Abstract
The impact of heavy-metal contamination on archaean communities was studied in soils amended with sewage sludge contaminated with heavy metals to varying extents. Fluorescent in situ hybridization showed a decrease in the percentage of Archaea from 1.3% ± 0.3% of 4′,6-diamidino-2-phenylindole-stained cells in untreated soil to below the detection limit in soils amended with heavy metals. A comparison of the archaean communities of the different plots by denaturing gradient gel electrophoresis revealed differences in the structure of the archaean communities in soils with increasing heavy-metal contamination. Analysis of cloned 16S ribosomal DNA showed close similarities to a unique and globally distributed lineage of the kingdom Crenarchaeota that is phylogenetically distinct from currently characterized crenarchaeotal species.
The presence of heavy metals in sewage sludge is often the main determinant restricting the application of sewage sludge to agricultural soil (28). After detailed assessments of the uptake and transfer of heavy metals into the food chain via crops, the Commission of the European Communities has set limits on the amount of selected heavy metals that can be added to agricultural soils receiving sewage sludge (8). However, even heavy-metal contamination that is below the upper limit set by the European Commission can have an effect on microbial community structure (1, 12, 13, 16, 32).
Many studies have focused on the effects of heavy metal on bacterial community structure (1, 16, 20, 30, 32, 34). However, no investigations have studied the effect of heavy-metal discharge on the archaean community. Until a few years ago, the domain Archaea was considered to consist only of methanogens that live under strict anoxic conditions and extremophiles that inhabit inhospitable environments (36, 39). Recent studies have shown that Archaea are also present in nonextreme environments, including marine (10, 11, 14, 27, 38a), freshwater (18), and terrestrial (3, 4, 21) ecosystems. This suggests that the Archaea also have ecological significance in nonextreme environments. Phylogenetic analysis of Archaea from nonextreme environments has revealed that they form a new cluster within the kingdom Crenarchaeota and are only distantly related to hitherto-described crenarchaea, as determined by 16S rRNA gene sequences. This group is now called the nonthermophilic Crenarchaeota and consists of at least four distinct phylogenetic clusters (4). All gene sequences isolated from soil are found in one of these clusters.
In this study, we analyzed the long-term effect of heavy metals on the archaean community in contaminated samples from five experimental field plots (Braunschweig, Germany) that received, between 1980 and 1991, different amounts of sludge and heavy metals (6). Using molecular methods such as fluorescent in situ hybridization (FISH) and denaturing gradient gel electrophoresis (DGGE), we analyzed differences in the abundance and diversity of Archaea in both heavy-metal-contaminated soil and uncontaminated soil. A comparative sequence analysis of 16S ribosomal DNA (rDNA) libraries showed that Archaea from heavy-metal-contaminated soils cluster within the group of terrestrial nonthermophilic Crenarchaeota.
MATERIALS AND METHODS
Soil characteristics and samples.
Soil samples, down to a 10-cm depth, were collected at the end of November 1994 from an experimental field site in Braunschweig, Germany. The plots were characterized by the following amendments: (i) N fertilization, (ii) low sludge and low metal, (iii) low sludge and high metal, (iv) high sludge and low metal, and (v) high sludge and high metal (Table 1). All plots had been planted with spring rape. The soil, with a matrix consisting of 5% clay, 50% silt, and 45% sand, received either 100 or 300 m3 of unamended or amended sludge ha−1 year−1 between 1980 and 1991 (Table 1). The amended sludge was spiked with heavy metals (Cd, Cu, Ni, and Zn) to increase the heavy-metal load to the upper limit set by the European Commission (6). During sampling, no pronounced differences in organic C (Table 1) were noticed among these soils, although the increase in total concentrations of heavy metals Cd, Cu, Ni, and Zn from the original, noncontaminated soil to the soil with the highest metal amendment was accompanied by a decrease in pH from 7.1 to 5.3 (Table 1).
TABLE 1.
Soil characteristics
| Soil (characteristic) | Treatment (ha−1 yr−1) | No. (107) of bacteria and archaea (g of soil [dry wt]−1) | % Organic C | pH | Heavy-metal content (mg kg of soil [dry wt]−1)a
|
|||
|---|---|---|---|---|---|---|---|---|
| Cd | Cu | Ni | Zn | |||||
| Control | 180 kg of N | 260 ± 10 | 0.88 | 7.1 | 0.4 | 12 | 9 | 56 |
| A (low sludge-low metal) | 100 m3 of sludge | 240 ± 9 | 1.07 | 6.8 | 0.6 | 21 | 11 | 102 |
| B (low sludge-high metal) | 100 m3 of sludge | 284 ± 12 | 1.10 | 6.9 | 0.8 | 43 | 16 | 163 |
| C (high sludge-low metal) | 300 m3 of sludge | 297 ± 12 | 1.48 | 5.5 | 1.1 | 37 | 16 | 216 |
| D (high sludge-high metal) | 300 m3 of sludge | 200 ± 9 | 1.51 | 5.3 | 2.7 | 94 | 32 | 359 |
Aqua regia digest (25).
Four samples from each of the five plots were sieved, pooled, and mixed before analysis. All five plots were investigated by DGGE analysis and FISH. For Archaea 16S rDNA library construction, two plots were studied, the low-sludge–low-metal- and high-sludge–high-metal-contaminated plots.
FISH.
Soil samples of 5 g each were fixed in 4% paraformaldehyde–phosphate-buffered saline (0.13 M NaCl, 7 mM Na2HPO4, and 3 mM NaH2PO4 [pH 7.2] in water) on ice for 3 h (17). The suspensions were centrifuged at 4,000 × g for 5 min, and the pellets were subsequently washed with phosphate-buffered saline, resuspended in 20 ml of 96% ethanol, and stored at −20°C at a concentration of 25 mg of soil (wet weight) ml−1. Before application to slides, 40 μl of the cell suspension was dispersed in 960 μl of 0.1% sodium pyrophosphate in distilled water by mild sonication for 30 s at a setting of 8 (B-12 sonifier; Branson, Danbury, Conn.) (40). Twenty microliters was subsequently spotted onto gelatin-coated slides [0.1% gelatin, 0.01% KCr(SO4)2], dried at 45°C for 30 min, and finally dehydrated sequentially in 50, 80, and 96% ethanol for 3 min (each).
Hybridization was performed with a fluorescent probe designed for the kingdom Archaea, ARCH915 (35). The oligonucleotide probe was synthesized with a Cy3 reactive fluorescent dye at the 5′ end (MWG Biotech, Ebersberg, Germany). The labelled oligonucleotide was diluted in distilled water to a concentration of 25 ng μl−1 and stored at −20°C. Hybridization was performed in 9 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl, 5 mM EDTA, 0.01% sodium dodecyl sulfate [SDS] [pH 7.2]) in the presence of 20% formamide, 1 μl of the probe (25 ng μl−1), and 1 μl of 4′,6-diamidino-2-phenylindole (DAPI) (200 ng/μl) at 42°C for 2 h (40). The slides were washed in buffer containing 20 mM Tris-HCl, 10 mM EDTA, 0.01% SDS, and 308 mM NaCl for 15 min at 48°C, rinsed in distilled water, and air dried (40).
Slides were mounted with AF1 solution (Citifluor, Canterbury, United Kingdom), and the preparations were examined with a Zeiss Axiophot microscope fitted for epifluorescence with a high-pressure mercury bulb (50 W), Zeiss 02 filter sets (G365, FT395, and LP420), and HQ-Cy3 filters (G535/50, FT565, and BT 610175; AHF Analysentechnik, Tübingen, Germany) at 1,000× magnification. DAPI-stained bacteria, Archaea, and cells hybridizing with the probe in 20 fields, selected at random and covering an area of 0.01 mm2 each, were counted. The fields were selected from a sample distributed over eight circular areas of 53 mm2 each.
DNA extraction.
DNA used for PCR amplification and cloning was isolated by direct lysis of the Archaea in the soil. Ten grams of soil was homogenized for 1 min in a Waring blender with 100 ml of Crombach buffer (9). Lysis was performed according to the method of Torsvik et al. (38). Ten milliliters of the soil homogenate was lysed by the addition of 5 mg of lysozyme (Sigma Chemical Co.) ml−1. After 1 h at 37°C, 0.2 mg of proteinase K (Sigma) ml−1 was added, and the suspension was further incubated at 37°C for 0.5 h. Thereafter, SDS was added to a final concentration of 1%. The mixture was then heated to 65°C for 15 min before the lysate was cleared by centrifugation at 10,000 × g for 10 min. The lysate was purified with the Wizard total DNA cleanup system from Promega (Madison, Wisc.).
PCR.
Part of the 16S rDNA was amplified from the total archaean DNA by PCR, with the GeneAmp 9600 thermocycler from Perkin-Elmer (Norwalk, Conn.). The primer sequences used for amplification were PRA46f (37) and PRU517r (22). The 100-μl reaction mixture contained the following: sterile distilled water, PCR buffer (standard 10× PCR buffer; Perkin-Elmer), 0.16% bovine serum albumin, deoxynucleoside triphosphates (each at 200 nM), primers (each at 100 μM), 2.5 U of Taq DNA polymerase (Perkin-Elmer), and template amplicon (1.9 ng). After an initial denaturation step at 95°C for 5 min, the reaction mixture was run for 30 cycles of 95°C for 0.5 min, 53°C for 0.5 min, and 72°C for 1.0 min, followed by 72°C for 6 min.
For DGGE analysis, amplicons from the PCR described above were used as templates in a new PCR with the primer PARC340f (complementing a region conserved among the Archaea [29]) and the universal primer PARC519r (29) with a GC clamp at the 5′ end. This seminested PCR format was necessary to obtain a sufficient amount of product for the DGGE. The 50-μl reaction mixture contained the following: sterile distilled water, PCR buffer (standard 10× PCR buffer; Perkin-Elmer), 0.16% bovine serum albumin, deoxynucleoside triphosphates (each at 200 nM), primers (each at 1 μM), 2.5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer), and template amplicon (1.9 ng). After an initial denaturation step at 96°C for 9 min, the reaction mixture was run for 35 cycles of 95°C for 0.5 min, 53°C for 1.3 min, and 72°C for 0.5 min, followed by 72°C for 6 min.
DGGE.
DGGE was performed with a Hoefer SE600 gel electrophoresis unit. PCR products were loaded onto 8% acrylamide gels (bisacrylamide gel stock solution, 37:5:1; Bio-Rad Laboratories, Inc.) and run with 0.5× TAE buffer (1× TAE is 0.04 M Tris base, 0.02 M sodium acetate, and 1.0 mM EDTA [pH adjusted to 7.4]). DGGE gels contained a 20 to 60% gradient of urea-formamide solution increasing in the direction of electrophoresis. A 100% urea-formamide solution is defined as 40% (vol/vol) formamide plus 7.0 M urea. DGGE was conducted at 60°C at a voltage of 20 V for 10 min and thereafter at 200 V for 3 h. The gels were stained for 1 h with a 1:10,000 dilution of SYBR Green II (Molecular Probes, Eugene, Oreg.) in 0.5× TAE buffer before photographing.
Cloning and restriction fragment length polymorphism.
PCR products from the primary PCRs were purified by preparative gel electrophoresis (1% low-melting-point NuSieve agar; FMC Bioproducts, Rockland, Maine), followed by purification with the PCR cleanup kit from Promega. The products were concentrated by precipitation by a standard procedure with sodium acetate (3 M) and 70% ethanol (31). Cloning into the pMOSBlue T vector was performed as described by the manufacturer (Amersham International Plc., Little Chalfont, Buckinghamshire, England). The clones were screened for α complementation by using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside) (31). One hundred micrograms of ampicillin per milliliter was added to Luria agar plates (10 g of tryptone, 5 g of yeast extract, 10 g of NaCl, 15 g of agar, 1,000 ml of distilled water) for the selection of positive clones. Approximately 150 blue colonies were picked from each cloning reaction from the two soil treatments. A total of 300 blue colonies were screened for positive inserts by PCR with T7 (TAATACGACTCACTATAGGG) and U-19mer (GTTTTCCCAGTCACGACGT) primers (Amersham). PCR was performed as described above but in a total volume of 50 μl with 1.25 U of Taq DNA polymerase (Perkin-Elmer). Template was added as whole cells. The products were verified by gel electrophoresis.
Amplified cloned inserts were cut with the restriction enzymes CfoI and HpaII (Promega). Restriction was carried out for 2 h at 37°C in a total volume of 10 μl containing 2 U of restriction enzymes and 9 μl of PCR product and restriction buffer. The resulting restriction fragment length polymorphism products were separated by gel electrophoresis at 7 V/cm for 2 h in a 2% agar containing 0.1 μg of ethidium bromide ml−1. The restriction products were visualized by UV excitation (Electronic Dual Light transilluminator; Ultra Lum, Carson, Calif.) and a charge-coupled device camera (Ultra Lum).
Sequence analysis.
A total of 11 of the cloned amplicons with different restriction patterns and containing 16S rDNA from the low-sludge–low-metal and high-sludge–high-metal soils were sequenced. Plasmid DNA was prepared from the clones by using the Qiaprep Plasmid Miniprep kit (Qiagen, Inc., Chatsworth, Calif.). Sequencing was performed by the Advanced Biotechnology Centre (Charing Cross and Westminster Centre, London, England). The whole insert, approximately 450 bp, was sequenced by using the T7 primer as a sequencing primer. These sequences were checked for chimeric artifacts by the CHECK-CHIMERA service provided by the Ribosomal Database Project (23). Sequences were aligned with the PILEUP program of the Genetics Computer Group Software package. Distance matrices and phylogenetic trees were made by using the CLUSTAL W program. The sequences were taxonomically assigned by using the nucleotide database at GenBank and BLAST (National Center for Biotechnology Information).
RESULTS AND DISCUSSION
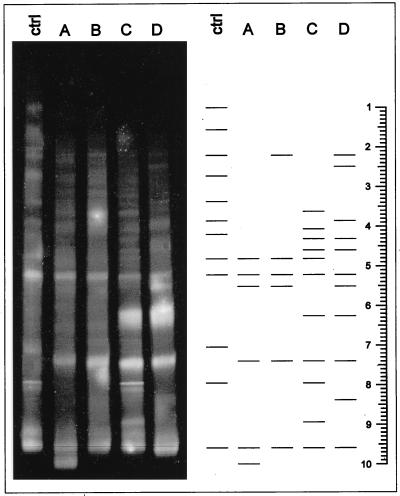
Application of DGGE revealed differences in the archaean community structure with increased heavy-metal contamination (Fig. 1). While the control soil resulted in a banding pattern of 12 bands, the low-metal soils, A and B, gave patterns of 6 bands each. However, soils amended with large amounts of heavy metals (C and D) gave banding patterns of 11 bands each. Two distinct bands (positions 5.2 and 9.6) were detected in all five plots (Fig. 1). The A and B soils showed similar banding patterns, except for one band that was unique to soil A (position 10.0) and another band that was unique to soil B (position 2.2) (Fig. 1). Likewise, the C and D soils also showed some similarities in DGGE banding patterns (Fig. 1). Three distinct bands (positions 4.3, 4.6, and 6.2) were detected only in these two plots. In addition, soils C and D each showed some faint, but unique, bands (positions 2.5, 3.1, 4.1, 8.4, and 8.9). Only one distinct band was detected in the metal-contaminated soil (position 7.4). The banding patterns from the low-sludge–low-metal plot (A) and the low-sludge–high-metal plot (B) were different from the control soil. The control, A, and B soils were somewhat similar with respect to pH (Table 1). Thus, the differences detected in the archaean community structure between the control soil and soils A and B could in part be attributed to the increase in the concentration of the heavy metals or the addition of sludge. These results were confirmed by another study showing a substantial reduction in total bacterial diversity between the control soil and soil A (low sludge-low metal) (32).
FIG. 1.
DGGE profile and schematic representations of the main bands of the DGGE gel of PCR-amplified 16S rDNA from Archaea in soil with different amendments. ctrl, soil with N fertilization; A, low sludge-low metal; B, low sludge-high metal; C, high sludge-low metal; D, high sludge-high metal.
The pHs of the A and B soils were in the same range as that of the control soil, while the pHs of the C and D soils were substantially lower (Table 1). The overall differences in the banding patterns between the control and A and B soils on one hand and the C and D soils on the other hand might therefore not be the effect of heavy metals alone but also that of an increase in the amount of sludge or a lowering in the pH (Table 1). pH has a great effect on the solubility of heavy metals (15). A twofold increase in heavy metals (Cd, Ni, and Zn) in solution, for example, has been observed after a one-unit decrease in pH (7, 33). However, differences in the banding patterns between the two low-sludge soils (A and B) and between the two high-sludge soils (C and D) must be attributed to the effect of heavy metals alone (Fig. 1).
FISH with probe ARCH915 showed that the number of Archaea belonging to this domain decreased from 1.3% ± 0.3% of the DAPI-stained cells in the control soil to numbers below the detection limit set at <1% of the DAPI-stained cells in contaminated soils (5, 40). This indicates that the heavy metals were somewhat toxic to the growth or survival of some of the Archaea. The numbers of Archaea observed in this study are in accordance with numbers found in other studies of soil ecosystems (5, 40).
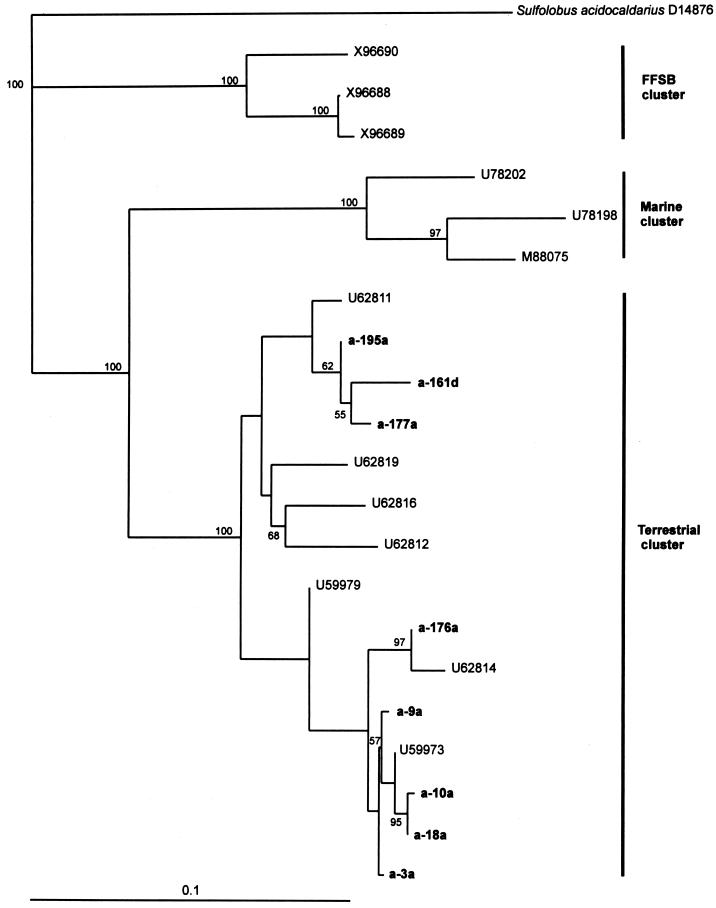
Phylogenetic analysis of the Archaea 16S rDNA clone libraries revealed from 95 to 98% sequence similarity to a novel group within the kingdom Crenarchaeota (2), also known as the nonthermophilic Crenarchaeota (4). Members within this group of Archaea have recently been detected from different environmental sources (4, 18, 19, 24, 26, 27), and they were shown to be phylogenetically distinct from currently characterized crenarchaeotal species. Phylogenetic analysis of the clones from these studies has shown that the clones belong to at least four groups, which appear to have a common ancestry (4). The sequences in this study feel into three groups within the terrestrial cluster of Crenarchaeota. Two groups (clones a-195a, a-161d, a-177a, and a-176a) were similar to clones from an agricultural soil in Wisconsin (U62811 and U62814) (2), while the third group (a-9a, a-10a, a-18a, and a-3a) showed sequence similarities to clones found in freshwater sediments (U59973) (18) (Fig. 2). In another study, sequences from freshwater sediments were also shown to group within the terrestrial cluster of nonthermophilic Crenarchaeota (4).
FIG. 2.
Neighbor-joining analysis showing the relationship of eight partial sequences from a 16S rDNA library from low-sludge–low-metal soil (with suffix “a”) and high-sludge–high-metal soil (with suffix “d”) to nonthermophilic Crenarchaeota 16S rDNA sequences. The FFSB cluster is a crenarchaeotal cluster from a forest soil in Finland (X96690, X96688, and X96689) (19). The marine cluster consists of the clones U78202, U78198 (24), and M88075 (10), while the terrestrial cluster contains clones from agricultural soil (U62811, U62812, U62814, U62816, and U62819) (2) and fresh water sediment (U59979 and U59973) (18). The numbers are GenBank accession numbers. Sequences determined in this study are in boldface type. Numbers at branching points show results from bootstrap analysis. The median number of changes per sequence position is shown on the scale bar.
Compared to noncontaminated soil, a significant reduction in the percentage of Archaea, as well as qualitative differences in the archaean community structure, was observed even at heavy-metal concentrations below the upper limit set by the European Commission. Molecular approaches have demonstrated that nonthermophilic Crenarchaeota are found in diverse environments and are globally distributed (2–4, 10, 18, 21, 24). Nonthermophilic Crenarchaeota have not been reported to occur in heavy-metal-contaminated soil. The findings in this study, that heavy metals were somewhat toxic to the growth or survival of some of the Archaea, may contribute to our knowledge about the ecological role of this novel group of organisms. Little is still known about their physiological characteristics and ecological significance, and further investigations are needed to fully comprehend their ecological importance.
ACKNOWLEDGMENTS
This work was supported by grants from the EC projects Microbial Diversity and Functions in Metal Contaminated Soils and High Resolution Automated Microbial Identification-2.
We also thank Bruce Knight (IACR-Rothamsted, Harpenden, Herts, United Kingdom) for measuring the heavy-metal concentrations in the soils used in the present study.
REFERENCES
- 1.Bååth E, Díaz-Raviña M, Frostegård ÅA, Campbell C D. Effect of metal-rich sludge amendments on the soil microbial community. Appl Environ Microbiol. 1998;64:238–245. doi: 10.1128/aem.64.1.238-245.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birtrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckley D H, Graber J R, Schmidt T M. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Appl Environ Microbiol. 1998;64:4333–4339. doi: 10.1128/aem.64.11.4333-4339.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatzinotas A, Sandaa R-A, Schönhuber W, Amann R I, Daae L F, Torsvik V, Zeyer J, Hahn D. Analysis of broad-scale differences in microbial communities of two pristine forest soils. Syst Appl Microbiol. 1998;21:588–592. doi: 10.1016/S0723-2020(98)80070-2. [DOI] [PubMed] [Google Scholar]
- 6.Chaudri A M, McGrath S P, Giller K E, Rietz E, Sauerbeck D R. Enumeration of indigenous Rhizobium leguminosarum biovar trifolii in soils previously treated with metal-contaminated sewage sludge. Soil Biol Biochem. 1993;25:301–309. [Google Scholar]
- 7.Christensen T H. Cadmium soil sorption at low concentrations. I. Effect of time, cadmium load, pH and calcium. Water Air Soil Pollut. 1984;21:105–114. [Google Scholar]
- 8.Commission of the European Communities. 1986. Council directive of 12 June 1986 on the protection of the environment, and in particular of the soil, when sewage sludge is used in agriculture. Journal of the European Communities No. L 181/6 (86/278/EEC).
- 9.Crombach W H J. DNA base composition of soil arthrobacters and other coryneforms from cheese and sea fish. Antonie Leeuwenhoek J Microbiol. 1972;38:105–120. doi: 10.1007/BF02328082. [DOI] [PubMed] [Google Scholar]
- 10.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong E F, Wu K Y, Prézelin B B, Jovine R V M. High abundance of archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 12.Frostegåard Å, Tunlid A, Bååth E. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl Environ Microbiol. 1993;59:3605–3617. doi: 10.1128/aem.59.11.3605-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frostegåard A, Tunlid A, Bååth E. Changes in microbial community structure during long-term incubation in two soils experimentally contaminated with metals. Soil Biol Biochem. 1996;28:55–63. [Google Scholar]
- 14.Fuhrman J A, McCallum K, Davis A A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 15.Giller K E, Witter E, McGrath S P. Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils: a review. Soil Biol Biochem. 1998;30:1389–1414. [Google Scholar]
- 16.Griffiths B S, Diaz-Raviña M, Ritz K, McNicol J W, Ebblewhite N, Bååth E. Community DNA hybridization and %G+C profiles of microbial communities from heavy metal polluted soils. FEMS Microbiol Ecol. 1997;24:103–112. [Google Scholar]
- 17.Hahn D, Amann R I, Ludwig W, Akkermans A D L, Schleifer K-H. Detection of microorganisms in soil after in situ hybridization with rRNA-targeted, fluorescently labelled oligonucleotides. J Gen Microbiol. 1992;138:879–887. doi: 10.1099/00221287-138-5-879. [DOI] [PubMed] [Google Scholar]
- 18.Hershberger K L, Barns S M, Reysenbach A-L, Dawson S C, Pace N R. Wide diversity of Crenarchaeota. Nature. 1996;384:420. doi: 10.1038/384420a0. [DOI] [PubMed] [Google Scholar]
- 19.Jurgens G, Lindström K, Saano A. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl Environ Microbiol. 1997;63:803–805. doi: 10.1128/aem.63.2.803-805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kandeler E, Kampichler C, Horak O. Influence of heavy metals on the functional diversity of soil microbial communities. Biol Fertil Soils. 1996;23:299–306. [Google Scholar]
- 21.Kudo Y, Shibata S, Miyaki T, Aono T, Oyaizu H. Peculiar archaea found in Japanese paddy soils. Biosci Biotechnol Biochem. 1997;61:917–920. [PubMed] [Google Scholar]
- 22.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massana R, Murray A E, Preston C M, DeLong E F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGrath S P, Cunliffe C H. A simplified method for the extraction of the metals Fe, Zn, Cu, Ni, Cd, Pb, Cr, Co and Mn from soils and sewage sludges. J Sci Food Agric. 1985;36:794–798. [Google Scholar]
- 26.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. Diversity of deep-sea hydrothermal vent Archaea at Loihi Seamount, Hawaii. Deep-Sea Res. 1998;45:303–317. [Google Scholar]
- 27.Murray A E, Preston C M, Massana R, Taylor L T, Blakis A, Wu K, DeLong E F. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obbard J P, Jones K C. The use of the cotton-strip assay to assess cellulose decomposition in heavy metal-contaminated sewage sludge-amended soils. Environ Pollut. 1993;81:173–178. doi: 10.1016/0269-7491(93)90083-z. [DOI] [PubMed] [Google Scholar]
- 29.Øvreåas L, Forney L, Daae F L, Torsvik V. Distribution of bacterioplankton in meromictic Lake Sælenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roane T M, Kellogg S T. Characterization of bacterial communities in heavy metal contaminated soils. Can J Microbiol. 1996;42:593–603. doi: 10.1139/m96-080. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Sandaa, R.-A., V. Torsvik, Ø. Enger, F. L. Daae, T. Castberg, and D. Hahn. Unpublished data. [DOI] [PubMed]
- 33.Sanders J R, McGrath S P, Adams T M. Zinc, copper and nickel concentrations in ryegrass grown on sludge-contaminated soils of different pH. J Sci Food Agric. 1986;37:961–968. [Google Scholar]
- 34.Smit E, Leeflang P, Wernars K. Detection of shifts in microbial community structure and diversity in soil caused by copper contamination using amplified ribosomal DNA restriction analysis. FEMS Microbiol Ecol. 1997;23:249–261. [Google Scholar]
- 35.Stahl D A, Amann R I. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1991. pp. 205–248. [Google Scholar]
- 36.Tindal B J. The archaebacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 677–808. [Google Scholar]
- 37.Torsvik, T., V. Torsvik, J. Keswani, and W. B. Whitman. Unpublished data.
- 38.Torsvik V, Goksøyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.van der Maarel M J E C, Artz R R E, Haanstra R, Forney L J. Association of marine archaea with the digestive tracts of two marine fish species. Appl Environ Microbiol. 1998;64:2894–2898. doi: 10.1128/aem.64.8.2894-2898.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woese C R, Kandler O, Wheelis L M. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zarda B, Hahn D, Chatzinotas A, Schönhuber W, Neef A, Amann R I, Zeyer J. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch Microbiol. 1997;168:185–192. [Google Scholar]