Abstract
Purpose of review
This article reviews the latest proteomic research on uveal melanoma (UM).
Recent Findings
Proteomic analysis of UM cell lines and tissue specimens has improved our understanding of the pathophysiology of UM and helped identify potential prognostic biomarkers. Circulating proteins in patient serum may aid in the surveillance of metastatic disease. The proteomes of aqueous and vitreous biopsy specimens may provide safer biomarkers for metastatic risk and candidate therapeutic targets in UM. Proteomic analysis has the potential to benefit patient outcomes by improving diagnosis, prognostication, surveillance, and treatment of UM.
Summary
These recent findings demonstrate that proteomic analysis is an important area of research to better understand the pathophysiology of UM and improve the personalized management of our patients.
Keywords: Proteomics, Uveal melanoma, Biomarker, Aqueous humor, Vitreous, Liquid biopsy
Introduction
Uveal melanoma (UM) remains the most common primary intraocular malignancy in adults [1]. Patients with UM continue to have a poor prognosis with an overall mortality of approximately 50% at fifteen years due to late diagnosis, limited systemic surveillance sensitivity, and suboptimal treatments for metastatic disease [2]. Recent advances in genetic analysis have led to the development of gene expression profiling which classifies patient prognosis based on RNA sequencing from direct intraocular tumor biopsy specimens [3, 4]. Genetic analysis has limitations in understanding tumor phenotype due to the inability to predict protein expression and post-translational modifications. Therefore, proteomic analysis provides a complementary technique to direct genetic analysis and has potential applications to improve the prognosis of patients with UM.
Proteomics is the large-scale study of the protein expression profile of an organism, tissue, cell, or fluid where thousands of proteins can be measured simultaneously. In the setting of UM, proteomics has the potential to benefit patient outcomes by improving diagnosis, prognostication, surveillance, and treatment (Figure 1). A better understanding of tumor pathophysiology and mechanisms of metastasis can also help identify therapeutic targets. Biomarkers for diagnosis, prognosis, metastasis, and treatment susceptibility can aid in the personalized management of patients with UM.
Figure 1. Tissue protein expression and proteomic analysis as a tool for discovery.
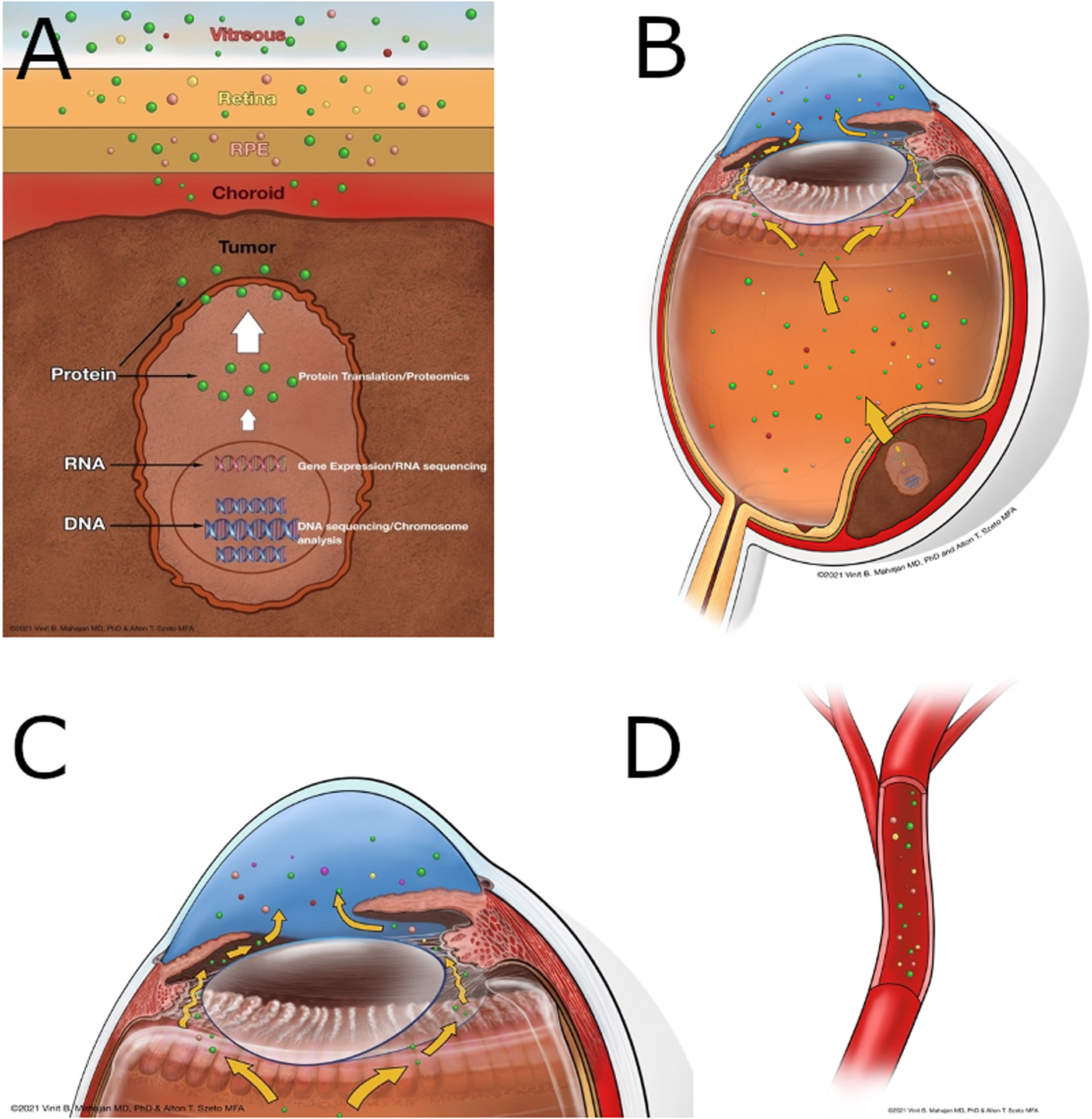
A. Proteins from tumor cells migrate through the choroid, retinal pigmented epithelium, and retina. B. A subset of these proteins can be found in the vitreous due to intrinsic proteases. C. Proteins in the vitreous spread to the aqueous humor in the anterior chamber. D. Tumor associated proteins can also be found in the blood stream.
Proteomics in cancer research
Proteomic analysis is an important tool for understanding cancer development and progression. Characterizing the protein profile of cancer tissue aids in mapping the molecular mechanisms that lead to carcinogenesis and metastatic transformation. These molecular mechanisms include protein modification and protein-protein interaction that are critical to tumor development. Understanding the function of proteins that make up these pathways is important to the discovery of biomarkers and identifying novel therapeutic targets. Proteomic analysis measures expression levels that can vary by disease severity and response to therapy. Current trends in cancer proteomics includes the development of protein databases consisting of comprehensive analysis of various cancer types. Numerous prognostic biomarkers and therapeutic targets have been identified using proteomics and represent a promising area to improve patient outcomes.
Utility of proteomics in ocular disease
The variety of tissue structures that form the eye make proteomic analysis a particular robust area of research in ophthalmology [5]. Evaluating the protein profile of tears is a non-invasive technique to study ocular surface diseases. Proteomic analysis of intraocular fluids has led to a better understanding of ocular diseases. The aqueous humor has been well studied in the field of glaucoma to learn about the pathologic changes at the trabecular meshwork. The vitreous is an attractive subject of study given its relatively less complex protein profile in healthy eyes [6]. The protein content of the cornea, lens, and retina have been well characterized in healthy subjects which has paved the way to identifying biomarkers and therapeutic targets in the diverse pathologies that affect these tissues [7–9].
Proteomics of UM cells
The first study of proteomics in UM cells was conducted in a primary UM cell culture (UM-A) from a 22-year-old patient who underwent enucleation for a spindle type UM with a basal diameter of 19.1 mm and thickness of 12.41 mm (Table 1) [10]. From these cells, Pardo et al. identified 683 proteins of which 96% had not been previously reported in UM and 18% were known to be related to cancer processes [11]. Many of the proteins identified had been studied in other cancers and play roles in cell proliferation, invasion, metastasis, oncogenesis, and drug resistance including MUC18 and HMG-1. MUC18 is associated with increased metastatic potential in prostate and bladder cancer while HMG-1 regulates transcription of genes involved in cell proliferation and metastasis [12, 13]. In a subsequent study, Pardo et al. compared the proteomes of cultured cells and multiple cell lines with different invasive potential [14]. They found that MUC18, HMG-1, and DJ-1 correlated with cell invasiveness. DJ-1 has been implicated in tumorigenesis of breast, non-small cell lung, and prostate cancer [15–17]. Subsequently, DJ-1 has been shown to be elevated in the serum of patients with high-risk choroidal nevi, UM, and metastatic UM [18, 19].
Table 1.
Proteomic studies on uveal melanoma specimens.
| Year | Study | Platform | Tissue | UM Sample Size | Differentially expressed proteins | Control group |
|---|---|---|---|---|---|---|
| 2004 | Pardo et al. | Immunoblotting (9 proteins) | Primary Tumor Cell Culture | 1 | cyclin D1, cyclin E, p16, p27 | “high level” but no control documented |
| 2006 | Pardo et al. | 2D-DIGE, LC-MS/MS | Primary Tumor Cell Culture | 2 | Ezrin, HSP90, S100, PARK7, VIM, Cytokeratins 8 and 18 | Invasive phenotype biomarker, no controls |
| 2010 | Coupland et al. | 2D-Gel Electrophoresis, Multiplex MPLA | Primary Tumors | 48 | HSP27, vimentin, PDHB | Chromosome 3 status, no controls |
| 2013 | Yan et al. | SILAC, 2D-LC-MS/MS | Primary Tumor Cell Culture | 1 | Many | Before and after irradiating cells |
| 2013 | Wang et al. | SILAC, 2D-LC-MS/MS | Primary Tumor Cell Culture | 1 | Many | Before and after irradiating cells |
| 2006 | Zuidervaart et al. | 2D-DIGE, LC-MS/MS | Primary Tumor and Metastasis Cell Culture | 1 | HSP27, Galectin-1, Cofilin, GST, β-hexosaminidase | Primary vs metastasis |
| 2020 | Slater et al. | ELISA (17 proteins) | Primary Tumor and Metastasis Cell Secretomes | 2 | IL-1B, IL-2, IL-6, IL-8, IL-10, IL12p70, IL-13 | Before and after quininib treatment |
| 2007 | Pardo et al. | 2D-DIGE, LC-MS/MS | Cell Culture Secretomes | 1 | Cathepsin D, Syntenin, gp100, HGFR, IGF-2R | Normal melanocytes |
| 2016 | Angi et al. | nano-LC-MS/MS | Cell Culture Secretomes | 14 | PDIA3, VIM/HEXA, SELENBP1, ERP29, TPI1, PARK7, EIF2S1 and RPSA | Normal melanocytes |
| 2003 | Missotten et al. | Immunohistochemistry (4 proteins) | Primary Tumors | 38 | Heat Shock Proteins; Did not compare to controls | Enucleated normal eyes or eyes without retinal problems |
| 2012 | Jmor et al. | Immunohistochemistry (1 protein) | Primary Tumors | 99 | HSP-27 | Chromosome 3 status, no controls |
| 2021 | Jang et al. | LC-MS/MS | Primary Tumor | 100 | CDH1, FYN, HLA-DPA1, HLA-DPB1, HLA-DQB1, HMGB1, LYN, PPP2CA, PPP2R1A, PPP2R5C, and PTPN6 | Primary vs metastasis |
| 2012 | Linge, et al. | 2D-DIGE, LC-MS/MS | Primary Tumors | 25 | FABP3, TPI1, PDIA3, VIM, SELENBP1, ENO1, CAPZA1, ERP29, PARK7 | Primary vs metastasis |
| 2015 | Crabb, et al. | iTRAQ | Primary Tumors | 15 | Collagen alpha-3 (VI), HSP27, FABP3, β-hexosaminidase | Primary vs metastasis |
Abbreviations: LC-MS/MS = liquid chromatography-tandem mass spectrometry; 2D-DIGE = two-dimensional difference gel electrophoresis; iTRAQ = isobaric tags for relative and absolute quantitation.
Zuidervaart et al. compared the proteome of a primary UM cell line (Mel 270) to two UM liver metastasis derived cell lines (Omm 1.3 and Omm 1.5), all from the same patient. They identified 24 proteins that were differently expressed in the primary UM cell line compared to the two metastatic cell lines. These proteins were variably involved in motility, adhesion, invasion, apoptosis, and cellular defense. One of these proteins, HSP-27, is expressed when cells are exposed to heat or stress and has been implicated as a prognostic biomarker in other cancers [20, 21]. HSP-27 levels have been shown to be expressed in a high percentage of UM cells in enucleated eyes [22]. Coupland et al. studied HSP-27 in three UM tissue specimens with disomy 3 and four specimens with monosomy 3 [23]. HSP-27 was found to have negative correlations with mitotic rate and monosomy 3 suggesting this protein as a possible prognostic biomarker in patients with UM. The same group went on to validate HSP-27 in a larger sample of 99 UM tumor specimens [24]. HSP-27 was measured using immunohistochemical staining and positively correlated with both disomy 3 and patient survival. Cellular proteins may serve as a surrogate for genetic analysis in UM prognostication.
Proteomics of UM secretome
A secretome describes the milieu of proteins secreted or shed from cells. These proteins are often important in intercellular communication and can contribute to cancer development and progression [25]. The first study of the UM secretome evaluated cell cultures and cell lines to identify potential targets for indirect tissue diagnosis (Table 1) [26]. More than half of the secretome proteins identified in the UM-A cell culture were not present in the corresponding cell proteosome. Cancer-related proteins of interest included Syntenin, Cathepsin D, and gp100 with the latter two also identified in the serum of patients with UM. Angi et al. compared the secretome of primary UM cell lines stratified based on chromosome 3 status and normal choroidal melanocytes (NCM) [27]. The secretomes in NCM and UM cells differed but there was no significant difference between the monosomy 3 and disomy 3 cell lines. One proposed reason for this finding was the disomy 3 cells were derived from surgical samples of large tumors which is also a known risk factor for metastasis. Proteins elevated in the UM secretome compared to NCM included MIA and GDF15. Both proteins have been shown to be elevated in the sera of patients with metastatic UM compared to locally controlled UM [28, 29]. MIA is an attachment regulating protein thought to promote detachment of melanoma cells from the extracellular matrix. MIA has proved particularly promising as a serum tumor marker in UM [30–32].
Proteomics of serum in UM
Biomarkers for the early detection of metastasis in patients with UM are important due to the high rate of liver metastasis around 30% at five years (Table 2) [33, 34]. Serological biomarkers are particularly promising due to the accessibility of patient serum for frequent screening and surveillance. Liver function tests were recommended by COMS as serological biomarkers for metastasis but have poor sensitivity and specificity for liver metastasis compared to more costly body imaging techniques [34–37]. Potential serum biomarkers, including MIA, OPN and S100-β, were tested in patients with metastatic UM compared to locally controlled cases with a minimum disease-free period of ten years [31, 38–41]. All three proteins correlated with the presence of liver metastasis and together had a high sensitivity compared to liver function tests. Haritoglou et al. confirmed these findings using the serum levels of MIA and OPN in UM patients with and without metastatic disease [42]. They did not find a correlation between MIA and OPN levels with primary tumor size suggesting an inability to diagnose or stage primary UM using these proteins alone.
Table 2.
Proteomic studies on serum from patients with uveal melanoma.
| Year | Study | Platform | UM Sample Size | Differentially expressed proteins | Control group |
|---|---|---|---|---|---|
| 2012 | Suesskind et al. | ELISA (1 protein) | 188 | GDF-15 | Healthy controls |
| 2002 | Schaller et al. | ELISA (1 protein) | 137 | MIA | Primary vs metastasis |
| 2005 | Reiniger et al. | ELISA (1 protein) | 305 | MIA | Primary vs metastasis |
| 2006 | Reiniger et al. | ELISA (1 protein) | 27 | Osteopontin | Healthy controls |
| 2009 | Haritoglou et al. | ELISA (2 protein) | 32 | Osteopontin, MIA | Primary vs metastasis |
| 2017 | Shi, et al. | Magnetic bead capture, MALDI-TOF MS | 18 | Fibrinogen | Healthy controls |
| 2007 | Song, et al. | Multiplex ELISA (7 proteins) | 48 | Osteopontin, MIA, HSP7 | Healthy controls |
Abbreviations: MALDI-TOF MS= matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
The diagnosis of UM is primarily based on clinical examination and imaging. However, diagnostic biomarkers may be useful for indeterminate lesions, screening patients with tumor predisposition syndromes, or the rare diagnostic dilemma. Song et al. recently tested seven biomarkers, including OPN, MIA, and HSP-27, in their ability to detect UM [43]. A two-marker panel of OPN and HSP-27 was significantly different between patients with UM compared to controls. The two-marker panel that best discriminated between metastatic UM and locally controlled patients was MIA and MIC-1 although this panel was not statistically significant. Additionally, the OPN and HSP-27 panel correlated with tumor histological type, but the study was under powered to validate this finding. Similarly, Shi et al. developed diagnostic panels from 49 serum proteins that were significantly different between patients with UM and healthy controls [44]. Many of these proteins demonstrated high sensitivity and specificity when paired in two-marker panels. Interestingly, most of these serum proteins normalized to healthy control levels after primary tumor removal (local resection or enucleation).
Proteomics of UM tissue
Given the utility of gene expression profiling and PRAME status, proteomic analysis of UM tissue is a promising area of study. The first proteomic studies of UM tissue were focused on HSP-27 as a prognostic biomarker (Table 1) [23, 24]. Linge et al. went on to compare the proteomes of primary UM tumors in nine metastatic disease cases to 16 locally controlled cases with a minimum disease-free period of seven years [45]. They identified 14 proteins that were differently expressed between the two groups. Two of these proteins, FABP3 and TPI1, were confirmed to be elevated in the metastatic group using immunohistochemistry. In the same study, a primary UM cell line (92.1) was used to show that siRNA knockdown of these two proteins led to decreased cell motility and invasion. In a follow up study of the same tissue specimens, liquid chromatography-mass spectrometry was used to identify 50 proteins that were differently expressed including FAP3, TPI1, HSP-27, and DJ-1 [46].
Crabb et al. also identified FAP3 and HSP-27 using iTRAQ (isobaric tags for relative and absolute quantitation) technology in a training set of ten primary UM tumors with five metastatic and five locally controlled cases [47]. Conversely, TPI1 and DJ-1 were found to be elevated in all specimens regardless of metastatic status. HSP-27 was one of only two proteins, along with collagen alpha-3(VI), that correctly classified the metastatic status in the independent test set of five UM specimens. Five of the proteins evaluated in this study are related products of the 12 genes analyzed in gene expression profiling [3]. One of those proteins, EIF4A1, was detected in nine of fifteen total specimens, and only elevated in the metastatic cases. The other four proteins were detected in only a small fraction of the specimens evaluated.
In a follow up study by the same group, 100 primary UM tumors underwent iTRAQ analysis including 53 metastatic and 47 non-metastatic cases [48]. Of the 3935 proteins quantified, 402 had differences in expression between the two groups. Pathway analysis of these 402 proteins revealed high representation of immune-related proteins in the metastatic cases and metabolic proteins in the non-metastatic cases. Based on the proteomic data, the authors proposed two candidates for immunotherapy, CDH1 and HLA-DPA1. CDH1 is one of the 12 genes analyzed in gene expression profiling and was detected in all 100 specimens. CDH1 showed no difference between the metastatic and non-metastatic cases but was significantly elevated in the metastatic specimens compared to same-eye choroid controls. HLA-DPA1 was found in 70% of UM specimens and elevated in the metastatic group compared to both the choroid controls and non-metastatic specimens.
Proteomics of aqueous humor
Cytokines in aqueous humor (AH) are potential diagnostic biomarkers and a useful tool to better understand the pathophysiology of UM. In 2003, Missotten et al. published the first proteomic study in UM human tissue (Table 3) [49]. They analyzed AH specimens from 24 patients with UM compared to 24 control eyes undergoing routine cataract surgery. Using mass spectrometry, they identified two unknown proteins that could distinguish eyes with UM with a sensitivity of 89%. The assay was unable to detect adequate protein content in five of the UM patients and ten of the controls. Despite this limitation, the study provided proof of concept for proteomics to aid in the molecular characterization of UM.
Table 3.
Proteomic studies on aqueous humor and vitreous in eyes with uveal melanoma.
| Year | Study | Platform | Tissue | UM Sample Size | Differentially expressed proteins | Control group |
|---|---|---|---|---|---|---|
| 2020 | Chen et al. | Multiplex ELISA (9 proteins) | AH | 18 | VEGF-A, PLGF | Before and after brachytherapy |
| 2012 | Lee et al. | Multiplex ELISA (12 proteins) | AH | 20 | IL-6, IL-8, IFN-g, MCP-1, IL-2, IL-10, TNF-a | Before and after brachytherapy |
| 2003 | Missotten et al. | SELDI-TOF MS | AH | 24 | Two proteins (4543.43 and 6853.30 kDa) | Age matched healthy controls |
| 2019 | Wierenga et al. | Multiplex ELISA (92 proteins) | AH | 108 | Galectin-9, TRAIL, Fas-L, ADA, CD244, CD40, MCP-3, PD-L1 | Prognostic biomarkers, no controls |
| 2017 | Usui et al. | Cytometric bead array (35 proteins) | AH | 13 | Angiogenin, IL-8, MCP-1 | Benign pigmented tumors |
| 2010 | Ly et al. | Multiplex bead array (15 proteins) | AH | 37 | IL-6, MCP-1, MIF, bFGF, RANTES, GM-CSF, VEGF, ICAM-11, VCAM-1, IP-10 | CEIOL healthy controls |
| 2018 | Cheng et al. | Multiplex bead array (10 proteins) | AH | 38 | Primary Tumor Cell Culture | CEIOL healthy controls |
| 2020 | Midena et al. | Multiplex ELISA (8 proteins) | AH | 35 | IL-6, IL-8, RANTES, EGF, bFGF, MIF, MCP | CEIOL healthy controls |
| 2021 | Velez, et al. | Multiplex ELISA (1,000 proteins) | Vitreous and plasma | 19 | HGF, HGFR/c-Met, SCFR/c-Kit, Autotaxin, Arginase-1, and KLK7 | GEP/PRAME status, no controls |
| 2012 | Dunavoelgyi et al. | Multiplex bead array | Vitreous and AH | 34 | Flt-3 ligand, IL-1a, IL-6, IL-7 IL-8, (IFN-g IP-10), MCP-1, MIP-1a, PDGF-AA, VEGF, RANTES | Vitreomacular traction |
| 2012 | Nagarkatti-Gude et al. | Multiplex bead array (27 proteins) | Vitreous | 33 | IL-1ra, IL-6, IL-8, IP-10, MCP-1, MIP-1a, MIP-1b, TNF-a, and RANTES | Healthy eyes |
Abbreviations: SELDI-TOF MS: surface-enhanced laser desorption/ionization time-of-flight mass spectrometry
More recently, Usui et al. compared the AH of 13 eyes with UM to 13 eyes with benign intraocular tumors undergoing cataract surgery or local resection [50]. They found elevated levels of angiogenin, IL-8, and MCP-1 in the UM group compared to the benign tumor group. IL-8 and MCP-1 are pro-inflammatory molecules involved in the recruitment of tumor associated macrophages [51, 52]. These two proteins were confirmed to be highly expressed in the AH of eyes with UM compared to eyes undergoing routine cataract surgery by other groups [53–56]. IL-8 was also shown to be acutely elevated after brachytherapy with adjunct transpupillary thermotherapy [55]. IL-8 plays a role in direct tumor angiogenesis and stimulates expression of VEGF which has also been shown to increase in AH after brachytherapy [57–59]. Further studies are necessary to better understand the relationship between inflammation and angiogenesis in UM.
Ly et al. evaluated 15 cytokines as potential prognostic biomarkers in AH specimens from 37 enucleated eyes with UM and 37 control eyes undergoing routine cataract surgery. They found elevated expression in almost all the fifteen cytokines evaluated in the UM eyes compared to control eyes. However, correlation with the clinical and histopathologic parameters evaluated were minimal. In a follow up study, AH from 90 eyes with UM were analyzed using a proximity extension assay (PEA) measuring 92 pre-selected proteins in 1 μL of each specimen [60]. The cytokine profiles were used to separate the specimens into three clusters. These clusters were found to correlate with ciliary body involvement, monosomy 3, and gain of 8q. This finding led the group to study the inflammatory phenotype further by measuring the soluble form of cell-bound HLA Class I which was found in 19 of 108 AH specimens [61]. Of these 19 patients, the level of HLA expression correlated with tumor size, monosomy 3, gain of 8q, and loss of BAP1. These studies provide evidence that proteomic analysis of AH in patients with UM correlates with clinical and genetic parameters and may provide complementary prognostic information.
Proteomics of vitreous
Proteomic analysis of the vitreous can detect biomarkers and therapeutic protein targets for adjacent tissue disease (Table 3) [5]. Many of the same proteins increased in AH of patients with UM have been shown to be increased in the vitreous including IL-8 and MCP-1 [56, 62]. These two proteins also correlate with regulatory T-cell infiltration while IL-6 expression level correlates with macrophage infiltration [62]. These findings suggest that proteins in the vitreous can serve as a surrogate biomarker for inflammatory changes associated with UM.
Transvitreal tumor sampling for genetic analysis provides an opportunity to study vitreous biopsy in patients with UM. Vitreous biopsy is less invasive than direct tumor biopsy and may provide safer prognostic biomarkers. Our group performed a proteomic analysis of vitreous biopsies in search of candidate biomarkers to serve as surrogates for tumor genes and potential therapeutic targets [63]. We identified multiple proteins that correlated with gene expression patterns for UM metastatic risk, including HGF, HGFR, Autotaxin, and SCFR, that were then prospectively validated in an independent cohort of patients. We also identified SCFR amongst the differentially expressed proteins as a candidate therapeutic target by the FDA-approved drug imatinib [64]. This finding demonstrates that proteomic analysis of the vitreous can identify candidate therapeutic targets.
Conclusion
Proteomic analysis in UM can be used to better understand the tumor microenvironment and the pathophysiology of tumor metastasis. Proteins in aqueous, vitreous, and tumor biopsies may serve as diagnostic and prognostic biomarkers that are substitutes for or complementary to current prognostic genetic analysis. These biopsies can also be used to identify candidate therapeutic targets for adjuvant or metastatic therapies. Continued proteomic research is needed to improve the prognosis for patients with UM.
Key Points.
Proteomic analysis in UM may be used to better understand the tumor microenvironment and the pathophysiology of tumor metastasis.
Proteins in aqueous, vitreous, and tumor biopsies may serve as diagnostic and prognostic biomarkers that are substitutes or complementary to current prognostic genetic analysis.
Biopsy of UM may be used to identify candidate therapeutic targets for adjuvant or metastatic therapies.
Acknowledgements
The authors thank Gabriel Velez, PhD and Jennifer Vu, BS for their assistance with this manuscript.
Financial support and sponsorship
VBM and PM are supported by a NIH grant P30EY026877, The Cancer League Research Grant, and Research to Prevent Blindness (RPB), New York, NY. PM is supported by a Retina Research Foundation/Macula Society Research Grant. VBM is supported by NIH grants [R01EY026682, R01EY024665, R01EY025225, R01EY024698, and R01EY03195201], Stanford ChEM-H IMA, the Stanford Center for Optic Disc Drusen.
Footnotes
Conflicts of Interest
None
References
Papers of particular interest, published within the annual period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Singh AD, Turell ME, Topham AK. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118(9):1881–5. [DOI] [PubMed] [Google Scholar]
- 2.Kaliki S, Shields CL, Shields JA. Uveal melanoma: estimating prognosis. Indian J Ophthalmol. 2015;63(2):93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12(4):461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onken MD, Worley LA, Char DH, et al. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119(8):1596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velez G, Tang PH, Cabral T, et al. Personalized Proteomics for Precision Health: Identifying Biomarkers of Vitreoretinal Disease. Transl Vis Sci Technol. 2018;7(5):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velez G, Mahajan VB. Molecular Surgery: Proteomics of a Rare Genetic Disease Gives Insight into Common Causes of Blindness. iScience. 2020;23(11):101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velez G, Machlab DA, Tang PH, et al. Proteomic analysis of the human retina reveals region-specific susceptibilities to metabolic- and oxidative stress-related diseases. PLoS One. 2018;13(2):e0193250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C, Liu P, Wang N, et al. Comparison of two tandem mass spectrometry-based methods for analyzing the proteome of healthy human lens fibers. Mol Vis. 2007;13:1873–7. [PubMed] [Google Scholar]
- 9.Karring H, Thogersen IB, Klintworth GK, et al. A dataset of human cornea proteins identified by Peptide mass fingerprinting and tandem mass spectrometry. Mol Cell Proteomics. 2005;4(9):1406–8. [DOI] [PubMed] [Google Scholar]
- 10.Pardo M, Pineiro A, de la Fuente M, et al. Abnormal cell cycle regulation in primary human uveal melanoma cultures. J Cell Biochem. 2004;93(4):708–20. [DOI] [PubMed] [Google Scholar]
- 11.Pardo M, Garcia A, Thomas B, et al. Proteome analysis of a human uveal melanoma primary cell culture by 2-DE and MS. Proteomics. 2005;5(18):4980–93. [DOI] [PubMed] [Google Scholar]
- 12.Wu GJ, Wu MW, Wang SW, et al. Isolation and characterization of the major form of human MUC18 cDNA gene and correlation of MUC18 over-expression in prostate cancer cell lines and tissues with malignant progression. Gene. 2001;279(1):17–31. [DOI] [PubMed] [Google Scholar]
- 13.Reeves R, Edberg DD, Li Y. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol Cell Biol. 2001;21(2):575–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardo M, Garcia A, Thomas B, et al. The characterization of the invasion phenotype of uveal melanoma tumour cells shows the presence of MUC18 and HMG-1 metastasis markers and leads to the identification of DJ-1 as a potential serum biomarker. Int J Cancer. 2006;119(5):1014–22. [DOI] [PubMed] [Google Scholar]
- 15.Le Naour F, Misek DE, Krause MC, et al. Proteomics-based identification of RS/DJ-1 as a novel circulating tumor antigen in breast cancer. Clin Cancer Res. 2001;7(11):3328–35. [PubMed] [Google Scholar]
- 16.MacKeigan JP, Clements CM, Lich JD, et al. Proteomic profiling drug-induced apoptosis in non-small cell lung carcinoma: identification of RS/DJ-1 and RhoGDIalpha. Cancer Res. 2003;63(20):6928–34. [PubMed] [Google Scholar]
- 17.Hod Y Differential control of apoptosis by DJ-1 in prostate benign and cancer cells. J Cell Biochem. 2004;92(6):1221–33. [DOI] [PubMed] [Google Scholar]
- 18.Bande MF, Santiago M, Blanco MJ, et al. Serum DJ-1/PARK 7 is a potential biomarker of choroidal nevi transformation. Invest Ophthalmol Vis Sci. 2012;53(1):62–7. [DOI] [PubMed] [Google Scholar]
- 19.Chen LL, Tian JJ, Su L, et al. DJ-1: a promising marker in metastatic uveal melanoma. J Cancer Res Clin Oncol. 2015;141(2):315–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisler JP, Tammela JE, Manahan KJ, et al. HSP27 in patients with ovarian carcinoma: still an independent prognostic indicator at 60 months follow-up. Eur J Gynaecol Oncol. 2004;25(2):165–8. [PubMed] [Google Scholar]
- 21.Malusecka E, Krzyzowska-Gruca S, Gawrychowski J, et al. Stress proteins HSP27 and HSP70i predict survival in non-small cell lung carcinoma. Anticancer Res. 2008;28(1B):501–6. [PubMed] [Google Scholar]
- 22.Missotten GS, Journee-de Korver JG, de Wolff-Rouendaal D, et al. Heat shock protein expression in the eye and in uveal melanoma. Invest Ophthalmol Vis Sci. 2003;44(7):3059–65. [DOI] [PubMed] [Google Scholar]
- 23.Coupland SE, Vorum H, Mandal N, et al. Proteomics of uveal melanomas suggests HSP-27 as a possible surrogate marker of chromosome 3 loss. Invest Ophthalmol Vis Sci. 2010;51(1):12–20. [DOI] [PubMed] [Google Scholar]
- 24.Jmor F, Kalirai H, Taktak A, et al. HSP-27 protein expression in uveal melanoma: correlation with predicted survival. Acta Ophthalmol. 2012;90(6):534–9. [DOI] [PubMed] [Google Scholar]
- 25.Caccia D, Zanetti Domingues L, Micciche F, et al. Secretome compartment is a valuable source of biomarkers for cancer-relevant pathways. J Proteome Res. 2011;10(9):4196–207. [DOI] [PubMed] [Google Scholar]
- 26.Pardo M, Garcia A, Antrobus R, et al. Biomarker discovery from uveal melanoma secretomes: identification of gp100 and cathepsin D in patient serum. J Proteome Res. 2007;6(7):2802–11. [DOI] [PubMed] [Google Scholar]
- 27.Angi M, Kalirai H, Prendergast S, et al. In-depth proteomic profiling of the uveal melanoma secretome. Oncotarget. 2016;7(31):49623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaller UC, Bosserhoff AK, Neubauer AS, et al. Melanoma inhibitory activity: a novel serum marker for uveal melanoma. Melanoma Res. 2002;12(6):593–9. [DOI] [PubMed] [Google Scholar]
- 29.Suesskind D, Schatz A, Schnichels S, et al. GDF-15: a novel serum marker for metastases in uveal melanoma patients. Graefes Arch Clin Exp Ophthalmol. 2012;250(6):887–95. [DOI] [PubMed] [Google Scholar]
- 30.Reiniger IW, Schaller UC, Haritoglou C, et al. “Melanoma inhibitory activity” (MIA): a promising serological tumour marker in metastatic uveal melanoma. Graefes Arch Clin Exp Ophthalmol. 2005;243(11):1161–6. [DOI] [PubMed] [Google Scholar]
- 31.Barak V, Frenkel S, Kalickman I, et al. Serum markers to detect metastatic uveal melanoma. Anticancer Res. 2007;27(4A):1897–900. [PMC free article] [PubMed] [Google Scholar]
- 32.Klingenstein A, Haritoglou I, Schaumberger MM, et al. Receiver operating characteristic analysis: calculation for the marker ‘melanoma inhibitory activity’ in metastatic uveal melanoma patients. Melanoma Res. 2011;21(4):352–6. [DOI] [PubMed] [Google Scholar]
- 33.Kujala E, Makitie T, Kivela T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci. 2003;44(11):4651–9. [DOI] [PubMed] [Google Scholar]
- 34.Diener-West M, Reynolds SM, Agugliaro DJ, et al. Screening for metastasis from choroidal melanoma: the Collaborative Ocular Melanoma Study Group Report 23. J Clin Oncol. 2004;22(12):2438–44. [DOI] [PubMed] [Google Scholar]
- 35.Kaiserman I, Amer R, Pe’er J. Liver function tests in metastatic uveal melanoma. Am J Ophthalmol. 2004;137(2):236–43. [DOI] [PubMed] [Google Scholar]
- 36.Choudhary MM, Gupta A, Bena J, et al. Hepatic Ultrasonography for Surveillance in Patients With Uveal Melanoma. JAMA Ophthalmol. 2016;134(2):174–80. [DOI] [PubMed] [Google Scholar]
- 37.Marshall E, Romaniuk C, Ghaneh P, et al. MRI in the detection of hepatic metastases from high-risk uveal melanoma: a prospective study in 188 patients. Br J Ophthalmol. 2013;97(2):159–63. [DOI] [PubMed] [Google Scholar]
- 38.Kadkol SS, Lin AY, Barak V, et al. Osteopontin expression and serum levels in metastatic uveal melanoma: a pilot study. Invest Ophthalmol Vis Sci. 2006;47(3):802–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiniger IW, Wolf A, Welge-Lussen U, et al. Osteopontin as a serologic marker for metastatic uveal melanoma: results of a pilot study. Am J Ophthalmol. 2007;143(4):705–7. [DOI] [PubMed] [Google Scholar]
- 40.Missotten GS, Tang NE, Korse CM, et al. Prognostic value of S-100-beta serum concentration in patients with uveal melanoma. Arch Ophthalmol. 2003;121(8):1117–9. [DOI] [PubMed] [Google Scholar]
- 41.Missotten GS, Korse CM, van Dehn C, et al. S-100B protein and melanoma inhibitory activity protein in uveal melanoma screening. A comparison with liver function tests. Tumour Biol. 2007;28(2):63–9. [DOI] [PubMed] [Google Scholar]
- 42.Haritoglou I, Wolf A, Maier T, et al. Osteopontin and ‘melanoma inhibitory activity’: comparison of two serological tumor markers in metastatic uveal melanoma patients. Ophthalmologica. 2009;223(4):239–43. [DOI] [PubMed] [Google Scholar]
- 43.Song J, Merbs SL, Sokoll LJ, et al. A multiplex immunoassay of serum biomarkers for the detection of uveal melanoma. Clin Proteomics. 2019;16:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi XY, Li Q, Wei WB, Tao LM. Peptidome profiling of human serum of uveal melanoma patients based on magnetic bead fractionation and mass spectrometry. Int J Ophthalmol. 2017;10(6):939–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linge A, Kennedy S, O’Flynn D, et al. Differential expression of fourteen proteins between uveal melanoma from patients who subsequently developed distant metastases versus those who did Not. Invest Ophthalmol Vis Sci. 2012;53(8):4634–43. [DOI] [PubMed] [Google Scholar]
- 46.Ramasamy P, Murphy CC, Clynes M, et al. Proteomics in uveal melanoma. Exp Eye Res. 2014;118:1–12. [DOI] [PubMed] [Google Scholar]
- 47.Crabb JW, Hu B, Crabb JS, et al. iTRAQ Quantitative Proteomic Comparison of Metastatic and Non-Metastatic Uveal Melanoma Tumors. PLoS One. 2015;10(8):e0135543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **48.Jang GF, Crabb JS, Hu B, et al. Proteomics of Primary Uveal Melanoma: Insights into Metastasis and Protein Biomarkers. Cancers (Basel). 2021;13(14). [DOI] [PMC free article] [PubMed] [Google Scholar]; The largest proteomic analysis of UM tissue to identify prognostic biomarkers and elucidate the mechanism of metastasis.
- 49.Missotten GS, Beijnen JH, Keunen JE, Bonfrer JM. Proteomics in uveal melanoma. Melanoma Res. 2003;13(6):627–9. [DOI] [PubMed] [Google Scholar]
- 50.Usui Y, Tsubota K, Agawa T, et al. Aqueous immune mediators in malignant uveal melanomas in comparison to benign pigmented intraocular tumors. Graefes Arch Clin Exp Ophthalmol. 2017;255(2):393–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McClellan JL, Davis JM, Steiner JL, et al. Linking tumor-associated macrophages, inflammation, and intestinal tumorigenesis: role of MCP-1. Am J Physiol Gastrointest Liver Physiol. 2012;303(10):G1087–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarode P, Schaefer MB, Grimminger F, et al. Macrophage and Tumor Cell Cross-Talk Is Fundamental for Lung Tumor Progression: We Need to Talk. Front Oncol. 2020;10:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng Y, Feng J, Zhu X, Liang J. Cytokines concentrations in aqueous humor of eyes with uveal melanoma. Medicine (Baltimore). 2019;98(5):e14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Midena E, Parrozzani R, Midena G, et al. In vivo intraocular biomarkers: Changes of aqueous humor cytokines and chemokines in patients affected by uveal melanoma. Medicine (Baltimore). 2020;99(38):e22091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee CS, Jun IH, Kim TI, et al. Expression of 12 cytokines in aqueous humour of uveal melanoma before and after combined Ruthenium-106 brachytherapy and transpupillary thermotherapy. Acta Ophthalmol. 2012;90(4):e314–20. [DOI] [PubMed] [Google Scholar]
- 56.Dunavoelgyi R, Funk M, Sacu S, et al. Intraocular activation of angiogenic and inflammatory pathways in uveal melanoma. Retina. 2012;32(7):1373–84. [DOI] [PubMed] [Google Scholar]
- 57.Li A, Dubey S, Varney ML, et al. IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J Immunol. 2003;170(6):3369–76. [DOI] [PubMed] [Google Scholar]
- 58.Martin D, Galisteo R, Gutkind JS. CXCL8/IL8 stimulates vascular endothelial growth factor (VEGF) expression and the autocrine activation of VEGFR2 in endothelial cells by activating NFkappaB through the CBM (Carma3/Bcl10/Malt1) complex. J Biol Chem. 2009;284(10):6038–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *59.Chen MX, Liu YM, Li Y, et al. Elevated VEGF-A & PLGF concentration in aqueous humor of patients with uveal melanoma following Iodine-125 plaque radiotherapy. Int J Ophthalmol. 2020;13(4):599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]; Proteomic analysis of AH in patients with UM before and after plaque brachytherapy. Aqueous proteins may provide molecular insights into the mechanism of radiation treatment and the development of radiation retinopathy in patients with UM.
- **60.Wierenga APA, Cao J, Mouthaan H, et al. Aqueous Humor Biomarkers Identify Three Prognostic Groups in Uveal Melanoma. Invest Ophthalmol Vis Sci. 2019;60(14):4740–7. [DOI] [PubMed] [Google Scholar]; A proteomic analysis of AH in patients with UM. This study provides evidence that proteomic analysis of AH in patients with UM correlates with clinical and genetic parameters and may provide complementary prognostic information.
- 61.Wierenga APA, Gezgin G, van Beelen E, et al. Soluble HLA in the Aqueous Humour of Uveal Melanoma Is Associated with Unfavourable Tumour Characteristics. Cancers (Basel). 2019;11(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagarkatti-Gude N, Bronkhorst IH, van Duinen SG, et al. Cytokines and chemokines in the vitreous fluid of eyes with uveal melanoma. Invest Ophthalmol Vis Sci. 2012;53(11):6748–55. [DOI] [PubMed] [Google Scholar]
- *63.Velez G, Nguyen HV, Chemudupati T, et al. Liquid biopsy proteomics of uveal melanoma reveals biomarkers associated with metastatic risk. Mol Cancer. 2021;20(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]; A proteomic analysis of vitreous biopsies in search of candidate biomarkers to serve as surrogates for tumor genes and potential therapeutic target. This study demonstrates that proteomic analysis of the vitreous can identify candidate therapeutic targets.
- 64.Jiao Q, Bi L, Ren Y, et al. Advances in studies of tyrosine kinase inhibitors and their acquired resistance. Mol Cancer. 2018;17(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]


