Abstract
The PAH1-encoded phosphatidate phosphatase, which catalyzes the dephosphorylation of phosphatidate to produce diacylglycerol, controls the divergence of phosphatidate into triacylglycerol synthesis and phospholipid synthesis. Pah1 is inactive in the cytosol as a phosphorylated form and becomes active on the nuclear/endoplasmic reticulum membrane as a dephosphorylated form by the Nem1-Spo7 protein phosphatase complex. The phosphorylation of Pah1 by protein kinases, which include casein kinases I and II, Pho85-Pho80, Cdc28-cyclin B, and protein kinases A and B, controls its cellular location, catalytic activity, and susceptibility to proteasomal degradation. Nem1 (catalytic subunit) and Spo7 (regulatory subunit), which form a protein phosphatase complex catalyzing the dephosphorylation of Pah1 for its activation, are phosphorylated by protein kinases A and C. In this review, we discuss the functions and interrelationships of the protein kinases in the control of the Nem1-Spo7/Pah1 phosphatase cascade and lipid synthesis.
Keywords: phospholipid, phosphatidic acid, triacylglycerol, diacylglycerol, Pah1 PA phosphatase, Nem1-Spo7 protein phosphatase, protein kinase, yeast
1. Introduction and background
In the model eukaryote yeast Saccharomyces cerevisiae1, the Nem1-Spo7/Pah1 phosphatase cascade has emerged as one of highly regulated processes in lipid metabolism (Carman, 2018; Carman and Han, 2019; Carman and Han, 2009; Hennessy et al., 2019; Pascual and Carman, 2013). Pah12 is a Mg2+-dependent phosphatidate (PA)3 phosphatase (PAP) that catalyzes the dephosphorylation of PA to yield diacylglycerol (DAG) (Fig. 1) (Han et al., 2006; Lin and Carman, 1989; Smith et al., 1957). The discovery of yeast PAH1 revealed that the PAP-encoding gene is conserved in eukaryotes, including fungi (Liu et al., 2019), plants (Eastmond et al., 2010; Nakamura et al., 2009), worms (Golden et al., 2009), flies (Ugrankar et al., 2011; Valente et al., 2010), mice (Donkor et al., 2007; Péterfy et al., 2001), and humans (Han et al., 2006; Han and Carman, 2010)2.
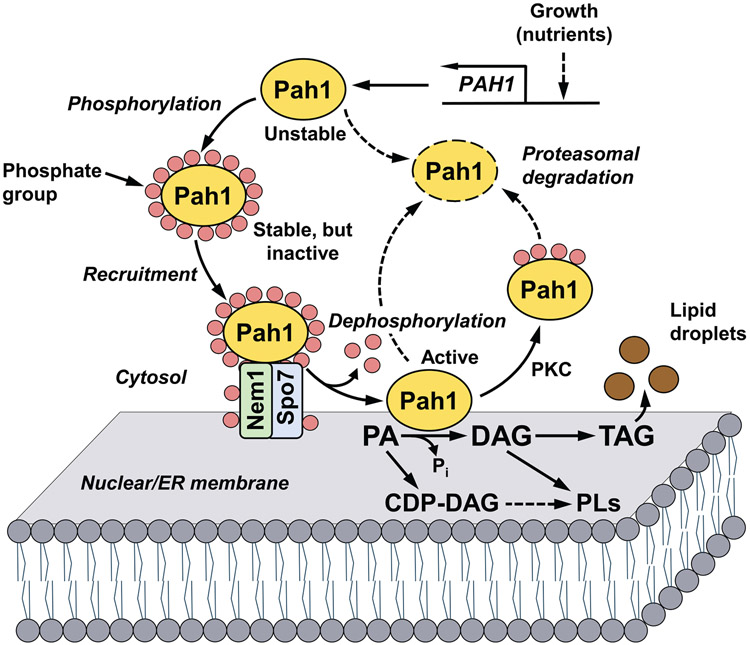
Fig. 1.
Model for the phosphorylation/dephosphorylation-mediated regulation of Pah1 PAP in lipid synthesis. Expression of the PAP-encoding gene PAH1 is regulated during growth by nutrient status. After expression, its product Pah1 in the cytosol is phosphorylated by multiple protein kinases. Phosphorylated Pah1 translocates to the nuclear/ER membrane through its recruitment and dephosphorylation by the Nem1-Spo7 protein phosphatase complex, which itself is subject to phosphorylation. Dephosphorylated Pah1 that is associated with the membrane catalyzes the conversion of PA to DAG, which is then acylated to form TAG that is stored in lipid droplets. Dephosphorylated Pah1 or PKC-phosphorylated Pah1 that is not phosphorylated at the target sites for Pho85-Pho80/Cdc28-cyclin B is degraded by the proteasome (indicated by the dashed line arrows and ellipse). The PAP substrate PA may also be converted to CDP-DAG, which is then used for the synthesis of the membrane phospholipids phosphatidylserine, phosphatidylethanolamine, phosphatidylcholine, phosphatidylinositol, phosphatidylglycerol, and cardiolipin. The PAP product DAG may also be converted to phosphatidylcholine and phosphatidylethanolamine via the Kennedy pathway. Greater details of the yeast phospholipid synthetic pathways may be found elsewhere (Carman and Han, 2011; Kwiatek et al., 2020).
Pah1 PAP largely governs whether yeast cells convert PA to DAG for the synthesis of triacylglycerol (TAG) or to the liponucleotide CDP-DAG for the synthesis of membrane phospholipids (Fig. 1) (Carman, 2018; Carman and Han, 2019; Carman and Han, 2009; Kwiatek et al., 2020; Pascual and Carman, 2013). PAP activity elevated in the cells entering the stationary phase of growth is associated with the increased production of TAG, whereas the enzyme activity reduced during exponential phase of growth is associated with the higher production of membrane phospholipids (Carman, 2018; Carman and Han, 2019; Carman and Han, 2009; Kwiatek et al., 2020; Pascual and Carman, 2013). PAP activity is the primary regulator of this metabolic switch (Carman, 2018; Carman and Han, 2019; Carman and Han, 2009; Kwiatek et al., 2020). The PAP-derived DAG can also be converted to phosphatidylcholine and phosphatidylethanolamine by an auxiliary (Kennedy) pathway when the cells are supplemented with choline and ethanolamine, respectively (Carman and Han, 2011; Kwiatek et al., 2020).
The impact of Pah1 PAP on lipid metabolism extends beyond converting PA to DAG (Kwiatek et al., 2020). By controlling the levels of PA and its derivatives (Fakas et al., 2011; Han et al., 2006; Hassaninasab et al., 2017), the enzyme affects the expression of many lipid synthesis genes (Han and Carman, 2017; Santos-Rosa et al., 2005), phospholipid synthesis (Pascual et al., 2013), nuclear/ER membrane growth (Santos-Rosa et al., 2005), and lipid droplet formation (Adeyo et al., 2011). The PAP activity is also required for growth on non-fermentable carbon sources (Han et al., 2006; Han et al., 2007), vacuole fusion (Sasser et al., 2012), cell wall integrity (Lussier et al., 1997; Ruiz et al., 1999), autophagy induction (Rahman et al., 2018), and resistance to stresses caused by fatty acids (Fakas et al., 2011), oxidizing agents (Park et al., 2015), heat (Han et al., 2006; Han et al., 2008; Irie et al., 1993; Santos-Rosa et al., 2005), and cold (Corcoles-Saez et al., 2016). Yeast cells lacking the enzyme have a shortened chronological life span (Park et al., 2015) and exhibit apoptotic cell death in the stationary phase (Fakas et al., 2011). Many of the pah1Δ phenotypes are linked to the elevation of PA levels, and require Dgk1 DAG kinase activity (Adeyo et al., 2011; Fakas et al., 2011; Han et al., 2008; Park et al., 2015). In mice and humans, the lack of lipin 1 PAP function is associated with a variety of lipid-based syndromes that include lipodystrophy, peripheral neuropathy, and rhabdomyolysis (Nadra et al., 2008; Péterfy et al., 2001; Phan and Reue, 2005; Wiedmann et al., 2008; Zeharia et al., 2008; Zhang et al., 2014).
Great strides have been made using yeast as a model system to understand the mode of action and regulation of the PAP enzyme (Carman, 2018; Carman and Han, 2019; Kwiatek et al., 2020). As depicted in Fig. 1, the function of Pah1 as a lipid biosynthetic enzyme is mainly controlled by its localization. Following its expression, which is regulated at the level of transcription by nutrient status (Pascual et al., 2013; Soto-Cardalda et al., 2011), Pah1 in the cytosol is phosphorylated on serine and threonine residues (Albuquerque et al., 2008; Bodenmiller et al., 2010; Chi et al., 2007; Gnad et al., 2009; Gruhler et al., 2005; Helbig et al., 2010; Lanz et al., 2021; Li et al., 2007; MacGilvray et al., 2020; O'Hara et al., 2006; Smolka et al., 2007; Soufi et al., 2009; Soulard et al., 2010; Swaney et al., 2013) by multiple protein kinases (Choi et al., 2011; Choi et al., 2012; Hassaninasab et al., 2019; Hsieh et al., 2016; Su et al., 2012; Su et al., 2014a). Phosphorylated Pah1 is non-functional because it is sequestered in the cytosol apart from its membrane-associated substrate PA. However, the phosphorylated protein translocates to the nuclear/ER membrane through its dephosphorylation by the Nem1 (catalytic)-Spo7 (regulatory) protein phosphatase complex (Barbosa et al., 2015; Choi et al., 2011; Choi et al., 2012; Karanasios et al., 2010; Karanasios et al., 2013; O'Hara et al., 2006; Santos-Rosa et al., 2005; Siniossoglou et al., 1998; Su et al., 2012; Xu et al., 2012)4. Following its dephosphorylation, Pah1 hops onto the membrane and catalyzes the conversion of PA to DAG that is acylated to TAG stored in lipid droplets (Fig. 1). Pah1 may subsequently scoot on the membrane for additional rounds of catalysis (Kwiatek and Carman, 2020). The dephosphorylated (or unphosphorylated) form of Pah1 is susceptible to degradation by the 20S proteasome, whereas the phosphorylated form is stable against the proteasomal degradation (Hsieh et al., 2015; Pascual et al., 2014). An exception to the phosphorylation effect is shown by protein kinase C (PKC), which stimulates the proteasomal degradation of Pah1 in the absence of prephosphorylation by the Pho85-Pho80 protein kinase, (Su et al., 2014a). Whereas the Nem1-Spo7 complex functions to dephosphorylate Pah1, both Nem1 and Spo7 are themselves subject to phosphorylation (Dey et al., 2017; Dubots et al., 2014; Holt et al., 2009; Su et al., 2018; Swaney et al., 2013), and these modifications add yet another layer of complexity to the regulation of Pah1 PAP function. Overall, the posttranslational modifications of phosphorylation and dephosphorylation are a major mechanism for regulating the membrane localization of Pah1 and its PAP activity for lipid synthesis. In this review, we summarize current knowledge on the phosphorylation of Pah1, Nem1, and Spo7, the functions and interrelationships of the protein kinases that phosphorylate these proteins, and how this knowledge might be used to develop pharmacological reagents to fine-tune the Nem1-Spo7/Pah1 phosphatase cascade and PAP function.
2. Domains/regions and phosphorylation sites in Pah1, Nem1, and Spo7
a. Pah1
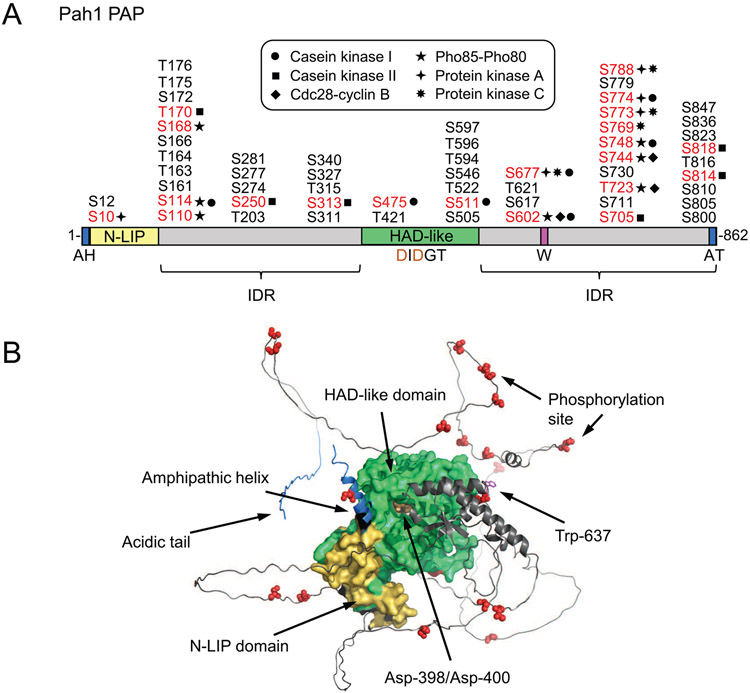
A linear diagram of the domains/regions of Pah1 and its phosphorylation sites are shown in Fig. 2A. The conserved N-LIP and haloacid dehalogenase (HAD)-like domains (Han et al., 2006; Péterfy et al., 2001) are required for PAP activity (Han et al., 2007; Park et al., 2017). In all organisms studied thus far, PAP activity is dependent on the DXDX(T/V) (e.g., DIDGT) catalytic motif in the HAD-like domain (Han et al., 2007; Khayyo et al., 2020; Péterfy et al., 2001). In the membrane translocation of Pah1, its C-terminal acidic tail interacts with the Nem1-Spo7 protein phosphatase complex (Karanasios et al., 2013), and the N-terminal amphipathic helix is required for membrane interaction following the Nem1-Spo7-mediated dephosphorylation (Karanasios et al., 2010). Pah1 is phosphorylated to control its subcellular location, catalytic activity, and protein stability (Carman and Han, 2019), and its phosphorylation sites are concentrated in the intrinsically disordered regions (IDRs) (Fig. 2A) located between the two conserved domains and at the C-terminal region (Hsieh et al., 2015). Many of those sites have been identified as target residues for specific protein kinases (e.g., casein kinase I (CKI) (Hassaninasab et al., 2019), casein kinase II (CKII) (Hsieh et al., 2016), Cdc28-cyclin B (Choi et al., 2011), Pho85-Pho80 (Choi et al., 2012), protein kinase A (PKA) (Su et al., 2012), and PKC (Su et al., 2014a)).
Fig. 2.
Domains/regions and phosphorylation sites in Pah1. A, the diagram shows the positions of the N-terminal amphipathic helix (AH), the conserved N-LIP and HAD-like catalytic domains, the conserved tryptophan (W) residue, the C-terminal acidic tail (AT), and the intrinsically disordered regions (IDR). The serine (S) and threonine (T) residues known to be phosphorylated are grouped at their approximate positions, and marked in red color for the known responsible protein kinases. B, the AlphaFold (Jumper et al., 2021) structure prediction of Pah1 was visualized with the PyMol program.
A conserved tryptophan residue (Trp-637) of Pah1 (Park et al., 2017), which is located in the C-terminal IDR, plays a role in the enzyme phosphorylation, the Nem1-Spo7 complex-mediated cytosol-to-membrane translocation, and enzyme function in TAG synthesis (Park et al., 2017; Park et al., 2022). This residue, however, is not essential for inherent PAP activity (Park et al., 2017; Park et al., 2022). The AlphaFold (Jumper et al., 2021) model of Pah1 shows that Trp-637 and the catalytic residues (i.e., Asp-398 and Asp-400) contained within the HAD-like domain almost lie in the same plane (Fig. 2B), suggesting that the proper alignment of the residues are important for the enzyme to recognize its substrate at the membrane surface in vivo (Park et al., 2022). The AlphaFold (Jumper et al., 2021) model also shows the close interactions between the structured N-LIP and HAD-like catalytic domains and the phosphorylation sites of known protein kinases within the IDRs of the protein (Fig. 2B)
b. Nem1 and Spo7
The Nem1-Spo7 protein phosphatase complex (Siniossoglou et al., 1998) is a major regulator of Pah1 function; it is responsible for recruiting and dephosphorylating Pah1 at the nuclear/ER membrane, and for stimulating PAP activity (Karanasios et al., 2010; Karanasios et al., 2013; O'Hara et al., 2006) (Fig. 1). Of the known protein kinase-phosphorylation site relationships in Pah1 (Choi et al., 2011; Choi et al., 2012; Hsieh et al., 2016; Su et al., 2012; Su et al., 2014a), the specificity of the Nem1-Spo7 protein phosphatase-mediated dephosphorylation is in the order of the sites phosphorylated by Pho85-Pho80 > PKA = CKII > Cdc28-cyclin B > PKC (Hsieh et al., 2016; Su et al., 2014b). Given the function of the Nem1-Spo7 protein phosphatase complex to activate Pah1, it is not surprising that the nem1Δ and spo7Δ mutants exhibit the same phenotypes of the pah1Δ, mutant (Mirheydari et al., 2020; Pascual et al., 2013; Rahman et al., 2018; Santos-Rosa et al., 2005; Siniossoglou et al., 1998; Xu and Okamoto, 2018).
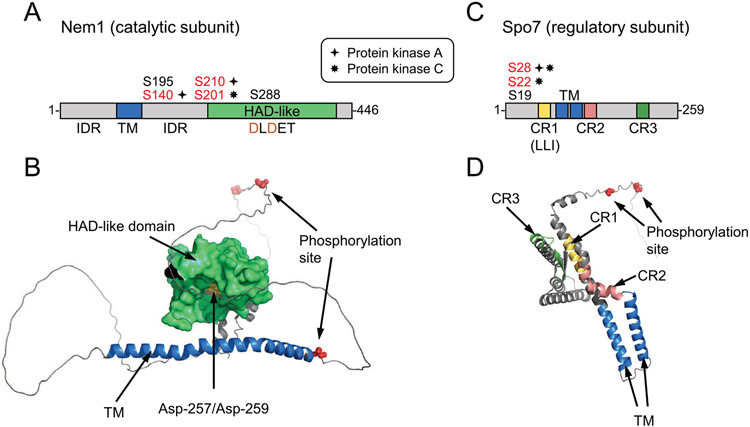
The domains/regions and phosphorylation sites of Nem1 and Spo7 are depicted in Fig. 3A. Nem1 and Spo7 are integral nuclear/ER membrane proteins possessing transmembrane spanning domains (Siniossoglou et al., 1998). Nem1, which serves as the catalytic subunit, is a member of the haloacid dehalogenase superfamily (Koonin and Tatusov, 1994; Madera et al., 2004); its phosphatase activity is conferred by the DXDX(T/V) (e.g., DLDET) catalytic motif within its HAD-like domain (Santos-Rosa et al., 2005; Siniossoglou et al., 1998). Nem1 binds to Spo7 through its conserved C-terminal domain, and this association is responsible for the formation of the complex (Siniossoglou et al., 1998). Spo7, which serves as the regulatory subunit of the phosphate complex (Siniossoglou et al., 1998), facilitates the recognition of its substrate Pah1 (Dubots et al., 2014; Karanasios et al., 2013). The diagram shows conserved regions (CR) 1, 2, 3, and an LLI sequence within CR1 that is required for Spo7 interaction with Nem1 (Mirheydari et al., 2020). The phosphorylation sites for PKA (Su et al., 2018) and PKC (Dey et al., 2019) in Nem1 and Spo7 are shown in red color. The protein kinases that phosphorylate residues shown in black have not been identified. The AlphaFold (Jumper et al., 2021) models for the structures of Nem1 and Spo7 are shown in Fig. 3B. Interestingly, the phosphorylation sites in Nem1 and Spo7, like most of those in Pah1, are located in the intrinsically disordered regions of the proteins (Fig. 3B). Insufficient information is available to predict how Nem1 forms a complex with Spo7 and how the phosphatase complex interacts with Pah1.
Fig. 3.
Domains/regions and phosphorylation sites in Nem1 and Spo7. A, the diagram denotes the catalytic HAD-like domain, transmembrane (TM) region, and intrinsically disordered regions (IDR) in Nem1. C, the diagram shows the conserved regions (CR) 1, 2, and 3 and transmembrane regions of Spo7. The serine residues phosphorylated (red color) by PKA or PKC in Nem1 and Spo7 are also indicated. B and D, the AlphaFold (Jumper et al., 2021) structure predictions of Nem1 and Spo7 were visualized with the PyMol program.
3. Functions and interrelationships of the protein kinases that phosphorylate Pah1, Nem1, and Spo7
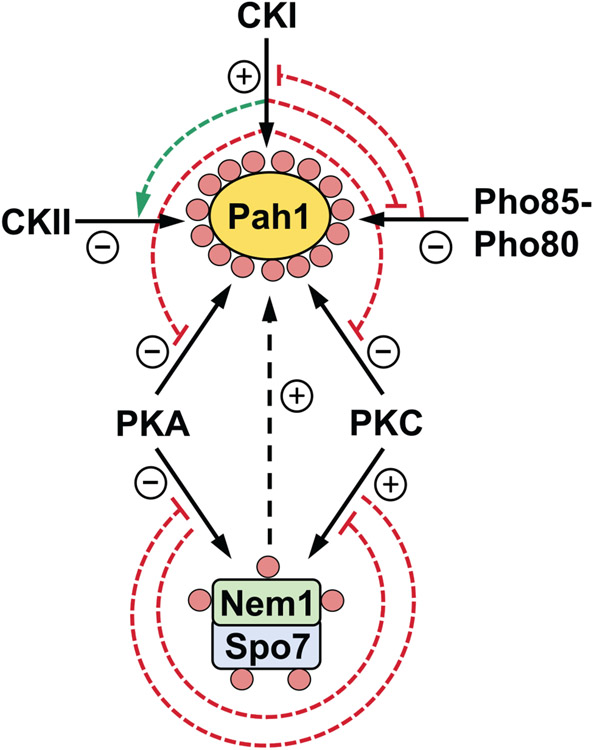
The protein kinase-mediated phosphorylations of Pah1, Nem1, and Spo7, along with the interrelationships of their phosphorylations are summarized in Table 1 and Fig. 4, respectively.
Table 1.
Protein kinases that phosphorylate Pah1, Nem1, and Spo7
| Protein kinase | Phosphorylation sites | Consequences | Reference |
|---|---|---|---|
| Pah1 | |||
| CKI | Ser-114, Ser-475, Ser-511, Ser-602, Ser-677, Ser-748, Ser-774 | Stimulates PAP activity, stimulates phosphorylation by CKII, inhibits phosphorylations by Pho85-Pho80, PKA, and PKC | (Hassaninasab et al., 2019) |
| CKII | Thr-170, Ser-250, Ser-313, Ser-705, Ser-814, Ser-818 | Stimulates TAG synthesis when the Pho85-Pho80 sites are not phosphorylated, inhibits phosphorylation by PKA | (Hsieh et al., 2016) |
| Cdc28-cyclin B | Ser-602, Thr-723, Thr-744 | Inhibits PAP activity and TAG synthesis, attenuates membrane association, stabilizes Pah1 abundance | (Choi et al., 2011) |
| Pho85-Pho80 | Ser-110, Ser-114, Ser-168, Ser-602, Thr-723, Ser-744, Ser-748 | Inhibits PAP activity and TAG synthesis, attenuates membrane association, stabilizes Pah1 abundance, inhibits phosphorylations by CKI and PKC | (Choi et al., 2012; Hassaninasab et al., 2019; Hsieh et al., 2015; Karanasios et al., 2010; Karanasios et al., 2013; O'Hara et al., 2006; Su et al., 2014a) |
| PKA | Ser-10, Ser-677, Ser-773, Ser-774, Ser-788 | Functions in conjunction with Pho85-Pho80 and Cdc28-cyclin B to regulate lipid synthesis, inhibits phosphorylation by CKII | (Hsieh et al., 2016; Su et al., 2012) |
| PKC | Ser-677, Ser-769, Ser-773, Ser-788 | Destabilizes Pah1 abundance when Pho85-Pho80/Cdc28-cyclin B sites are not phosphorylated, inhibits phosphorylation by CKII | (Hsieh et al., 2016; Su et al., 2014a) |
| Nem1 | |||
| PKA | Ser-140, Ser-210 | Inhibits Nem1-Spo7 protein phosphatase activity and TAG synthesis, phosphorylation of the Nem1-Spo7 complex inhibits phosphorylation of Spo7 by PKC | (Dey et al., 2019; Su et al., 2018) |
| PKC | Ser-201 | Stimulates Nem1-Spo7 activity and TAG synthesis, phosphorylation of the Nem1-Spo7 complex inhibits phosphorylation of Nem1 by PKA | (Dey et al., 2019) |
| Spo7 | |||
| PKA | Ser-28 | Inhibits Nem1-Spo7 protein phosphatase activity and TAG synthesis, phosphorylation of the Nem1-Spo7 complex inhibits phosphorylation of Spo7 by PKC | (Dey et al., 2019; Su et al., 2018) |
| PKC | Ser-22 | Phosphorylation of the Nem1-Spo7 complex inhibits phosphorylation of Nem1 by PKA | (Dey et al., 2019) |
Fig. 4.
Interrelationships between the phosphorylations of Pah1, Nem1, and Spo7. Pah1 is phosphorylated by CKI, CKII, Pho85-Pho80, PKA, and PKC (solid arrows, upper diagram). The CKI phosphorylation of Pah1 stimulates its subsequent phosphorylation by CKII (dashed green arrow), but inhibits its subsequent phosphorylations by Pho85-Pho80, PKA, and PKC (dashed blunted red line). The Pho85-Pho80 phosphorylation of Pah1 inhibits its subsequent phosphorylation by CKI (dashed blunted red line). Nem1 and Spo7 are phosphorylated by PKA and PKC (solid arrows, lower diagram). The phosphorylation of the Nem1-Spo7 complex by PKA inhibits phosphorylation of Spo7 by PKC, whereas the phosphorylation of the complex by PKC inhibits phosphorylation of Nem1 by PKA (dashed blunted red lines). Phosphorylated Pah1, Nem1, and Spo7 are indicated by the small pink circles, and the effects of the phosphorylations by the various protein kinases are indicated by + and − symbols.
a. Pah1
Pho85, which is a multifunctional cyclin (e.g., Pho80)-dependent protein kinase involved in several signal transduction pathways that affect cell cycle progression and the metabolism of nutrients (Carroll and O'Shea, 2002; Huang et al., 2007; Moffat et al., 2000), has strong regulatory effects on the location, enzyme activity, and protein stability of Pah1 (Choi et al., 2012; Hsieh et al., 2015; Karanasios et al., 2010; O'Hara et al., 2006). The seven target sites of Pho85-Pho80 in Pah1 are distributed at the N- and C-termini of the protein (Fig. 2A). Three of the seven sites are also targets for the Cdc28 protein kinase (Choi et al., 2011; O'Hara et al., 2006), a master regulator of cell-cycle transitions whose activity is governed by interaction with various G1 and B-type cyclins (Enserink and Kolodner, 2010). Pah1 phosphorylated by Pho85-Pho80 (Choi et al., 2012) and Cdc28-cyclin B (Choi et al., 2011) is not active in the cell due to its sequestration in the cytosol apart from the substrate PA in the nuclear/ER membrane (Fig. 1). In addition, Pah1 PAP activity is reduced through its phosphorylation at the seven sites by Pho85-Pho80 (Choi et al., 2011; Choi et al., 2012).
In conjunction with Pho85-Pho80 and Cdc28-cyclin B, PKA, a cAMP-dependent protein kinase whose activity is associated with active cell growth and increased metabolic activity, and an increase in membrane phospholipid synthesis (Broach and Deschenes, 1990; Carman and Han, 2011; Thevelein, 1994), functions to regulate the location and activity of Pah1 through its phosphorylation at five sites located at the beginning and end of the protein (Su et al., 2012) (Fig. 2A). Pah1 phosphorylated by the Pho85-Pho80, Cdc28-cyclin B, and PKA is protected against its degradation by the 20S proteasome (Hsieh et al., 2015; Pascual et al., 2014).
In contrast to the protective phosphorylation effect, Pah1 phosphorylated by PKC, which occurs on four sites at the C-terminus (Fig. 2A), is susceptible to the proteasomal degradation (Su et al., 2014a). PKC is a lipid-dependent protein kinase (Dey et al., 2017; Kamada et al., 1996; Nishizuka, 1984; Nishizuka, 1992), and the enzyme in yeast is required for cell cycle regulation and plays a role in regulating phospholipid synthesis (Choi et al., 2003; Choi et al., 2005; Park et al., 2003; Sreenivas et al., 2001; Yang et al., 1996; Yang and Carman, 1995) and in maintaining cell wall integrity (Kamada et al., 1995; Levin et al., 1990; Levin and Bartlett-Heubusch, 1992).
The phosphorylation of Pah1 by CKII, a highly conserved serine/threonine protein kinase that is essential for cell viability in yeast (Glover, III, 1998; Guerra and Issinger, 1999; Litchfield, 2003; Poole et al., 2005), occurs on six sites at the N- and C-termini (Fig. 2A). The CKII phosphorylation of Pah1 prevents its subsequent phosphorylation by PKA, and stimulates its catalytic activity (Hsieh et al., 2016). The regulatory effects of Pah1 phosphorylation by PKC (Su et al., 2014a) and CKII (Hsieh et al., 2016) on its stability and function are shown when it is not prephosphorylated at the seven sites by Pho85-Pho80/Cdc28-cyclin B. The phosphorylation of Pah1 by Pho85-Pho80/Cdc28-cyclin B exerts an inhibitory effect on its phosphorylation by PKC (Su et al., 2014a), but has no effect on its phosphorylation by CKII (Hsieh et al., 2016). Unlike their effects on Pah1 phosphorylation, PKC (Su et al., 2014a) and CKII (Hsieh et al., 2016) have little effect on its PAP activity.
CKI is a constitutively active serine/threonine protein kinase that plays a key role in nutrient-mediated cell morphogenesis, cytokinesis, secretion, and endocytosis (Robinson et al., 1992; Robinson et al., 1993; Snowdon and Johnston, 2016; Stalder and Novick, 2016; Wang et al., 1992). The protein kinase phosphorylates Pah1 at eight sites scattered throughout the protein (Fig. 2A). The CKI phosphorylation of Pah1 has two major effects; it stimulates PAP activity and regulates the phosphorylation of Pah1 by other protein kinases (Hassaninasab et al., 2019). The CKI phosphorylation of Pah1 stimulates its phosphorylation by CKII, but inhibits its phosphorylation by Pho85-Pho80, PKA, and PKC.
The six sites phosphorylated by CKI are also targets for other protein kinases (Fig. 2A), and thus, CKI could phosphorylate the common sites when those protein kinases are inactive. For example, Pho85-Pho80 and PKA are most active during the exponential phase of growth when phospholipid synthesis is favored over TAG synthesis (Pascual et al., 2013). Therefore, at any point during growth, lipid synthesis might be regulated via the CKI-mediated phosphorylation of Pah1 at a site(s) that is phosphorylated by Pho85-Pho80 and PKA when they are not active.
Ser-475 and Ser-511, which are located within the HAD-like domain of Pah1 (Fig. 2A), are CKI-specific phosphorylation sites (Hassaninasab et al., 2019). The phosphorylations of these sites play roles in the subsequent phosphorylations by other protein kinases. The CKI phosphorylation of Ser-475 and Ser-511 are required for the stimulation of Pah1 phosphorylation by CKII (Hassaninasab et al., 2019). As discussed above, the CKII phosphorylation of Pah1 stimulates the synthesis of TAG, but this occurs when the enzyme is not phosphorylated by Pho85-Pho80 (Hsieh et al., 2016). Thus, the CKI-mediated phosphorylation of Ser-475 and Ser-511 would be expected to have a positive impact on Pah1 function as mediated by CKII. Yet, the phosphorylation of Ser-475 and Ser-511 are required for the CKI-mediated inhibition of the phosphorylation by PKA and PKC (Hassaninasab et al., 2019). As discussed above, PKA works in conjunction with Pho85-Pho80 to attenuate Pah1 interaction with the ER membrane and inhibit PAP activity, but at the same time, stabilize the enzyme to proteasomal degradation (Hsieh et al., 2015; Su et al., 2012). Moreover, as PKC facilitates the proteasomal degradation of Pah1 when the seven Pho85-Pho80 sites are not phosphorylated (Hsieh et al., 2015; Su et al., 2014a), the phosphorylations of Ser-475 and Ser-511 would be expected to sustain the PKA- and PKC-mediated regulations of Pah1.
The phosphorylation of Pah1 by CKI inhibits the subsequent phosphorylation by Pho85-Pho80, but Ser-475 and Ser-511 are not involved in this regulation (Hassaninasab et al., 2019). These sites are not involved in the stimulatory effect of CKI on PAP activity. In fact, the S475A and S511A mutations enhance the stimulation of PAP activity by the CKI-mediated phosphorylation (Hassaninasab et al., 2019), suggesting that the phosphorylation of these sites inhibit the phosphorylation of another site(s) that must be responsible for the stimulation of activity. The phosphorylation of Pah1 by Pho85-Pho80 inhibits PAP activity, and the alanine mutations of its seven target sites augments the stimulatory effect CKI has on PAP activity. Clearly, the stimulation of the PAP activity caused by its phosphorylation by CKI is complex.
Pho85-Pho80, Cdc28-cyclin B, PKA, PKC, and CKII are all associated with the cytosolic fraction of the cell (Huh et al., 2003). Accordingly, it is presumed that the phosphorylation of Pah1 by most protein kinases occurs in cytosol (Fig. 1). However, CKI associates with the plasma membrane through the posttranslational modification of palmitoylation (Babu et al., 2002), and this association is required for its known physiological functions (Wang et al., 1996). Whether a soluble unmodified form of CKI phosphorylates Pah1 in the cytosol is unknown. Contact sites between the ER membrane and plasma membrane exist (Quon et al., 2018), and so CKI associated with the plasma membrane could phosphorylate Pah1 associated with the ER membrane.
b. Nem1 and Spo7
Nem1 and Spo7 are both phosphorylated by PKA and PKC at their N-terminal regions (Fig. 3A). For Nem1, the sites phosphorylated by PKA and PKC are unique, whereas for Spo7, one site (i.e., Ser-28) is phosphorylated by both protein kinases (Dey et al., 2019; Su et al., 2018). Data indicate that PKA has a negative impact on the activity of Nem1-Spo7 and TAG synthesis (Su et al., 2018), whereas the phosphorylation of the complex by PKC has the opposite effect (Dey et al., 2019). Additionally, the prephosphorylation of the Nem1-Spo7 complex by PKC has an inhibitory effect on the phosphorylation of Nem1 by PKA, whereas the prephosphorylation of the complex by PKA has an inhibitory effect on the phosphorylation of Spo7 by PKC.
4. How might knowledge of phosphorylation be used to control the Nem1-Spo7/Pah1 phosphatase cascade and lipid synthesis in vivo?
The lack of PAP activity in yeast, as well as in humans, leads to aberrant lipid metabolism and cell physiology (Carman and Han, 2019; Reue and Wang, 2019). Too much PAP activity is also detrimental (e.g., obesity in the mouse model) (Phan and Reue, 2005). Thus, understanding the phosphorylation of the Nem1-Spo7/Pah1 phosphatase cascade proteins may lead to the identification of effector molecules to fine-tune PAP activity and/or the Pah1 cellular location. Peptide-based protein kinase inhibitors are attractive because they may block phosphorylation or disrupt protein-protein interactions (Eldar-Finkelman and Eisenstein, 2009; Jenardhanan et al., 2019). Most peptide inhibitors of protein kinases are based on the consensus phosphorylation motif (Eldar-Finkelman and Eisenstein, 2009; Jenardhanan et al., 2019). However, those inhibitors have a drawback of not having specificity for a particular substrate target of phosphorylation. A peptide sequence unique to a specific phosphorylation site would provide specificity without off-target side effects.
A peptide containing a specific phosphorylation site would be expected to serve as a substrate for the protein kinase that phosphorylates that site. In fact, Pah1 peptides that contain a specific phosphorylation site for CKI (Ser-511, residues 506-LYFEDSDNEVDT-517) and PKC (Ser-769, residues 763-NYNRTKSRRA-772) are substrates for CKI and PKC, respectively (Dey et al., 2017; Hassaninasab et al., 2019). Likewise, Nem1 peptides with specific phosphorylation sites for PKA (Ser-140, residues 135-KRNRGSNASEN-145 and Ser-210, residues 205-RPRSYSKSELS-215) and PKC (Ser-201, residues 197-LRAQSVKSRPR-207) are substrates for PKA and PKC, respectively (Dey et al., 2019; Su et al., 2018). Substitution of a non-phosphorylatable alanine residue for the phosphorylatable serine residues (shown in bold red color) in these peptide substrates obviates the phosphorylation by the protein kinase involved (Dey et al., 2019; Hassaninasab et al., 2019; Su et al., 2018).
A peptide containing a specific phosphorylation site should inhibit the phosphorylation of that site in the full-length protein. Indeed, in a preliminary experiment we have shown that the Pah1 peptide 506-LYFEDSDNEVDT-517, which contains the major CKI phosphorylation site Ser-511, competitively inhibits (IC50 = 80 μM) the phosphorylation of full-length Pah1 by CKI (Hassaninasab et al., 2019). This sequence is specific to Pah1 as it does not align with any other proteins in yeast, and thus, the peptide should not cause off-target effects. This provides a proof-of-concept that knowledge of phosphorylation sites in Pah1, Nem1, or Spo7 could theoretically be used to design peptides that prevent phosphorylation of specific sites to control the Pah1/Nem1-Spo7 phosphatase cascade and lipid synthesis. Most pharmacological inhibitors work in the nM range, and thus, strategies (e.g., peptide cyclization and modification of backbone structure (Wojcik and Berlicki, 2016)) would need to be developed to enhance the inhibitory activity and effectiveness of a Pah1 peptide if it were to work in vivo. Nonetheless, additional studies with the Ser-511-containing peptide, as well as peptides that target other phosphorylation sites in Pah1, Nem1, and Spo7 are warranted to test the notion that peptides might be used to fine-tune the Nem1-Spo7/Pah1 phosphatase cascade and lipid synthesis in cells.
5. Concluding comments
In this review, we have summarized current knowledge of the mode of action and the phosphorylation-mediated regulation of components of the Nem1-Spo7/Pah1 phosphatase cascade in yeast. The posttranslational modification of phosphorylation/dephosphorylation is an important regulatory mechanism to control the location, activity, and stability of Pah1 PAP. The phosphorylation of Pah1 is particularly complex as the protein is phosphorylated on at least 56 residues. Some of the phosphorylations are hierarchical in nature (e.g., phosphorylation on one site affects phosphorylation on another site), whereas other phosphorylations occur on common sites by different protein kinases. Moreover, the protein kinase-phosphorylation site relationships of all the sites have yet to be determined. In this regard, phosphoproteomics and bioinformatics, coupled with site-specific mutagenesis and old fashion biochemistry should provide information on all the protein kinases that regulate the enzyme. Moreover, it is the mutagenesis studies of specific sites of phosphorylation that have yielded the most fruitful information on the regulation. We propose that knowledge of specific phosphorylations might be used to develop pharmacological peptides to fine-tune the Nem1-Spo7/Pah1 phosphatase cascade and lipid synthesis in vivo. Furthermore, such an approach might be applicable to higher eukaryotes owing that this cascade and its regulation by phosphorylation is conserved.
Acknowledgments
This work was supported, in whole or in part, by United States Public Health Service Grants GM028140, GM050679, and GM136128 from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
In this review, Saccharomyces cerevisiae is used interchangeably with yeast.
The PAP orthologs in various organisms are known by different acronyms that are based on the names of genes that encode the enzyme. For S. cerevisiae, the protein product of the PAH1 gene is known as Pah1 (Han et al., 2006), whereas in human and mouse, the protein products of the LPIN1 and Lpin1 genes, respectively, are known as lipin 1 (Péterfy et al., 2001). The PAP encoded by PAH1 differs from the PAP enzymes encoded by APP1 (Chae et al., 2012; Chae and Carman, 2013), DPP1 (Toke et al., 1998) and LPP1 (Toke et al., 1999), which dephosphorylate a broad spectrum of substrates (e.g., phosphatidate, lysophosphatidate, diacylglycerol pyrophosphate) and are not involved in de novo lipid synthesis (Chae et al., 2012).
Abbreviations: PA, phosphatidate; PAP, PA phosphatase; DAG, diacylglycerol; TAG, triacylglycerol; HAD, haloacid dehalogenase; PKA, protein kinase A; PKC, protein kinase C; CK, casein kinase; IDR, intrinsically disordered region;
The orthologous components of yeast Nem1-Spo7 protein phosphatase complex in higher eukaryotes consist of CTDNEP1 (catalytic subunit) and NEP1-R1 (regulatory subunit) (Han et al., 2012; Kim et al., 2007).
Conflicts of interest
None.
Data availability
All data are contained within the manuscript
References
- Adeyo O, Horn PJ, Lee S, Binns DD, Chandrahas A, Chapman KD, Goodman JM, 2011. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol 192, 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H, 2008. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell Proteomics 7, 1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu P, Bryan JD, Panek HR, Jordan SL, Forbrich BM, Kelley SC, Colvin RT, Robinson LC, 2002. Plasma membrane localization of the Yck2p yeast casein kinase 1 isoform requires the C-terminal extension and secretory pathway function. J. Cell Sci 115, 4957–4968. [DOI] [PubMed] [Google Scholar]
- Barbosa AD, Sembongi H, Su W-M, Abreu S, Reggiori F, Carman GM, Siniossoglou S, 2015. Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol. Biol. Cell 26, 3641–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, Brusniak MY, Roschitzki B, Zhang C, Shokat KM, Schlapbach R, Colman-Lerner A, Nolan GP, Nesvizhskii AI, Peter M, Loewith R, von MC, Aebersold R, 2010. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal 3, rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach JR, Deschenes RJ, 1990. The function of RAS genes in Saccharomyces cerevisiae. Adv. Cancer Res 54, 79–139. [DOI] [PubMed] [Google Scholar]
- Carman GM, 2018. Discoveries of the phosphatidate phosphatase genes in yeast published in the Journal of Biological Chemistry. J. Biol. Chem 294, 1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Han GS, 2019. Fat-regulating phosphatidic acid phosphatase: a review of its roles and regulation in lipid homeostasis. J. Lipid Res 60, 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Han G-S, 2009. Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem 284, 2593–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Han G-S, 2011. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Ann. Rev. Biochem 80, 859–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AS, O'Shea EK, 2002. Pho85 and signaling environmental conditions. Trends Biochem. Sci 27, 87–93. [DOI] [PubMed] [Google Scholar]
- Chae M, Carman GM, 2013. Characterization of the yeast actin patch protein App1p phosphatidate phosphatase. J. Biol. Chem 288, 6427–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae M, Han G-S, Carman GM, 2012. The Saccharomyces cerevisiae actin patch protein App1p is a phosphatidate phosphatase enzyme. J. Biol. Chem 287, 40186–40196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF, 2007. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U. S. A 104, 2193–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H-S, Su W-M, Han G-S, Plote D, Xu Z, Carman GM, 2012. Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J. Biol. Chem 287, 11290–11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H-S, Su W-M, Morgan JM, Han G-S, Xu Z, Karanasios E, Siniossoglou S, Carman GM, 2011. Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of Ser602, Thr723, and Ser744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem 286, 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M-G, Kurnov V, Kersting MC, Sreenivas A, Carman GM, 2005. Phosphorylation of the yeast choline kinase by protein kinase C. Identification of Ser25 and Ser30 as major sites of phosphorylation. J Biol. Chem 280, 26105–26112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M-G, Park TS, Carman GM, 2003. Phosphorylation of Saccharomyces cerevisiae CTP synthetase at Ser424 by protein kinases A and C regulates phosphatidylcholine synthesis by the CDP-choline pathway. J. Biol. Chem 278, 23610–23616. [DOI] [PubMed] [Google Scholar]
- Corcoles-Saez I, Hernandez ML, Martinez-Rivas JM, Prieto JA, Randez-Gil F, 2016. Characterization of the S. cerevisiae inp51 mutant links phosphatidylinositol 4,5-bisphosphate levels with lipid content, membrane fluidity and cold growth. Biochim. Biophys. Acta 1861, 213–226. [DOI] [PubMed] [Google Scholar]
- Dey P, Su WM, Han GS, Carman GM, 2017. Phosphorylation of lipid metabolic enzymes by yeast Pkc1 protein kinase C requires phosphatidylserine and diacylglycerol. J. Lipid Res 58, 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P, Su WM, Mirheydari M, Han GS, Carman GM, 2019. Protein kinase C mediates the phosphorylation of the Nem1-Spo7 protein phosphatase complex in yeast. J. Biol. Chem 294, 15997–16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor J, Sariahmetoglu M, Dewald J, Brindley DN, Reue K, 2007. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J Biol. Chem 282, 3450–3457. [DOI] [PubMed] [Google Scholar]
- Dubots E, Cottier S, Peli-Gulli MP, Jaquenoud M, Bontron S, Schneiter R, De Virgilio C, 2014. TORC1 regulates Pah1 phosphatidate phosphatase activity via the Nem1/Spo7 protein phosphatase complex. PLoS. One 9, e104194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Quettier AL, Kroon JT, Craddock C, Adams N, Slabas AR, 2010. Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 22, 2796–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar-Finkelman H, Eisenstein M, 2009. Peptide inhibitors targeting protein kinases. Curr. Pharm. Des 15, 2463–2470. [DOI] [PubMed] [Google Scholar]
- Enserink JM, Kolodner RD, 2010. An overview of Cdk1-controlled targets and processes. Cell Div. 5, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakas S, Qiu Y, Dixon JL, Han G-S, Ruggles KV, Garbarino J, Sturley SL, Carman GM, 2011. Phosphatidate phosphatase activity plays a key role in protection against fatty acid-induced toxicity in yeast. J. Biol. Chem 286, 29074–29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover CV III., 1998. On the physiological role of casein kinase II in Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol 59, 95–133. [DOI] [PubMed] [Google Scholar]
- Gnad F, de Godoy LM, Cox J, Neuhauser N, Ren S, Olsen JV, Mann M, 2009. High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics. 9, 4642–4652. [DOI] [PubMed] [Google Scholar]
- Golden A, Liu J, Cohen-Fix O, 2009. Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J. Cell Sci 122, 1970–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON, 2005. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell Proteomics 4, 310–327. [DOI] [PubMed] [Google Scholar]
- Guerra B, Issinger OG, 1999. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis 20, 391–408. [DOI] [PubMed] [Google Scholar]
- Han G-S, Carman GM, 2010. Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J. Biol. Chem 285, 14628–14638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G-S, Carman GM, 2017. Yeast PAH1-encoded phosphatidate phosphatase controls the expression of CHO1-encoded phosphatidylserine synthase for membrane phospholipid synthesis. J. Biol. Chem 292, 13230–13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G-S, O'Hara L, Carman GM, Siniossoglou S, 2008. An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J. Biol. Chem 283, 20433–20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G-S, Siniossoglou S, Carman GM, 2007. The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem 282, 37026–37035. [DOI] [PubMed] [Google Scholar]
- Han G-S, Wu W-I, Carman GM, 2006. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem 281, 9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Bahmanyar S, Zhang P, Grishin N, Oegema K, Crooke R, Graham M, Reue K, Dixon JE, Goodman JM, 2012. Nuclear envelope phosphatase 1-regulatory subunit 1 (formerly TMEM188) Is the metazoan Spo7p ortholog and functions in the lipin activation pathway. J. Biol. Chem 287, 3123–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassaninasab A, Han G-S, Carman GM, 2017. Tips on the analysis of phosphatidic acid by the fluorometric coupled enzyme assay. Anal. Biochem 526, 69–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassaninasab A, Hsieh LS, Su WM, Han GS, Carman GM, 2019. Yck1 casein kinase I regulates the activity and phosphorylation of Pah1 phosphatidate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem 294, 18256–18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig AO, Rosati S, Pijnappel PW, van BB, Timmers MH, Mohammed S, Slijper M, Heck AJ, 2010. Perturbation of the yeast N-acetyltransferase NatB induces elevation of protein phosphorylation levels. BMC. Genomics 11, 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy M, Granade ME, Hassaninasab A, Wang D, Kwiatek JM, Han G-S, Harris TE, Carman GM, 2019. Casein kinase II-mediated phosphorylation of lipin 1β phosphatidate phosphatase at Ser-285 and Ser-287 regulates its interaction with 14-3-3β protein. J. Biol. Chem 294, 2365–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO, 2009. Global analysis of Cdkl substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh L-S, Su W-M, Han G-S, Carman GM, 2015. Phosphorylation regulates the ubiquitin-independent degradation of yeast Pah1 phosphatidate phosphatase by the 20S proteasome. J. Biol. Chem 290, 11467–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh L-S, Su W-M, Han G-S, Carman GM, 2016. Phosphorylation of yeast Pah1 phosphatidate phosphatase by casein kinase II regulates its function in lipid metabolism. J. Biol. Chem 291, 9974–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Friesen H, Andrews B, 2007. Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Mol. Microbiol 66, 303–314. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK, 2003. Global analysis of protein localization in budding yeast. Nature 425, 686–691. [DOI] [PubMed] [Google Scholar]
- Irie K, Takase M, Araki H, Oshima Y, 1993. A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol. Gen. Genet 236, 283–288. [DOI] [PubMed] [Google Scholar]
- Jenardhanan P, Panneerselvam M, Mathur PP, 2019. Targeting Kinase Interaction Networks: A New Paradigm in PPI Based Design of Kinase Inhibitors. Curr. Top. Med. Chem 19, 467–485. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D, 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Jung US, Piotrowski J, Levin DE, 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9, 1559–1571. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Qadota H, Python CP, Anraku Y, Ohya Y, Levin DE, 1996. Activation of yeast protein kinase C by Rhol GTPase. J. Biol. Chem 271, 9193–9196. [DOI] [PubMed] [Google Scholar]
- Karanasios E, Barbosa AD, Sembongi H, Mari M, Han G-S, Reggiori F, Carman GM, Siniossoglou S, 2013. Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol. Biol. Cell 24, 2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E, Han G-S, Xu Z, Carman GM, Siniossoglou S, 2010. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. U. S. A 107, 17539–17544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayyo VI, Hoffmann RM, Wang H, Bell JA, Burke JE, Reue K, Airola MVV, 2020. Crystal structure of a lipin/Pah phosphatidic acid phosphatase. Nat. Commun 11, 1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Gentry MS, Harris TE, Wiley SE, Lawrence JC Jr., Dixon JE, 2007. A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc. Natl. Acad. Sci. U. S. A 104, 6596–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Tatusov RL, 1994. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J Mol. Biol 244, 125–132. [DOI] [PubMed] [Google Scholar]
- Kwiatek JM, Carman GM, 2020. Yeast phosphatidic acid phosphatase Pah1 hops and scoots along the membrane phospholipid bilayer. J. Lipid Res 61, 1232–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatek JM, Han GS, Carman GM, 2020. Phosphatidate-mediated regulation of lipid synthesis at the nuclear/endoplasmic reticulum membrane. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1865, 158434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz MC, Yugandhar K, Gupta S, Sanford EJ, Faca VM, Vega S, Joiner AMN, Fromme JC, Yu H, Smolka MB, 2021. In-depth and 3-dimensional exploration of the budding yeast phosphoproteome. EMBO Rep. 22, e51121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE, Bartlett-Heubusch E, 1992. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol 116, 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE, Fields FO, Kunisawa R, Bishop JM, Thorner J, 1990. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell 62, 213–224. [DOI] [PubMed] [Google Scholar]
- Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP, 2007. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome. Res 6, 1190–1197. [DOI] [PubMed] [Google Scholar]
- Lin Y-P, Carman GM, 1989. Purification and characterization of phosphatidate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem 264, 8641–8645. [PubMed] [Google Scholar]
- Litchfield DW, 2003. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J 369, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Yun Y, Yin Y, Hahn M, Ma Z, Chen Y, 2019. Lipid droplet biogenesis regulated by the FgNem1/Spo7-FgPah1 phosphatase cascade plays critical roles in fungal development and virulence in Fusarium graminearum. New Phytol. 223, 412–429. [DOI] [PubMed] [Google Scholar]
- Lussier M, White AM, Sheraton J, di PT, Treadwell J, Southard SB, Horenstein CI, Chen-Weiner J, Ram AF, Kapteyn JC, Roemer TW, Vo DH, Bondoc DC, Hall J, Zhong WW, Sdicu AM, Davies J, Klis FM, Robbins PW, Bussey H, 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGilvray ME, Shishkova E, Place M, Wagner ER, Coon JJ, Gasch AP, 2020. Phosphoproteome response to dithiothreitol reveals unique versus shared features of Saccharomyces cerevisiae stress responses. J. Proteome. Res 19, 3405–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madera M, Vogel C, Kummerfeld SK, Chothia C, Gough J, 2004. The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res. 32, D235–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirheydari M, Dey P, Stukey GJ, Park Y, Han GS, Carman GM, 2020. The Spo7 sequence LLI is required for Nem1-Spo7/Pah1 phosphatase cascade function in yeast lipid metabolism. J. Biol. Chem 295, 11473–11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Huang D, Andrews B, 2000. Functions of Pho85 cyclin-dependent kinases in budding yeast. Prog. Cell Cycle Res 4, 97–106. [DOI] [PubMed] [Google Scholar]
- Nadra K, De Preux Charles A-S, Medard J-J, Hendriks WT, Han G-S, Gres S, Carman GM, Saulnier-Blache J-S, Verheijen MHG, Chrast R, 2008. Phosphatidic acid mediates demyelination in Lpin1 mutant mice. Genes Dev. 22, 1647–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Koizumi R, Shui G, Shimojima M, Wenk MR, Ito T, Ohta H, 2009. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc. Natl. Acad. Sci. U. S. A 106, 20978–20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y, 1984. The role of protein kinase C in cell surface signal transduction and tumor promotion. Nature 308, 693–698. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y, 1992. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science 258, 607–614. [DOI] [PubMed] [Google Scholar]
- O’Hara L, Han G-S, Peak-Chew S, Grimsey N, Carman GM, Siniossoglou S, 2006. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem 281, 34537–34548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T-S, O'Brien DJ, Carman GM, 2003. Phosphorylation of CTP synthetase on Ser36, Ser330, Ser354, and Ser454 regulates the levels of CTP and phosphatidylcholine synthesis in Saccharomyces cerevisiae. J. Biol. Chem 278, 20785–20794. [DOI] [PubMed] [Google Scholar]
- Park Y, Han GS, Carman GM, 2017. A conserved tryptophan within the WRDPLVDID domain of yeast Pah1 phosphatidate phosphatase is required for its in vivo function in lipid metabolism. J. Biol. Chem 292, 19580–19589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Han GS, Mileykovskaya E, Garrett TA, Carman GM, 2015. Altered lipid synthesis by lack of yeast Pah1 phosphatidate phosphatase reduces chronological life span. J. Biol. Chem 290, 25382–25394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Stukey GJ, Jog R, Kwiatek JM, Han GS, Carman GM, 2022. Mutant phosphatidate phosphatase Pah1-W637A exhibits altered phosphorylation, membrane association, and enzyme function in yeast. J. Biol. Chem 101578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual F, Carman GM, 2013. Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim. Biophys. Acta 1831, 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual F, Hsieh L-S, Soto-Cardalda A, Carman GM, 2014. Yeast Pah1p phosphatidate phosphatase is regulated by proteasome-mediated degradation. J. Biol. Chem 289, 9811–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual F, Soto-Cardalda A, Carman GM, 2013. PAH1-encoded phosphatidate phosphatase plays a role in the growth phase- and inositol-mediated regulation of lipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem 288, 35781–35792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péterfy M, Phan J, Xu P, Reue K, 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet 27, 121–124. [DOI] [PubMed] [Google Scholar]
- Phan J, Reue K, 2005. Lipin, a lipodystrophy and obesity gene. Cell Metab. 1, 73–83. [DOI] [PubMed] [Google Scholar]
- Poole A, Poore T, Bandhakavi S, McCann RO, Hanna DE, Glover CV, 2005. A global view of CK2 function and regulation. Mol. Cell Biochem 274, 163–170. [DOI] [PubMed] [Google Scholar]
- Quon E, Sere YY, Chauhan N, Johansen J, Sullivan DP, Dittman JS, Rice WJ, Chan RB, Di PG, Beh CT, Menon AK, 2018. Endoplasmic reticulum-plasma membrane contact sites integrate sterol and phospholipid regulation. PLoS. Biol 16, e2003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MA, Mostofa MG, Ushimaru T, 2018. The Nem1/Spo7-Pah1/lipin axis is required for autophagy induction after TORC1 inactivation. FEBS J. 285, 1840–1860. [DOI] [PubMed] [Google Scholar]
- Reue K, Wang H, 2019. Mammalian lipin phosphatidic acid phosphatases in lipid synthesis and beyond: metabolic and inflammatory disorders. J. Lipid Res 60, 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LC, Hubbard EJ, Graves PR, Paoli-Roach AA, Roach PJ, Kung C, Haas DW, Hagedorn CH, Goebl M, Culbertson MR, ., 1992. Yeast casein kinase I homologues: an essential gene pair. Proc. Natl. Acad. Sci. U. S. A 89, 28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LC, Menold MM, Garrett S, Culbertson MR, 1993. Casein kinase I-like protein kinases encoded by YCK1 and YCK2 are required for yeast morphogenesis. Mol. Cell Biol 13, 2870–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz C, Cid VJ, Lussier M, Molina M, Nombela C, 1999. A large-scale sonication assay for cell wall mutant analysis in yeast. Yeast 15, 1001–1008. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S, 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasser T, Qiu QS, Karunakaran S, Padolina M, Reyes A, Flood B, Smith S, Gonzales C, Fratti RA, 2012. The yeast lipin 1 orthologue Pah1p regulates vacuole homeostasis and membrane fusion. J. Biol. Chem 287, 2221–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E, 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17, 6449–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SW, Weiss SB, Kennedy EP, 1957. The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem 228, 915–922. [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H, 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U. S. A 104, 10364–10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowdon C, Johnston M, 2016. A novel role for yeast casein kinases in glucose sensing and signaling. Mol. Biol. Cell 27, 3369–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Cardalda A, Fakas S, Pascual F, Choi HS, Carman GM, 2011. Phosphatidate phosphatase plays role in zinc-mediated regulation of phospholipid synthesis in yeast. J. Biol. Chem 287, 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi B, Kelstrup CD, Stoehr G, Frohlich F, Walther TC, Olsen JV, 2009. Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol. Biosyst 5, 1337–1346. [DOI] [PubMed] [Google Scholar]
- Soulard A, Cremonesi A, Moes S, Schutz F, Jeno P, Hall MN, 2010. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 21, 3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivas A, Villa-Garcia MJ, Henry SA, Carman GM, 2001. Phosphorylation of the yeast phospholipid synthesis regulatory protein Opi1p by protein kinase C. J. Biol. Chem 276, 29915–29923. [DOI] [PubMed] [Google Scholar]
- Stalder D, Novick PJ, 2016. The casein kinases Yck1p and Yck2p act in the secretory pathway, in part, by regulating the Rab exchange factor Sec2p. Mol. Biol. Cell 27, 686–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W-M, Han GS, Dey P, Carman GM, 2018. Protein kinase A phosphorylates the Nem1-Spo7 protein phosphatase complex that regulates the phosphorylation state of the phosphatidate phosphatase Pah1 in yeast. J. Biol. Chem 293, 15801–15814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W-M, Han G-S, Carman GM, 2014a. Cross-talk phosphorylations by protein kinase C and Pho85p-Pho80p protein kinase regulate Pah1p phosphatidate phosphatase abundance in Saccharomyces cerevisiae. J. Biol. Chem 289, 18818–18830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W-M, Han G-S, Carman GM, 2014b. Yeast Nem1-Spo7 protein phosphatase activity on Pah1 phosphatidate phosphatase is specific for the Pho85-Pho80 protein kinase phosphorylation sites. J. Biol. Chem 289, 34699–34708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W-M, Han G-S, Casciano J, Carman GM, 2012. Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p-Pho80p and Cdc28p-cyclin B kinases to regulate lipid synthesis in yeast. J. Biol. Chem 287, 33364–33376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villen J, 2013. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 10, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, 1994. Signal Transduction in Yeast. Yeast 10, 1753–1790. [DOI] [PubMed] [Google Scholar]
- Toke DA, Bennett WL, Dillon DA, Wu W-I, Chen X, Ostrander DB, Oshiro J, Cremesti A, Voelker DR, Fischl AS, Carman GM, 1998. Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding for diacylglycerol pyrophosphate phosphatase. J. Biol. Chem 273, 3278–3284. [DOI] [PubMed] [Google Scholar]
- Toke DA, Bennett WL, Oshiro J, Wu W-I, Voelker DR, Carman GM, 1999. Isolation and characterization of the Saccharomyces cerevisiae LPP1 gene encoding a Mg2+-independent phosphatidate phosphatase. J. Biol. Chem 273, 14331–14338. [DOI] [PubMed] [Google Scholar]
- Ugrankar R, Liu Y, Provaznik J, Schmitt S, Lehmann M, 2011. Lipin is a Central Regulator of Adipose Tissue Development and Function in Drosophila. Mol. Cell Biol 31, 1646–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente V, Maia RM, Vianna MC, Paco-Larson ML, 2010. Drosophila melanogaster lipins are tissue-regulated and developmentally regulated and present specific subcellular distributions. FEBS J. 277, 4775–4788. [DOI] [PubMed] [Google Scholar]
- Wang P-C, Vancura A, Mitcheson TGM, Kuret J, 1992. Two genes in Saccharomyces cerevisiae encode a membrane-bound form of casein kinase-1. Mol. Biol. Cell 3, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hoekstra MF, DeMaggio AJ, Dhillon N, Vancura A, Kuret J, Johnston GC, Singer RA, 1996. Prenylated isoforms of yeast casein kinase I, including the novel Yck3p, suppress the gcs1 blockage of cell proliferation from stationary phase. Mol. Cell Biol 16, 5375–5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann S, Fischer M, Koehler M, Neureuther K, Riegger G, Doering A, Schunkert H, Hengstenberg C, Baessler A, 2008. Genetic Variants Within the LPIN1 Gene, Encoding Lipin, Are Influencing Phenotypes of the Metabolic Syndrome in Humans. Diabetes 57, 209–217. [DOI] [PubMed] [Google Scholar]
- Wojcik P, Berlicki L, 2016. Peptide-based inhibitors of protein-protein interactions. Bioorg. Med. Chem. Lett 26, 707–713. [DOI] [PubMed] [Google Scholar]
- Xu X, Okamoto K, 2018. The Nem1-Spo7 protein phosphatase complex is required for efficient mitophagy in yeast. Biochem. Biophys. Res. Commun 496, 51–57. [DOI] [PubMed] [Google Scholar]
- Xu Z, Su W-M, Carman GM, 2012. Fluorescence spectroscopy measures yeast PAH1-encoded phosphatidate phosphatase interaction with liposome membranes. J. Lipid Res 53, 522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W-L, Bruno MEC, Carman GM, 1996. Regulation of yeast CTP synthetase activity by protein kinase C. J. Biol. Chem 271, 11113–11119. [DOI] [PubMed] [Google Scholar]
- Yang W-L, Carman GM, 1995. Phosphorylation of CTP synthetase from Saccharomyces cerevisiae by protein kinase C. J. Biol. Chem 270, 14983–14988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeharia A, Shaag A, Houtkooper RH, Hindi T, de LP, Erez G, Hubert L, Saada A, de KY, Eshel G, Vaz FM, Pines O, Elpeleg O, 2008. Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am. J. Hum. Genet 83, 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Verity MA, Reue K, 2014. Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab 20, 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript