes involved in flavonoid biosynthesis are also determined. The results show little difference in the enzyme activities of PAL and CHI in different samples of L. polystachyus at different periods (P < 0.05). There is a large difference in enzyme activities for 4CL, followed by CHS and C4H (Fig. 11H to L). In addition, the results of SPSS correlation analysis suggested that PAL, C4H, 4CL, CHS, and CHI activities are not correlated with the total content of flavonoids, naringenin, kaempferol, apigenin, trilobatin, phlorizin, and phloretin (Supplementary Table 5).
4. Discussion
One of the regulatory mechanisms of gene expression is the binding of TFs to DNA. TFs recognize specific sequences in the promoter region and activate or suppress their expression [31]. MYB proto-oncogene protein (CMYB) binds to Sirtuin 4 (SIRT4) promoter region and suppresses gene transcriptional activity, and nuclear respiratory factor 1 (NRF1) binds to the SIRT4 promoter region and suppresses gene transcriptional repression [32]. DNA methylation of promoter CpG islands causes silencing of gene expression by blocking TF binding [31]. However, recent studies have shown that three models of DNA methylation affect TF binding: DNA methylation suppresses TF binding, DNA methylation promotes TF binding, and DNA methylation does not affect TF binding [33]. Methylation of 5-methylcytosine within the AP-2, MYC binding sites can directly suppress TF binding to nucleotide sequence [34]. DNA methylation of Kaiso and C/EBPα binding sites facilitates the binding to recognition sequences [35,36]. CpG methylation does not affect the binding of YY1 protein to the bi-directional promoters of the Surf-1 and Surf-2 genes [37]. The core promoter region of the CHI gene in Paeonia lactiflora Pall has 16 CpG sites, with methylation levels ranging from 59.8% to 65.7% at different growth stages. The DNA methylation levels are negatively correlated with CHI expressions, and total flavonoid content in petals presents a similar trend to CHI expression at development stages, indicating that DNA methylation in the promoter region of CHI affects flavonoid content [38]. Previous studies have also shown that the methylation level of LpCHI gene promoter DNA is negatively correlated with the relative expression level of L. polystachyus [17]. In this study, the DNA methylation level of the LpCHI promoter is 37.5% for Type A, 68.75% for Type B, and 18.75% Type C. LpCHI expression is higher in Type A than in Type B, while there is no difference between Type B and Type C. Furthermore, no correlation is found between the DNA methylation level of LpCHI promoter and gene expression (P < 0.05). Therefore, we further analyzed the CpG-sites and found 3 different sites between the DNA methylation Type A and the Type C of the LpCHI gene promoter (-1469, -1480, -1380). The recognition sites for C2H2, MYB, and NAC TFs are at -1469 and ERF at -1469 and -1380, respectively (Fig. 4). We speculate that after DNA methylation, TFs are binding to the -1469 and -1380 sites that transcriptionally activate the expression of the LpCHI gene. Five different GpG-sites between the Type A and Type B, as well as -1797, -1783, -1735, -1723 sites, can bind to LBD, TCP, HSF, C2H2, GATA, ERF, and bZIP (Fig. 4). These 4 sites may contain TFs that bind DNA methylation to suppress and promote gene expression. The expression of the LpCHI gene is higher in Type A than in Type B. However, no significant difference is found between Type B and Type C, which can be reasonably explained when transcriptional inhibition is greater than transcriptional activation. The sensitivity of human TFs to DNA methylation by methyl-SELEX and bisulfite-SELEX show that most C2H2 and GATA family members preferentially recognize the binding sites of DNA methylation. Moreover, DNA methylation in the binding region decreases the binding of MYB, bZIP, and HSF families to the recognition sequences [39].
Secondary metabolites are important in the defense mechanisms of plants against abiotic and biotic stresses, improving flower colors and fruit quality and attracting pollinators and seed dispersers. They are defensive metabolites and toxins, which fight invasive pathogens and herbivores and protect plants from abiotic stresses, such as UV stress. In addition, secondary metabolites are a source of natural flavors and medicines [40]. TFs are DNA binding proteins with sequence specificity. It interacts with the regulatory regions of the target gene (usually promoters) to activate or suppress the expression patterns and levels of structural genes, regulating the synthesis of plant secondary metabolites. Several families of TFs do not directly bind to the target gene but interact with other cofactors to form complexes to regulate the expression of the target gene [40]. The MYB TF family is one of the largest families of transcription factors that regulate flavonoid biosynthesis in plants. The R2R3-MYB gene SbMYB8 of Scutellaria baicalensis can bind to the GmMYB92 BS3 sequence in the SbCHS promoter region, thereby regulating flavonoid biosynthesis [41]. TT2 (MYB), TT8 (bHLH), and TTG1 (WDR) form a stable ternary complex that can bind to the BANYULS promoter to directly regulate the expression of BANYULS, which in turn promotes the anthocyanin biosynthesis in A. thaliana [42]. Camalexin levels are much lower in mutants of ANAC042 plants than in the wild type, and NAC042 regulates the camalexin biosynthetic genes P450s then induce phytoalexin biosynthesis in A. thaliana [30]. TCP1 binds to the DWF4 promoter and regulates DWF4 expression, which modulates brassinosteroid biosynthesis in A. thaliana [43]. Three AP2/ERF TFs are identified as positive regulators of the CitCHIL1 expression. CitERF32 and CitERF33 activate the transcription by directly binding to the CitCHIL1 promoter, while CitRAV1 forms a transcriptional complex with CitERF33, strongly enhancing the activation efficiency and flavonoid accumulation in Ougan (Citrus reticulata cv. Suavissima) [44]. G-BOX BINDING FACTOR 1 (GBF1) of the bZIP family binds to CATALASE2 (CAT2) and inhibits CAT2 expression during senescence [45]. GBF1 and MYC2 (bHLH) physically interact in the nucleus, bind to HYH promoter, and mutually inhibit each other through non-DNA binding bHLH–bZIP heterodimers [46].
The CHI is one of the key enzymes required for flavonoid biosynthesis, regulating flavonoid content accumulation. Sun et al. reported that the expressing OjCHI of Ophiorrhiza japonica in Arabidopsis tt5 mutant restores the phenotype of anthocyanins and flavonols, indicating its function as a Type Ⅰ CHI for flavonoid biosynthesis in vivo [47]. In vitro functional demonstrates that DcCHI1 of Dracaena cambodiana catalyzes the formation of naringenin from naringenin chalcone, while DcCHI4 lacks this catalytic activity. Moreover, overexpression of DcCHI1 or DcCHI4 enhances flavonoid production in D. cambodiana and tobacco [13]. CHIL does not possess the activity of intramolecular cyclization of naringenin chalcone. However, studies have shown that CHIL functions as a flavonoid enhancer in plants, such as Japanese morning glory [48], Humulus lupulus Linn. [49], and Arabidopsis [50]. In this study, the comprehensive analysis of the transcriptomic and metabolomic data showed that the expression of LpCHI and LpCHIL are significantly and positively correlated with the total content of flavonoids, kaempferol, apigenin, trilobatin, phlorizin, and phloretin (P < 0.01). Yang et al. measured the total content of flavonoids, trilobatin, and phlorizin of L. polystachyus leaves at different development stages. Total flavonoid content is highest in the early stage and decreases at the mature stage [4]. Comprehensive transcriptome and metabolome analysis of L. polystachyus leaves revealed that the expression of key genes in flavonoid biosynthesis pathways is consistent with the content accumulation pattern of the flavonoid, with high expression in young leaves and low expression in mature leaves [1]. In this study, the total flavonoids content is determined by UV-spectrophotometry, and the 6 flavonoids are determined by UPLC. The results show that the total flavonoid content is highly accumulated at the early stage and less accumulated at the mature stage. The expression of key genes in flavonoid biosynthesis pathways at different stages is examined by qRT-PCR, indicating a higher expression in young leaves than in mature leaves. These results of this study are in line with previous results.
The environment where plants grow can influence enzyme activities in vivo and regulate metabolic activities. The CO2 stress enhances phenolic accumulation in the root of Panax ginseng through induction of glucose-6-phosphate (G6PDH) activity [51]. Blue light-treated strawberries induce increased activities of four key enzymes in anthocyanin production: PAL, TAL (tyrosine ammonia-lyase), C4H, and 4CL [51]. In this study, the enzyme activities of PAL, C4H, 4CL, CHS, and CHI involved in flavonoid synthesis are measured. We found some differences in enzyme activities of different samples and different stages. However, there is no correlation between enzyme activity and flavonoid content. The differences in flavonoid metabolites in different types of LpCHI promoter DNA methylation samples are mainly caused by differences in the expression levels of genes involved in flavonoid metabolism in different samples.
5. Conclusion
DNA methylation at a specific site in the LpCHI promoter CpG island region affects the activity of TFs bind to the promoter, which then regulates LpCHI expression. In this study, DNA methylation at specific sites in the promoter region is found to have a more significant regulatory effect on gene expression than the DNA methylation ratio. Among 16 CpG-sites, 9 TFs binding to them are detected. DNA methylation of -1469 and -1380 cytosines lead to greater transcriptional activation than transcriptional inhibition. In addition, DNA methylation of -1797, -1783, -1735, and -1723 cytosines resulted in more significant transcriptional inhibition than transcriptional activation. Since different TFs differ in their sensitivities to DNA methylation, further experiments are needed to identify the TFs causing differences in LpCHI transcription. Under the same growth environment, the enzyme activities of PAL, C4H, 4CL, CHS, and CHI are different in various samples and leaves at different growth and development stages. However, these activities are not the key factors affecting the accumulation of flavonoids. The transcriptional differences of genes involved in flavonoid biosynthesis are the main factors regulating the accumulation of flavonoids.
Data availability
The raw RNA-seq data supporting the findings of this study have been deposited in the NCBI SRA database under accession number PRJNA771767.
CRediT authorship contribution statement
Limei Lin: designed the research, conducted most of the experiments, analyzed the transcriptome and metabolome, Formal analysis, Writing – original draft, analyzed the data and drafted the manuscript. Shuqing Wang: conducted most of the experiments, revised and verified the manuscript. Jie Zhang: conducted most of the experiments, Formal analysis, analyzed the transcriptome and metabolome, and. Xin Song: conducted most of the experiments, Formal analysis, analyzed the transcriptome and metabolome. Duoduo Zhang: conducted some of the experiments, and. Wenwen Cheng: conducted some of the experiments. Yuehong Long: designed the research, revised and verified the manuscript, and. Zhaobin Xing: designed the research, revised and verified the manuscript.
Declaration of competing interest
The authors report no declarations of interest.
Acknowledgments
This work was supported by the Natural Science Foundation of Hebei Province (H2020209033) and the Science and Technology Project of Hebei Education Department (ZD2019075).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2022.05.003.
Contributor Information
Yuehong Long, Email: longyh@ncst.edu.cn.
Zhaobin Xing, Email: xingzb@ncst.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
Fig. S2.
Fig. S3.
Fig. S4.
References
- 1.Li K., Liu K., Chen Y., Huang X.L., Liang W.H., Li B.C., et al. Comprehensive transcriptome and metabolome analysis of Lithocarpus polystachyus leaf revealed key genes in flavonoid biosynthesis pathways. J Am Soc Hortic Sci. 2021;1(aop):1–11. doi: 10.21273/JASHS05020-20. [DOI] [Google Scholar]
- 2.Shang A., Liu H., Luo M., Xia Y., Yang X., Li H., et al. Sweet tea (Lithocarpus polystachyus rehd.) as a new natural source of bioactive dihydrochalcones with multiple health benefits. Crit Rev Food Sci Nutr. 2020;8:1–18. doi: 10.1080/10408398.2020.1830363. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y., Li X., Zeng X., Huang S., Hou S., Lai X. Characterization of phenolic constituents in Lithocarpus polystachyus. Analytical Methods. 2014;6(5):1359–1363. doi: 10.1039/C3AY41288A. [DOI] [Google Scholar]
- 4.Yang J., Huang Y., Yang Z., Zhou C., Hu X. Identification and quantitative evaluation of major sweet ingredients in sweet tea (Lithocarpus polystachyus Rehd.) based upon location, harvesting time, leaf age. J Chem Soc Pak. 2018;40:158. 01. [Google Scholar]
- 5.Jia Z., Xie Y., Wu H., Wang Z., Li A., Li Z., et al. Phlorizin from sweet tea inhibits the progress of esophageal cancer by antagonizing the JAK2/STAT3 signaling pathway. Oncology reports. 2021;46(1):137. doi: 10.3892/or.2021.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W., Chen Shiqi, Fu H., Shu G., Tang H., Zhao X., et al. Hypoglycemic and hypolipidemic activities of phlorizin from Lithocarpus polystachyus Rehd in diabetes rats. Foos Sci Nutr. 2021;9(4):1989–1996. doi: 10.1002/fsn3.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X., Li X., Fang H., Guo F., Li F., Chen A., et al. Flavonoids as inducers of white adipose tissue browning and thermogenesis: signalling pathways and molecular triggers. Nutr Metab (Lond) 2019;16(1) doi: 10.1186/s12986-019-0370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toheg T., Watanabe M., Hoefgen R., Fernie A.R. The evolution of phenylpropanoid metabolism in the green lineage. Crit Rev Biochem Mol Biol. 2013;48(2) doi: 10.3109/10409238.2012.758083. [DOI] [PubMed] [Google Scholar]
- 9.Ngaki M.N., Louie G.V., Philippe R.N., Manning G., Pojer F., Bowman M.E., et al. Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature. 2012;485(7399):530–533. doi: 10.1038/nature11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin Y., Zhang X., Gao Z., Hu T., Liu Y. The Research Progress of chalcone isomerase (CHI) in plants. Mol Biotechnol. 2018;61(1):32–52. doi: 10.1007/s12033-018-0130-3. [DOI] [PubMed] [Google Scholar]
- 11.Lan X., Quan H., Xia X., Yin W., Zheng W. Molecular cloning and transgenic characterization of the genes encoding chalcone synthase and chalcone isomerase from the Tibetan herbal plant Mirabilis himalaica. Biotechnol Appl Biochem. 2016;63(3):419–426. doi: 10.1002/bab.1376. [DOI] [PubMed] [Google Scholar]
- 12.Liu X., Ahmad N., Yang L., Fu T., Kong J., Yao N., et al. Molecular cloning and functional characterization of chalcone isomerase from Carthamus tinctorius. AMB Express. 2019;9(1):132. doi: 10.1186/s13568-019-0854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J., Zhao W., Li R., Guo D., Li H., Wang Y., et al. Identification and characterization of chalcone isomerase genes involved in flavonoid production in Dracaena cambodiana. Front Plant Sci. 2021;12:616396. doi: 10.3389/fpls.2021.616396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni R., Zhu T., Zhang X., Wang P., Sun C., Qiao Y., et al. Identification and evolutionary analysis of chalcone isomerase-fold proteins in ferns. J Exp Bot. 2019;71(1):290–304. doi: 10.1093/jxb/erz425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zu Q., Qu Y., Ni Z., Zheng K., Chen Q., Chen Q. The chalcone isomerase family in cotton: whole-genome bioinformatic and expression analyses of the Gossypium barbadense L. response to fusarium wilt infection. Genes (Basel) 2019;10(12):1006. doi: 10.3390/genes10121006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dastmalchi M., Dhaubhadel S. Soybean chalcone isomerase: evolution of the fold, and the differential expression and localization of the gene family. Planta. 2015;241(2):507–523. doi: 10.1007/s00425-014-2200-5. [DOI] [PubMed] [Google Scholar]
- 17.Lin L., Long Y., Wang Z., Guo H., Cui M., Huang J., et al. Correlation between DNA methylation of the chalcone isomerase gene in Lithocarpus polystachyus Rehd. (Fagaceae) and phlorizin accumulation. Pak J Bot. 2021;53(4):1247–1252. doi: 10.30848/PJB2021-4(36. [DOI] [Google Scholar]
- 18.Bartels A., Han Qiang, Nair Pooja, Stacey Liam, Gaynier Hannah, Mosley Matthew, et al. Dynamic DNA methylation in plant growth and development. Int J Mol Sci. 2018;19(7):2144. doi: 10.3390/ijms19072144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curradi M., Izzo A., Badaracco G., Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol. 2002;22(9):3157–3173. doi: 10.1128/mcb.22.9.3157-3173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Zhu Y., Yuan P., Song S., Duan H. Response of wheat DREB transcription factor to osmotic stress based on DNA Methylation. Int J Mol Sci. 2021;22(14):7670. doi: 10.3390/ijms22147670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng J., Fu Z., Chen S., Damaris Rebecca Njeri, Wang K., Li T., et al. Proteomic and epigenetic analyses of lotus (Nelumbo nucifera) petals between red and white cultivars. Plant Cell Physiol. 2015;56(8):1546–1555. doi: 10.1093/pcp/pcv077. [DOI] [PubMed] [Google Scholar]
- 22.Han M., Yin J., Zhao Y., Sun X., Meng J., Zhou J., et al. How the color fades from Malus halliana flowers: transcriptome sequencing and DNA methylation analysis. Front Plant Sci. 2020;11 doi: 10.3389/fpls.2020.576054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao S., Ma W., Lyu X., Cao X., Yao Y. Melatonin may increase disease resistance and flavonoid biosynthesis through effects on DNA methylation and gene expression in grape berries. BMC Plant Biol. 2020;20(1):231. doi: 10.1186/s12870-020-02445-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W., Ning G., Mao J., Guo Z., Zhou Q., Chen B. Whole-genome DNA methylation patterns and complex associations with gene expression associated with anthocyanin biosynthesis in apple fruit skin. Planta. 2019;250(6):1833–1847. doi: 10.1007/s00425-019-03266-4. [DOI] [PubMed] [Google Scholar]
- 25.Chen W., Gong L., Guo Z., Wang W.S., Zhang H., Liu X., et al. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol Plant. 2013;6(6):1769–1780. doi: 10.1093/mp/sst080. [DOI] [PubMed] [Google Scholar]
- 26.Basdeki L., Hagidimitriou M. The role of DNA methylation in perennial plants. Notulae Scientia Biologicae. 2019;11(1):1–11. doi: 10.15835/nsb11110446. [DOI] [Google Scholar]
- 27.Dare A.P., Tomes S., McGhie T.K., van Klink J.W., Sandanayaka M., Hallett I.C., et al. Overexpression of chalcone isomerase in apple reduces phloridzin accumulation and increases susceptibility to herbivory by two-spotted mites. Plant J. 2020;103(1):293–307. doi: 10.1111/tpj.14729. [DOI] [PubMed] [Google Scholar]
- 28.Lin L.M., Long Y.H., Feng R.X., Yin F., Huang J., Xing Z.B. Cloning and bioinformatic analysis of chalcone isomerase gene in Lithocarpus polystachyus. Chinese Trad & Herb Drugs. 2017;48(24):5080–5084. doi: 10.7501/j.issn.0253-2670.2017.24.004. [DOI] [Google Scholar]
- 29.Iglesias-Fernández R., Barrero-Sicilia C., Carrillo-Barral N., Oñate-Sánchez L., Carbonero P. Arabidopsis thaliana bZIP44: a transcription factor affecting seed germination and expression of the mannanase-encoding gene AtMAN7. Plant J. 2013;74(5):767–780. doi: 10.1111/tpj.12162. [DOI] [PubMed] [Google Scholar]
- 30.Saga H., Ogawa T., Kai K., Suzuki H., Ogata Y., Sakurai N., et al. Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of Camalexin biosynthesis in Arabidopsis. Mol Plant Microbe Interact. 2012;25(5):684–696. doi: 10.1094/mpmi-09-11-0244. [DOI] [PubMed] [Google Scholar]
- 31.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 32.Hong J., Wang X., Mei C., Wang H., Zan L. DNA Methylation and transcription factors competitively regulate SIRT4 promoter activity in bovine adipocytes: roles of NRF1 and CMYB. DNA Cell Biol. 2019;38(1):63–75. doi: 10.1089/dna.2018.4454. [DOI] [PubMed] [Google Scholar]
- 33.Héberlé É., Bardet A.F. Sensitivity of transcription factors to DNA methylation. Essays Biochem. 2019;63(6):727–741. doi: 10.1042/EBC20190033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tate P.H., Bird A.P. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3(2):226–231. doi: 10.1016/0959-437X(93)90027-M. [DOI] [PubMed] [Google Scholar]
- 35.Prokhortchouk A., Hendrich B., Jørgensen H., Ruzov A., Wilm M., Georgiev G., et al. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15(13):1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rishi V., Bhattacharya P., Chatterjee R., Rozenberg J., Zhao J., Glass K., et al. CpG methylation of half-CRE sequences creates C/EBPα binding sites that activate some tissue-specific genes. Proc Natl Acad Sci U S A. 2010;107(47):20311–20316. doi: 10.1105/tpc.109.069203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaston K., Fried M. CpG methylation has differential effects on the binding of YY1 and ETS proteins to the bi-directional promoter of the Surf-1 and Surf-2 genes. Nucleic Acids Res. 1995;23(6):901–909. doi: 10.1093/nar/23.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y., Liu L., Zhao D., Tao J. Age-associated methylation change of CHI promoter in herbaceous peony (Paeonia lactiflora Pall) Biosci Rep. 2018;38(5) doi: 10.1042/BSR20180482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356(6337) doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang C., Fang X., Wu X., Mao Y., Wang L., Chen X. Transcriptional regulation of plant secondary metabolism. J Integr Plant Biol. 2012;54(10):703–712. doi: 10.1111/j.1744-7909.2012.01161.x. [DOI] [PubMed] [Google Scholar]
- 41.Yuan Y., Qi L., Yang J., Wu C., Liu Y., Huang L. A Scutellaria baicalensis R2R3-MYB gene, SbMYB8, regulates flavonoid biosynthesis and improves drought stress tolerance in transgenic tobacco. Plant Cell Tiss Organ Cult. 2015;120(3) doi: 10.1007/s11240-014-0650-x. 973-973. [DOI] [Google Scholar]
- 42.Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39(3):366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 43.Guo Z., Fujioka S., Blancaflor E.B., Miao S., Gou X., Li J. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. The Plant Cell. 2010;22(4):1161–1173. doi: 10.1105/tpc.109.069203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao C., Liu X., Gong Q., Cao J., Shen W., Yin X., et al. Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol J. 2021;19(4):671–688. doi: 10.1111/pbi.13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giri M.K., Singh N., Banday Z.Z., Singh V., Ram H., Singh D., et al. GBF1 differentially regulates CAT2 and PAD4 transcription to promote pathogen defense in Arabidopsis thaliana. Plant J. 2017;91(5):802–815. doi: 10.1111/tpj.13608. [DOI] [PubMed] [Google Scholar]
- 46.Maurya J.P., Sethi V., Gangappa S.N., Gupta N., Chattopadhyay S. Interaction of MYC2 and GBF1 results in functional antagonism in blue light-mediated Arabidopsis seedling development. Plant J. 2015;83(3):439–450. doi: 10.1111/tpj.12899. [DOI] [PubMed] [Google Scholar]
- 47.Sun W., Shen H., Xu H., Tang X., Tang M., Ju Z., et al. Chalcone isomerase a key enzyme for anthocyanin biosynthesis in Ophiorrhiza japonica. Front Plant Sci. 2019;10(865) doi: 10.3389/fpls.2019.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morita Y., Takagi K., Fukuchi-Mizutani M., Ishiguro K., Tanaka Y., Nitasaka E., et al. A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. Plant J. 2014;78(2):294–304. doi: 10.1111/tpj.12469. [DOI] [PubMed] [Google Scholar]
- 49.Ban Z., Qin H., Mitchell A.J., Liu B., Zhang F., Weng J.-K., et al. Noncatalytic chalcone isomerase-fold proteins in Humulus lupulus are auxiliary components in prenylated flavonoid biosynthesis. Proc Natl Acad Sci U S A. 2018;115(22):E5223–E5232. doi: 10.1073/pnas.1802223115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang W., Yin Q., Wu R., Zheng G., Liu J., Richard A., Dixon, et al. Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. J Exp Bot. 2015;66(22):7165–7179. doi: 10.1093/jxb/erv413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu F., Cao S., Shi L., Chen W., Su X., Yang Z. Blue light irradiation affects anthocyanin content and enzyme activities involved in postharvest strawberry fruit. J Agric Food Chem. 2014;62(20):4778–4783. doi: 10.1021/jf501120u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-seq data supporting the findings of this study have been deposited in the NCBI SRA database under accession number PRJNA771767.