Abstract
A novel serine carboxypeptidase (EC 3.4.16.1) was found in an Aspergillus oryzae fermentation broth and was purified to homogeneity. This enzyme has a molecular weight of ca. 67,000, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and its specific activity is 21 U/mg for carbobenzoxy (Z)-Ala-Glu at pH 4.5 and 25°C. It has a ratio of bimolecular constants for Z-Ala-Lys and Z-Ala-Phe of 3.75. Optimal enzyme activity occurs at pH 4 to 4.5 and 58 to 60°C for Z-Ala-Ile. The N terminus of this carboxypeptidase is blocked. Internal fragments, obtained by cyanogen bromide digestion, were sequenced. PCR primers were then made based on the peptide sequence information, and the full-length gene sequence was obtained. An expression vector that contained the recombinant carboxypeptidase gene was used to transform a Fusarium venenatum host strain. The transformed strain of F. venenatum expressed an active recombinant carboxypeptidase. In F. venenatum, the recombinant carboxypeptidase produced two bands which had molecular weights greater than the molecular weight of the native carboxypeptidase from A. oryzae. Although the molecular weights of the native and recombinant enzymes differ, these enzymes have very similar kinetic parameters.
Commercial use of proteases having different origins for hydrolysis of food proteins appears to be very promising due to the nonchemical origin of enzymes. However, this technology has been hindered by the production of a bitter hydrolysate (6, 7). The bitterness has been ascribed to various peptides that are produced during enzymatic hydrolysis (8, 13, 14, 15). From the other side, free amino acids and short peptides play positive roles and elicit characteristic tastes of foods. Carboxypeptidases and aminopeptidases having different specificities, therefore, could be expected to be “natural” boosters of flavor during hydrolysis of food proteins. It has been shown that the debittering effects of both aminopeptidases (13) and carboxypeptidases (1, 23, 24) occur due to liberation of hydrophobic amino acids from the bitter peptides.
Serine carboxypeptidases (EC 3.4.16.1) are widely distributed in fungi, as well as in higher plants and animals, and they are particularly abundant in filamentous fungi (5). These enzymes are exopeptidases that specifically release amino acids from the C termini of peptides and proteins by utilizing a catalytic triad consisting of Ser, His, and Asp in their active sites. Their ability to cleave C-terminal amino acids during hydrolysis and their ability to replace C-terminal amino acid residues in transpeptidase reactions without internal cleavage of peptides have also attracted attention in peptide synthesis (3) and C-terminal sequencing (5) studies.
Here we describe a novel carboxypeptidase from Aspergillus oryzae which appears to be a major carboxypeptidase of Flavourzyme.
MATERIALS AND METHODS
Reagents.
The chemicals used as buffers and reagents were commercial products and were at least reagent grade. Flavourzyme, a commercial fungal extract obtained from A. oryzae fermentation broth, was obtained from Novo Nordisk A/S (Copenhagen, Denmark). Flavourzyme is produced by fermentation of a selected strain of A. oryzae and contains both endoprotease and exopeptidase activities. All peptide substrates were obtained from Sigma Chemical Co. (St. Louis, Mo.). Purification was performed with a Pharmacia fast protein liquid chromatography device with column supports and resins obtained from the same source. The Tricine gels and polyvinylidene difluoride membranes used for peptide separation and sequencing were obtained from Novex (San Diego, Calif.). All assays were performed with a Hitachi model U-2001 spectrophotometer. Quartz cuvettes with a 1-cm path length were used. The model 476A protein sequencer used was obtained from Applied Biosystems.
Purification of A. oryzae carboxypeptidase I.
Flavourzyme (20 ml) was diluted twofold in 20 mM sodium phosphate buffer (pH 7.0). Then the preparation was centrifuged at 11,000 × g for 10 min and filtered through a 0.45-μm-pore-size filter (Nalgene). The enzyme solution was diluted so that the conductivity was ∼2.4 mS and the pH was 7.0 by using 20 mM sodium phosphate buffer (pH 7.0) and then was loaded onto a Q-Sepharose Big Bead column (XK 26; ∼80 ml of resin) which had been preequilibrated with 20 mM sodium phosphate buffer (pH 7.0). The first gradient used for elution was a linear gradient from 100% 20 mM sodium phosphate buffer (pH 7.0) to 100% 20 mM sodium acetate buffer (pH 5.0) (flow rate, 5 ml/min; gradient volume, 300 ml; fraction size, 12.5 ml); and the final gradient was a linear pH gradient from 20 mM sodium acetate buffer (pH 4.3) to 100% 20 mM sodium acetate buffer (pH 3.5) (gradient volume, 400 ml; fraction size, 12.5 ml). The active fractions in the gradient from 20 mM sodium acetate buffer (pH 4.3) to 20 mM sodium acetate buffer (pH 3.5) were pooled, and the pH was adjusted to 7.0 with 20 mM dibasic sodium phosphate. A sample was then loaded onto a 20-ml Mono Q 16/10 column that had been preequilibrated with 20 mM sodium phosphate buffer (pH 7.0). The carboxypeptidase was eluted with a 0 to 0.3 M NaCl gradient at a flow rate of 5 ml/min (gradient volume, 500 ml; fraction size, 12.5 ml).
Physicochemical characterization.
Molecular mass and purity were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by using a minicell apparatus and 8 to 16% Tris–glycine gels (Novex). Isoelectric focusing (IEF) was performed by using a Novex pH 3 to 10 IEF gel. The manufacturer’s suggested method was used. Bio-Rad and Pharmacia IEF markers were used.
N-terminal sequencing of the purified carboxypeptidase was performed with an Applied Biosystems model 476A protein sequencer (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.) with on-line high-performance liquid chromatography and liquid phase trifluoroacetic acid delivery. Samples of the purified carboxypeptidase were transblotted onto polyvinylidene difluoride membranes (Novex) from SDS-PAGE gels, and the sequence was determined with a blot cartridge by using sequencing reagents (Perkin-Elmer, Applied Biosystems Division). Phenylthiohydantoin amino acids were detected by on-line high-performance liquid chromatography by using buffer A containing 3.5% tetrahydrofuran in water along with 15 to 30 ml of the Premix concentrate (Perkin-Elmer, Applied Biosystems Division) containing acetic acid, sodium acetate, and sodium hexanesulfonate and buffer B containing acetonitrile. Data were collected and analyzed by using a Macintosh IIsi computer and Applied Biosystems 610 data analysis software.
The purified carboxypeptidase was also cleaved with cyanogen bromide in order to generate peptide fragments of the enzyme for sequencing. The carboxypeptidase was digested with cyanogen bromide by reconstituting a dried sample of the purified carboxypeptidase in 70% formic acid with a few crystals of cyanogen bromide and incubating the preparation for 18 h at room temperature in the dark. The peptide fragments were separated by SDS-PAGE by using 10 to 20% Novex Tricine gels and were sequenced as described above.
Properties of the enzyme. (i) Carboxypeptidase assay.
Two methods were used for the carboxypeptidase assay. The first method was based on direct monitoring of decreases in the absorbance at 340 nm (A340) as a result of hydrolysis of N-(3-[furyl]acryloyl)-Ala-Lys (1 mM in 50 mM phosphate buffer, pH 6.0). The second method (the o-phthaldialdehyde [OPA] method) was based on detection of a free amino group. At 25°C, 50 μl of enzyme solution was added to 1 ml of a 0.5 mM carbobenzoxy (Z)-Ala-Glu solution in 20 mM sodium acetate buffer (pH 4.5). The release of free glutamic acid was monitored by using OPA and dithiothreitol (18). One unit of activity was defined as the quantity of enzyme which liberated 1 μmol of glutamic acid per min under the conditions described above for the second method.
(ii) Inhibition study.
The purified carboxypeptidase was incubated with 1,10-phenanthroline (which is known as an effective Zn chelator) at a concentration of 5 mM and phenylmethylsulfonyl fluoride at a concentration of 1 mM at pH 4.5 (20 mM acetate buffer). The residual activity was measured by using N-(3-[furyl]acryloyl)-Ala-Lys as a substrate at pH 4.5 in 20 mM acetate buffer. The release of free lysine was monitored by measuring the decrease in A340.
(iii) pH and temperature optima.
The optimal pH for enzymatic activity was determined by using a 50 mM sodium acetate–50 mM sodium phosphate–50 mM sodium citrate universal buffer at pH values ranging from 2.9 to 7.5. Z-Ala-Ile (concentration, 0.5 mM) was used as the substrate. The OPA method was used to quantitate the Ile that was released. The optimal temperature for enzymatic activity was determined by using 0.5 mM Z-Ala-Ile in 50 mM sodium acetate buffer (pH 4.0). One milliliter of substrate was preincubated for 30 min before the enzyme sample was added.
(iv) Thermal stability.
Thermal stability was studied by using 50 mM sodium acetate buffer (pH 4.0). A 50-μl enzyme solution was added to 450 μl of preheated buffer. The enzyme sample was incubated for 10 min, and residual activity was measured by using Z-Ala-Ile at pH 4.0. Incubations were performed at temperatures ranging from 35 to 75°C in 5°C increments.
(v) Substrate specificity.
The substrate specificity of carboxypeptidase was established by determining the bimolecular constants for a series of Z-Ala-X-OH compounds by using the carboxypeptidase assay method described above. The kcat/Km ratio was determined by using the Michaelis-Menten equation.
(vi) Sequential release of C-terminal amino acid residues from natural peptides.
Bradykinin and β-amyloid were dissolved in 1.4 and 1.0 ml of 50 mM acetate buffer (pH 4.5), respectively, to a final concentration of 0.55 mM. Carboxypeptidase was added to the peptide samples to final concentrations of 0.114 and 0.0878 U/ml for bradykinin and β-amyloid, respectively. After incubation at room temperature (21°C), 150-μl aliquots were added to 150-μl portions of 60 mM HCl to terminate the reactions. The resulting samples were then analyzed with a Beckman model 6300 automatic amino acid analyzer.
Cloning and expression. (i) Construction of cDNA library.
Total cellular RNA was extracted from A. oryzae mycelia harvested from a Flavourzyme production fermentation (Novo Nordisk A/S). Double-stranded cDNA was synthesized from 5 μg of poly(A)+ RNA as described by Kofod et al. (11). A library consisting of 106 independent clones was stored as individual pools (25,000 to 30,000 CFU/pool) in 20% glycerol at −80°C.
(ii) PCR amplification of carboxypeptidase I coding sequences.
Based on amino acid sequence data obtained from cyanogen bromide fragments of purified carboxypeptidase I, the following PCR primers were synthesized: forward primer 5′-dTAYGGNGGICAYTAYGGICCNG-3′ and reverse primer 5′-dATRAARTTIGGCCAIACRAARTC-3′ (R = A or G; Y = C or T; N = G, A, C, or T). PCR mixtures (100 μl) were prepared by using 1 μg of an A. oryzae cDNA library as the template. In addition, each reaction mixture contained the following components: 40 pmol of forward primer, 40 pmol of reverse primer, 200 μM dATP, 200 μM dCTP, 200 μM dGTP, 200 μM dTTP, 1× Taq polymerase buffer (Perkin-Elmer Corp., Branchburg, N.J.), and 2.5 U of Taq polymerase (Perkin-Elmer Corp.). The reaction mixtures were incubated in a Perkin-Elmer model 480 thermal cycler programmed as follows: cycle 1 consisting of 95°C for 5 min, 45°C for 2 min, and 67°C for 2 min; and cycles 2 through 30 consisting of 95°C for 2 min, 45°C for 1 min, and 67°C for 2 min. The reaction products were isolated on a 1% agarose gel and cloned into a pCRII vector (Invitrogen, San Diego, Calif.).
(iii) Identification of carboxypeptidase I clones.
The A. oryzae cDNA library was screened by colony hybridization (4) by using PCR-generated probes.
(iv) DNA sequence analysis of A. oryzae ATCC 20386 carboxypeptidase I gene.
DNA sequencing of one carboxypeptidase I clone (designated pEJG12) was performed with a model 373A automated DNA sequencer (Applied Biosystems, Inc., Foster City, Calif.); both strands were examined by using the primer walking technique with dye terminator chemistry (9). Oligonucleotide sequencing primers for complementary sequences in the carboxypeptidase I gene were designed and were synthesized with an Applied Biosystems model 394 DNA-RNA synthesizer used according to the manufacturer’s instructions.
(v) Construction of a carboxypeptidase I expression vector.
We designed the following two synthetic oligonucleotide primers to amplify the A. oryzae carboxypeptidase coding region from pEJG12 for subcloning into Fusarium expression vector pDM181: forward primer 5′-ATTTAAATATGCGTGGCTACGAATT-3′ and reverse primer 5′-GGGTTAATTAACTATGCCATACCAA-3′ (boldface type indicates the coding sequence).
In order to facilitate subcloning of the amplified fragment into pDM181, SwaI and PacI restriction enzyme sites were introduced at the 5′ and 3′ ends of the carboxypeptidase gene, respectively. Vector pDM181 contains the Fusarium oxysporum trypsinlike protease promoter and terminator as regulatory sequences (19). This plasmid also contains the bar gene (4) as a selectable marker for fungal transformation. The amplified carboxypeptidase cDNA fragment was isolated by gel electrophoresis and was purified by using QiaexII (Qiagen). The fragment was digested with SwaI and PacI and ligated with pDM181 that had been previously cut with SwaI and PacI, which resulted in expression plasmid pEJG15.
(vi) Transformation of Fusarium strain and analysis of transformants.
The recombinant DNA molecule (pEJG15) was introduced into the host organism by incubating the plasmid with protoplasts of Fusarium venenatum CC1-3. Transformants were selected by growing organisms on a medium containing phosphinothricin, the active ingredient in the herbicide BASTA (Hoechst). BASTA-resistant transformants were screened for carboxypeptidase activity. The vectors containing the F. oxysporum trypsinlike protease promoter and terminator, as well as the protocols used for transformation of F. venenatum, have been described previously (19).
Nucleotide sequence accession number.
The gene sequence which we determined has been deposited in the GENESEQN database under accession no. V28620.
RESULTS
Biochemical properties of native A. oryzae carboxypeptidase I.
The secreted carboxypeptidase obtained from A. oryzae was homogeneous after purification on two anion-exchange columns. When SDS-PAGE was used, a single component with a molecular weight of ca. 67,000 was detected. The protein sample also yielded a single band with a pI near 4.6 during IEF.
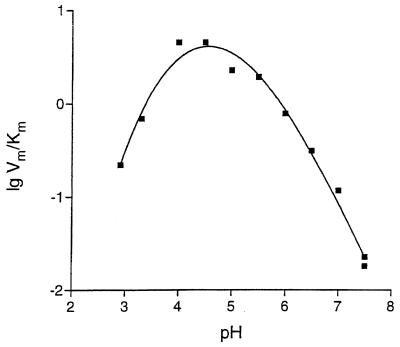
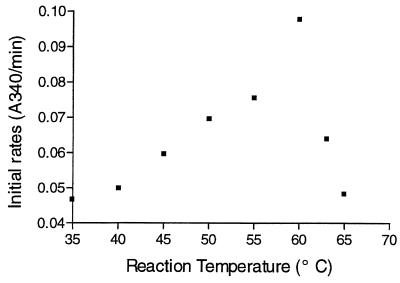
The theoretical extinction coefficient of the enzyme was calculated based on the known protein sequence (16). The specific activity at pH 4.5 for Z-Ala-Glu was 21 U/mg under the conditions described above; this value was calculated by assuming that the A280 of a 1-mg/ml solution of carboxypeptidase I was 1.74. The kcat/Km ratios for various Z-Ala-X compounds (Table 1) revealed no significant preference for any class of amino acids. This quality of the carboxypeptidase was also demonstrated by using natural peptides, such as bradykinin and β-amyloid, as substrates (Table 2). The pH dependence of the bimolecular constant kcat/Km at 25°C for Z-Ala-Ile in a Dixon-Webb plot resulted in a bell-shaped curve with a pronounced optimum at pH 4.0 to 4.5 (Fig. 1). The slopes of the right and left branches of the curve appeared to be −1 and 1, respectively. The curve could be described based on a simple rapid equilibrium diprotic model (20) with pK values of 3.3 and 5.7 for ionizable groups of the active site of an enzyme. The enzyme exhibited optimal activity for Z-Ala-Ile hydrolysis under the conditions used at temperatures ranging from 58 to 60°C (Fig. 2). Assuming that the level of activity for Z-Ala-Ile at 35°C and pH 4.0 was 100%, the residual activities at 55, 60, and 65°C after 10-min incubations were found to be 74, 65, and 34.5%, respectively.
TABLE 1.
Substrate specificities of A. oryzae native carboxypeptidase I at pH 4 and of P. janthinellum carboxypeptidase at pH 4.1, as determined with Z-Ala-X substratesa
| Substrate |
A. oryzae carboxypeptidase I
|
P. janthinellum carboxypeptidase kcat/Km (min−1 mM−1)b | ||
|---|---|---|---|---|
| Km (mM) | kcat (min−1) | kcat/Km (min−1 mM−1) | ||
| Z-Ala-Ile | 0.11 | 2,500 | 22,780 | 340 |
| Z-Ala-Glu | 0.10 | 3,340 | 33,410 | NDc |
| Z-Ala-Lys | 0.12 | 3,340 | 27,840 | 2,900 |
| Z-Ala-Arg | 0.11 | 2,820 | 25,620 | 3,400 |
| Z-Ala-Asp | 0.27 | 2,820 | 10,440 | 190 |
| Z-Ala-Asn | 0.09 | 1,980 | 22,040 | 230 |
| Z-Ala-Gly | 0.92 | 580 | 630 | 4 |
| Z-Ala-Phe | 0.24 | 1,770 | 7,400 | 1,600 |
| Z-Ala-Tyr | 0.09 | 1,830 | 20,330 | ND |
| Z-Glu-Tyr | 0.11 | 490 | 4,450 | ND |
A typical error in Km and kcat determinations did not exceed 11%.
Data from reference 2.
ND, not determined.
TABLE 2.
Release of C-terminal amino acids from bradykinin (5 nmol) and β-amyloid (5.25 nmol) by native A. oryzae carboxypeptidase Ia
| Substrate | Released amino acid | Amt of released amino acid (nmol) after incubation forb:
|
|||||
|---|---|---|---|---|---|---|---|
| 0 h | 0.5 h | 1 h | 3 h | 6 h | 24 h | ||
| Bradykininc | Arg | 0.78 | 4.92 | 4.89 | 4.94 | 4.92 | 5.00 |
| Phe | 0 | 1.29 | 2.39 | 4.99 | 6.80 | 9.60 | |
| Pro | 0 | 1.05 | 2.09 | 4.21 | 4.59 | 4.91 | |
| Ser | 0 | 1.00 | 2.04 | 4.12 | 4.45 | 4.95 | |
| Gly | 0 | 0 | 0 | 0 | 0 | 3.65 | |
| β-Amyloidd | Glu | 0.38 | 4.94 | 5.11 | 5.12 | 5.145 | 5.80 |
| Tyr | 0 | 2.24 | 4.02 | 5.09 | 5.075 | 5.25 | |
| Gly | 0 | 2.13 | 3.96 | 5.10 | 5.11 | 5.23 | |
| Ser | 0 | 1.68 | 3.76 | 5.10 | 5.09 | 5.24 | |
| Asp | 0 | 0.10 | 0.50 | 2.93 | 4.47 | 5.23 | |
| His | 0 | NAe | NA | NA | NA | 2.11 | |
| Arg | 0 | 0 | 0 | 0 | 0.61 | 2.09 | |
| Phe | 0 | 0 | 0 | 0 | 0.47 | 2.17 | |
The reaction conditions are described in the text.
Means based on three measurements. A typical error in a concentration determination did not exceed 5%.
The sequence of bradykinin is Arg-Pro-Pro-Gly-Phe-Ser-Pro-Phe-Arg.
The sequence of β-amyloid is Asp-Ala-Glu-Phe-Arg-His-Asp-Ser-Gly-Tyr-Glu.
NA, the values for histidine could not be determined precisely due to the presence of a coeluting peak.
FIG. 1.
pH dependence of hydrolysis of Z-Ala-Ile catalyzed by the carboxypeptidase (25°C, 50 mM sodium acetate–50 mM sodium phosphate–50 mM sodium citrate universal buffer). A typical error in Km and kcat determinations did not exceed 15%.
FIG. 2.
Temperature dependence of hydrolysis of Z-Ala-Ile catalyzed by the carboxypeptidase (50 mM acetate buffer, pH 4.0; [Z-Ala-Ile], 1.5 mM; [E], 0.15 mg/ml). The means shown were based on three measurements. A typical error in concentration determinations did not exceed 6%.
No decline in carboxypeptidase activity was observed after 110 min of incubation with 1,10-phenanthroline at a concentration of 5 mM, but 110 min of incubation with 1 mM phenylmethylsulfonyl fluoride resulted in a loss of all carboxypeptidase activity.
N-terminal sequencing of carboxypeptidase I revealed that the N terminus was blocked. One of the cyanogen bromide fragments had the following amino acid sequence (a question mark indicates that the residue could not be determined; underlined residues are identical to residues in carboxypeptidase S1 of Penicillium janthinellum): ?YGGHYGPAFFNHFYEQNE (peptide 1).
Sequencing of the cyanogen bromide fragments revealed the following peptide sequences which were 30 to 40% homologous to sequences in the carboxypeptidase of P. janthinellum (21) (residues in parentheses were not determined with absolute certainty): DAIGVNI?YTQ?NNDVYYA (peptide 2; 50 kDa); DAIGVNI(N)YTQSNN(D)VYYAFQQTGDFVWPNFIEDL (peptide 3; 42 kDa); and ?CRDNVEGP(?)YAFAGRGVYDIRHPYDPDT (peptide 4; 17 kDa).
Sequence analysis of cDNA encoding A. oryzae carboxypeptidase I.
A carboxypeptidase I gene segment consisting of approximately 186 codons (550 bp) was amplified from A. oryzae cDNA with the carboxypeptidase-specific PCR primers. Screening of the A. oryzae cDNA library resulted in six colonies that exhibited strong hybridization signals with the probe. The carboxypeptidase-encoding plasmids were confirmed by DNA sequencing.
Sequence analysis of the longest cloned insert revealed an open reading frame of 1,665 nucleotides (excluding the stop codon) that encoded a 555-amino-acid protein. The G+C content of this open reading frame was 52.1%. Based on the rules of von Heijne (25), the first 18 amino acids likely comprise a secretory signal peptide which directs the nascent polypeptide into the endoplasmic reticulum.
The deduced amino acid sequence of A. oryzae ATCC 20386 carboxypeptidase I (Fig. 3) indicates that the calculated molecular weight of the primary translation product is 61,200, which is consistent with the estimated molecular weight of 67,000, which was based on the mobility of the purified protein on SDS-PAGE gels. There are 12 potential sites for N-linked glycosylation in the deduced amino acid sequence. The amino acid sequences of peptides derived from purified carboxypeptidase I are consistent with the deduced amino acid sequence encoded by A. oryzae carboxypeptidase I cDNA.
FIG. 3.
Nucleotide sequence and deduced amino acid sequence of A. oryzae carboxypeptidase. The amino acid sequences of the peptide fragments obtained from cyanogen bromide cleavage of the wild-type carboxypeptidase are underlined. The arrows indicate PCR primers. The putative signal peptide is enclosed in a box. Boldface type indicates the amino acid residues that form the catalytic triad.
The deduced amino acid sequence of A. oryzae carboxypeptidase I has a catalytic triad consisting of Asp-His-Ser, which is conserved in serine carboxypeptidases.
The Clustal alignment program (10) was used to compare the amino acid sequence of our carboxypeptidase, deduced from the cloned gene, to the available amino acid sequences of other fungal carboxypeptidases. Two fungal carboxypeptidases, carboxypeptidases of P. janthinellum and Absidia zychae, exhibited the highest levels of sequence identity (∼40%) with carboxypeptidase I. Three other fungal carboxypeptidases, P. janthinellum carboxypeptidase S2, Aspergillus niger carboxypeptidase, and Aspergillus phoenicis carboxypeptidase, were 13.3, 14.7, and 16.4% identical, respectively.
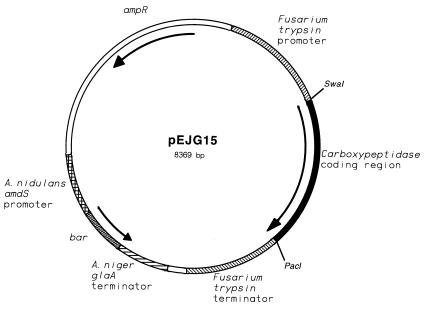
The coding region of carboxypeptidase I cDNA was amplified by performing PCR with primers which introduced a SwaI site at the start codon and a PacI site following the stop codon (see above). The amplified DNA segment was subcloned into an expression vector that contained the F. oxysporum trypsin promoter and terminator regions and a selectable marker (bar) that conferred resistance to phosphinothricin (Fig. 4). Transformants were subsequently screened for the ability to produce extracellular carboxypeptidase activity in shake flask cultures. The transformant that secreted the highest level of activity was grown in a shake flask, and the recombinant carboxypeptidase was purified from the culture broth. The purified recombinant carboxypeptidase was digested by using the cyanogen bromide method that was previously used to digest the wild-type carboxypeptidase. The N-terminal sequences of the resulting peptides revealed that the peptides were all predicted carboxypeptidase cyanogen bromide fragments.
FIG. 4.
Plasmid used for expression of the carboxypeptidase in F. venenatum. The F. oxysporum trypsinlike protease promoter and terminator regions were used for expression of the recombinant carboxypeptidase. The Aspergillus nidulans amdS promoter was used to express the bar gene. The bar gene codes for a Streptomyces hydroscopicus phosphinothricin acetyltransferase. The polyadenylation and transcription termination region was derived from the Aspergillus niger glaA (glucoamylase) gene.
Properties of recombinant carboxypeptidase I.
Purified F. venenatum carboxypeptidase I produced two bands at molecular weights near 110,000 and 90,000 on SDS-PAGE gels. This finding was attributed to the presence of two different glycoforms of recombinant carboxypeptidase I. Enzymatic deglycosylation of a preparation containing the two components of F. venenatum carboxypeptidase I with endoglycosidase F/N-glycosidase F resulted in a homogeneous protein with a molecular weight near 60,000 (as determined by SDS-PAGE).
The values of kinetic parameters of F. venenatum carboxypeptidase I for various Z-Ala-X substrates were very similar to the values obtained for wild-type carboxypeptidase I (data not shown) if it was assumed that the molecular weight of the recombinant protein was 60,000. It is important that the enzyme did not exhibit a significant preference for any class of amino acids.
DISCUSSION
Our inhibition study and sequence homology results unequivocally show that carboxypeptidase I is a serine carboxypeptidase. Based on preferences for hydrophobic (Phe) and basic (Lys) amino acids at the P1′ position, two subclasses (the carboxypeptidase C and carboxypeptidase D subclasses, respectively) were identified (17). The preferences are determined by examining the ratio of the bimolecular constant for Z-Ala-Lys to the bimolecular constant for Z-Ala-Phe. Carboxypeptidase Y from yeast has a low ratio (0.11) and therefore is considered a member of the carboxypeptidase C subclass. Carboxypeptidase AII, a member of the carboxypeptidase D subclass, has by far the highest ratio, 1,300. We found (Table 1) that the ratio of the bimolecular constants for Z-Ala-Lys and Z-Ala-Phe is ∼3.75. This finding does not definitively identify carboxypeptidase I as a member of the carboxypeptidase D subclass. P. janthinellum produces a carboxypeptidase (2) that has a ratio of 1.8 (Table 1). This enzyme also cannot be placed into either class definitively. It is reasonable to place these enzymes in a subclass of serine carboxypeptidases which do not exhibit a dramatic preference for basic amino acids over hydrophobic amino acids at the P1′ position or vice versa.
Carboxypeptidase I possesses unique properties. It is very favorable for protein hydrolysis that this enzyme does not exhibit a preference for any class of amino acids. Indeed, the Km values for the nonpolar amino acid Ile, the polar but not ionized amino acid Asn, and the oppositely charged ionogenic amino acids Glu and Arg during catalytic hydrolysis of Z-Ala-X dipeptides are comparable, as are the kcat values (Table 1). Apparently, Gly and Pro are less preferable amino acids for carboxypeptidase I due to the small size of Gly and the special structure of Pro. In spite of this, carboxypeptidase I effectively goes through both Gly and Pro in bradykinin and β-amyloid and even hydrolyzes a Pro-Gly peptide bond. A comparative study of the release of amino acids during hydrolysis of these natural substrates led to the conclusion that both Gly and Pro hinder hydrolysis of the C-terminal amino acid when they are present at the penultimate position.
A study of the pH dependence of catalytic hydrolysis of Z-Ala-Ile (Fig. 1) and Z-Ala-Glu (data not shown) revealed a peak of carboxypeptidase activity around pH 4.0 to 4.5. This pH is not optimal for hydrolysis of, for example, soy protein, which normally starts at neutral pH. However, Fig. 1 shows that the value of the bimolecular constant for hydrolysis of Z-Ala-Ile at pH 4.0 to 4.5 is less than sixfold greater than the value at pH 6.0.
Several other carboxypeptidases with molecular weights near 60,000 have been found in A. oryzae; these enzymes include carboxypeptidase O-1, carboxypeptidase O-2 (22), and carboxypeptidase III (15). Although the previously described data are not sufficient for detailed comparisons of these three enzymes and carboxypeptidase I to be made, the parameter values for hydrolysis of Z-Glu-Tyr by carboxypeptidases O-1 and O-2 obviously show that these two enzymes have much lower (approximately 25-fold lower) specific activities and narrower pH stabilities (pH 5.0 to 6.0) than carboxypeptidase I. In contrast to carboxypeptidase I, carboxypeptidase III (15) has a greater preference for Z-Glu-Tyr than for Z-Ala-Glu and a temperature optimum of 40°C, while carboxypeptidase I is most active at temperatures between 55 and 60°C.
F. venenatum A3/5 has been shown to be a promising host for production of fungal enzymes. Previous studies have demonstrated that this organism is able to produce a variety of recombinant enzymes, such as cellulase, protease, and lipase (19). In addition, its low background levels of proteases and other secreted proteins are advantageous for production of relatively pure enzyme products (19). In the present study, we employed F. venenatum as a host for expression of A. oryzae carboxypeptidase I. Expression of recombinant carboxypeptidase I was confirmed based on activity and protein sequence data. Carboxypeptidase I from F. venenatum and A. oryzae carboxypeptidase were found to be very similar in all parameters except molecular weight. The enzyme from F. venenatum is heavily glycosylated. Both carboxypeptidases exhibited broad substrate specificity, which is uncommon for serine carboxypeptidases (17).
The ability of carboxypeptidase I expressed in the edible fungus F. venenatum A3/5 to release all C-terminal amino acids, including Gly and Pro, and the relatively high specific activity and high thermostability of this enzyme make it a promising candidate for use in production of protein hydrolysates of different origins and in flavor-improving processes in the food industry.
ACKNOWLEDGMENTS
We thank Sakari Kauppinen for providing the library coding sequences. Alan Klotz and Randy Berka are acknowledged for helpful discussions and for critical reading of the manuscript.
REFERENCES
- 1.Arai S, Noguchi M, Kurosawa S, Kato H, Fujimaki M. Applying proteolytic enzymes on soybean. 6. Deodorization effect of aspergillopeptidase A and debittering effect of Aspergillus acid carboxypeptidase. J Food Sci. 1970;35:392–395. [Google Scholar]
- 2.Breddam K. Carboxypeptidase from Penicillium janthinellum: enzymatic properties in hydrolysis and aminolysis reactions. Carlsberg Res Commun. 1988;53:309–320. doi: 10.1007/BF02904436. [DOI] [PubMed] [Google Scholar]
- 3.Breddam K, Widmer F, Johansen J T. Carboxypeptidase Y catalyzed transpeptidations and enzymatic peptide synthesis. Carlsberg Res Commun. 1980;45:227–247. [Google Scholar]
- 4.de Block M, Botterman J, Vandewiele M, Dockx J, Thoen C, Gossele V, Movva N R, Thompson C, van Montagu M, Leemans J. Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J. 1987;6:2513–2518. doi: 10.1002/j.1460-2075.1987.tb02537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Degan F D, Ribadeau-Dumas B, Breddam K. Purification and characterization of two serine carboxypeptidases from Aspergillus niger and their use in C-terminal sequencing of proteins and peptide synthesis. Appl Environ Microbiol. 1992;58:2144–2152. doi: 10.1128/aem.58.7.2144-2152.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujimaki M, Arai S, Yamashita M, Kato H, Noguchi M. Taste peptide fraction from a fish protein hydrolysate. Agric Biol Chem. 1973;37:2891–2898. doi: 10.1021/jf60197a003. [DOI] [PubMed] [Google Scholar]
- 7.Fujimaki M, Kato H, Arai S, Tamaki E. Applying proteolytic enzymes on soybean. 1. Proteolytic enzyme treatment of soybean and its effect on the flavor. Food Technol. 1968;22:889–892. [Google Scholar]
- 8.Fujimaki M, Yamashita M, Okazawa Y, Arai S. Applying proteolytic enzymes on soybean. 3. Diffusable bitter peptides and free amino acids in peptic hydrolysate of soybean protein. J Food Sci. 1970;35:215–218. [Google Scholar]
- 9.Giesecke H, Obermaier B, Domdey H, Neubert W J. Rapid sequencing of the Sendai virus 6.8 kb large (L) gene through primer walking with an automated DNA sequencer. J Virol Methods. 1992;38:47–60. doi: 10.1016/0166-0934(92)90168-d. [DOI] [PubMed] [Google Scholar]
- 10.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 11.Kofod L, Kauppinen S, Christgau S, Andersen L N, Heldt-Hansen H P, Dörreich K, Dalbøge H. Cloning and characterization of two structurally and functionally divergent rhamnogalacturonases from Aspergillus aculeatus. J Biol Chem. 1994;269:29182–29189. [PubMed] [Google Scholar]
- 12.Matoba T, Hayashi R, Hata T. Isolation of bitter peptides from tryptic hydrolysate of casein and their chemical structure. Agric Biol Chem. 1970;34:1235–1243. [Google Scholar]
- 13.Matsuoka H, Fuke Y, Kaminogawa S, Yamauchi K. Purification and debittering effect of aminopeptidase II from Penicillium caseicolum. J Agric Food Chem. 1991;39:1392–1395. [Google Scholar]
- 14.Minagawa E, Kaminogawa S, Tsukasaki F, Yamauchi K. Debittering mechanism in bitter peptides of enzymatic hydrolysates from milk casein by aminopeptidase T. J Food Sci. 1989;54:1225–1229. [Google Scholar]
- 15.Nakadai T, Nasuno S, Iguchi N. Purification and properties of acid carboxypeptidase III from Aspergillus oryzae. Agric Biol Chem. 1972;36:1481–1488. [Google Scholar]
- 16.Pace C N, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2433. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Remington S J, Breddam K. Carboxypeptidases C and D. Methods Enzymol. 1994;244:231–248. doi: 10.1016/0076-6879(94)44020-4. [DOI] [PubMed] [Google Scholar]
- 18.Roth M. Fluorescence reaction for amino acids. Anal Chem. 1971;43:880–882. doi: 10.1021/ac60302a020. [DOI] [PubMed] [Google Scholar]
- 19.Royer J C, Moyer D M, Reiwitch S G, Madden M S, Jensen E B, Brown S H, Yonker C C, Johnstone J A, Golightly E J, Yoder W T, Shuster J R. Fusarium graminearum A 3/5 as a novel host for heterologous protein production. Bio/Technology. 1995;13:1479–1483. doi: 10.1038/nbt1295-1479. [DOI] [PubMed] [Google Scholar]
- 20.Segel I H. Enzyme kinetics. Behavior and analysis of rapid equilibrium and steady-state enzyme systems. New York, N.Y: John Wiley & Sons, Inc.; 1993. pp. 888–898. [Google Scholar]
- 21.Svendsen I, Hofmann T, Endrizzi J, Remington S J, Breddam K. The primary structure of carboxypeptidase S1 from Penicillium janthinellum. FEBS Lett. 1993;333:39–43. doi: 10.1016/0014-5793(93)80371-z. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi M, Ushijima T, Ichishima E. A new acid carboxypeptidase, O-1, from Aspergillus oryzae. Curr Microbiol. 1982;7:19–23. [Google Scholar]
- 23.Umetsu H, Ichishima E. Mechanism of digestion of bitter peptides from a fish protein concentrate by wheat carboxypeptidase. Nippon Shokuhin Kogyo Gakkaishi. 1985;32:281–287. [Google Scholar]
- 24.Umetsu H, Matsuoka H, Ichishima E J. Debittering mechanism of bitter peptides from milk casein by wheat carboxypeptidase. Agric Food Chem. 1983;31:50–53. [Google Scholar]
- 25.von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984;173:243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]