Abstract
The shift from outcrossing to self-fertilization (selfing) is considered one of the most prevalent evolutionary transitions in flowering plants. Selfing species tend to share similar reproductive traits in morphology and function, and such a set of traits is called the ‘selfing syndrome’. Although the genetic basis of the selfing syndrome has been of great interest to evolutionary biologists, knowledge of the causative genes or mutations was limited until recently. Thanks to advances in population genomic methodologies combined with high-throughput sequencing technologies, several studies have successfully unravelled the molecular and genetic basis for evolution of the selfing syndrome in Capsella, Arabidopsis, Solanum and other genera. Here we first introduce recent research examples that have explored the loci, genes and mutations responsible for the selfing syndrome traits, such as reductions in petal size or in pollen production, that are mainly relevant to pre-pollination processes. Second, we review the relationship between the evolution of selfing and interspecific pollen transfer, highlighting the findings of post-pollination reproductive barriers at the molecular level. We then discuss the emerging view of patterns in evolution of the selfing syndrome, such as the pervasive involvement of loss-of-function mutations and the relative importance of selection versus neutral degradation.
This article is part of the theme issue ‘Genetic basis of adaptation and speciation: from loci to causative mutations’.
Keywords: mating systems, selfing, selfing syndrome, interspecific pollen transfer, genome-wide association study, relaxed selection
1. Introduction
Ever since Charles Darwin's seminal works, one of the central challenges in evolutionary biology has been to unravel the general patterns and mechanisms underlying the evolution of adaptive traits. Identification of causal genes or mutations can help us in this understanding, thereby obtaining information on the number, dominance and effect size of responsible mutations as well as the timing, repeatability and strength of selection involved [1–3].
A major way to reveal general patterns is to focus on the recurrent evolution of the same traits across independent taxa or populations [3–5]. Parallel evolution of self-fertilization (selfing) in flowering plants has been an excellent platform for studies of recurrent patterns in adaptive evolution for many years [6,7]. First, selfing has evolved a number of times independently in angiosperms, and the shift from outcrossing to selfing is considered one of the most prevalent evolutionary transitions in flowering plants [8–10]. Second, selfing is suggested to be adaptive; although selfing is considered detrimental because of inbreeding depression, it can be advantageous because of its transmission advantage and reproductive assurance. The transmission advantage for selfers is its efficiency in transmitting gametes compared with outcrossers. Selfers can become both the ovule and pollen donor for their own progeny and the pollen donor for outcrossed progeny, whereas obligate outcrossers cannot be pollen donors for their selfed progeny [11]. Another advantage of selfers is reproductive assurance, by enabling reproduction when pollinators or mates are scarce [12].
Recurrent evolutionary patterns in selfing populations or species have been studied intensively for decades. Selfing species often share numbers of reproductive, life-history, genomic and population-level properties. First, the evolution of selfing is generally accompanied by the loss of self-incompatibility (SI), which is the major mechanism preventing selfing in flowering plants. Second, the evolution of selfing has a large effect on the various properties of genome-wide polymorphisms, including nucleotide diversity and the density of transposable elements. Third, selfing species tend to share similar reproductive traits in morphology and function, and such a set of traits is often called the ‘selfing syndrome’ [6,13,14]. The typical traits of the selfing syndrome include small flower size, reduced pollen number, smaller pollen-to-ovule (P/O) ratios, loss of herkogamy or dichogamy and reduced scent or nectar production.
Because of its ecological and evolutionary significance, the genetic basis of the selfing syndrome has been of great interest, but studies that have successfully identified genes or mutations involved in the selfing syndrome remain limited, mainly because these traits are generally quantitative and controlled by multiple genetic and environmental factors, making it difficult to pinpoint the causal genes and mutations. Nevertheless, thanks to advances in population genomic methodologies combined with high-throughput sequencing technologies [15–17], there are now several reports on the genes and mutations involved in the evolution of the selfing syndrome, which we highlight in this review (table 1).
Table 1.
Identified genes involved in the traits related to the selfing syndrome. QTL, quantitative trait loci; GWAS, genome-wide association study.
| trait | gene name | study species | methodology for identification | reference |
|---|---|---|---|---|
| reduction in petal size | STERILE APETALA (SAP) | Capsella rubella | QTL mapping | Sicard et al. [18] |
| reduction in petal size | CYP724A1 | Capsella rubella | QTL mapping | Fujikura et al. [19] |
| loss of floral scent emission (benzaldehyde) | Cinnamate : CoA ligase (CNL1) | Capsella rubella | QTL mapping | Sas et al. [20] |
| reduction in pollen number | Reduced Pollen Number 1 (RDP1) | Arabidopsis thaliana | GWAS | Tsuchimatsu et al. [21] |
| loss of interspecific incompatibility | Stigmatic Privacy 1 (SPRI1) | Arabidopsis thaliana | GWAS | Fujii et al. [22] |
| instability in petal number | APETALA1 (AP1) and QTLs | Cardamine hirsuta | QTL mapping | Monniaux et al. [23] |
| loss of herkogamy (change in style length) | Style2.1 | Solanum lycopersicum | QTL mapping | Chen et al. [24] |
| loss of herkogamy (change in style length) | SE3.1 | Solanum lycopersicum | GWAS | Shang et al. [25] |
While several traits are considered hallmarks of the selfing syndrome, these can be classified into a few categories. Shimizu & Tsuchimatsu [14] proposed the following categories: (1) reduced allocation to outcrossing-related traits, particularly those involved in pollinator attraction, such as flower number and size, nectar and scent; (2) reproductive traits that promote selfing, such as loss of heterogamy and dichogamy enabling auto-pollination by mechanical contact between mature stigmas and anthers; and (3) reduced allocation to male functions and increased allocation to female functions, as often represented by reduced P/O ratios or the dry weight ratios of male versus female organs. These traits, which typically have been interpreted as composing the selfing syndrome, are mostly relevant to pre-pollination processes, such as those involved in pollinator attraction, in resource allocation to male and female gametes, or in the efficiency of self-pollination. On the other hand, there are several traits involved in post-pollination processes that would also have evolved associated with the evolution of selfing, such as reproductive barriers between species, reduced parent–offspring conflict, or reductions in seed size. Here, we consider the meaning of the selfing syndrome more broadly and also feature the traits related to post-pollination processes (but see also reviews in [14,26] mentioning some of these traits as part of the selfing syndrome). We also discuss how these traits involved in post-pollination processes are related to the ‘weak inbreeder/strong outbreeder’ (WISO) hypothesis, postulating that outcrossing parents ‘overpower’ selfing parents because of higher parental conflicts [27].
The evolution of selfing from outcrossing has profound genomic consequences via decreased effective population size and changes in the efficacy and the mode of selection, leaving various molecular signatures on the genomes [28–31]. Although these genomic signatures are sometimes interpreted as part of the selfing syndrome (the genomic selfing syndrome’) [31,32], we will not focus on these aspects, because there are already outstanding reviews on the genomic consequences of mating system transitions [28–31,33].
In this review, we first introduce recent research examples that have explored the loci, genes and mutations responsible for the evolution of the traits involved in the selfing syndrome, featuring studies in the genera Capsella, Arabidopsis and Solanum. These include reductions in petal size, floral scent or pollen number, which are mainly relevant to pre-pollination processes. Second, we review the relationship between the evolution of selfing and interspecific pollen transfer (IPT), highlighting recent studies of post-pollination reproductive barriers at the molecular level. Third, we discuss other notable traits that are associated with the evolution of selfing. Based on these findings at the molecular levels, we then discuss the emerging view of the pattern in the evolution of selfing syndrome traits, such as the pervasive involvement of loss-of-function mutations, adaptive significance of the selfing syndrome and the relative importance of selection versus neutral degradation.
2. The genetic basis of the selfing syndrome: traits relevant to pre-pollination processes
The genetic basis of the selfing syndrome has been studied in multiple genera, including Leptosiphon, Mimulus, Solanum, Ipomoea, Capsella, Collinsia and Arabidopsis, mostly through transcriptome analysis or quantitative genetic approaches such as qualitative trait locus (QTL) mapping [24,34–39]. In general, QTL studies have identified several small- to large-effect loci for each trait, but in most cases, the causal genes underlying QTLs have not been determined until recently. Partly thanks to advances in genome sequencing technologies, several studies in Capsella, Arabidopsis and Solanum have successfully narrowed down the genes involved in the selfing syndrome, which we highlight in this section (table 1). This section aims to describe the background of each study system, methodologies of the gene identification and the molecular nature of genes involved in the selfing syndrome. Evolutionary implications emerging from these findings are discussed in a later section (§5).
(a) . Capsella
The genus Capsella (Brassicaceae) has been studied intensively in the context of the selfing syndrome for more than a decade. A series of studies on Capsella have identified causal mutations and genes responsible for the evolution of multiple traits involved in the selfing syndrome, unveiling how, when, and how many times these traits have evolved at the molecular level. Capsella comprises five species, including both outcrossers and selfers [40]. Capsella grandiflora is an outcrossing species with a sporophytic SI system [41], representing the ancestral mode of reproduction of the genus, but the SI trait has been lost independently in Capsella orientalis and Capsella rubella, leading to the evolution of predominant selfers [42,43]. These two species share typical features of the selfing syndrome, including reductions in petal size, as well as reduced pollen, nectar and scent production [40]. The divergence of C. rubella from C. grandiflora and the associated evolution of selfing occurred as recently as 20 000 years ago [44], and much of the genomic variation within C. rubella is also found within C. grandiflora, consistent with a recent evolutionary split between the two species [45]. Detailed population genomic analyses indicated that C. rubella was founded by a potentially large number of individuals, and has subsequently experienced a strong genetic bottleneck [45]. In addition, the divergence between the C. orientalis lineage and the C. grandiflora–C. rubella lineage is estimated to be more ancient, about 1–2 million years ago (Ma).
Given the quite recent split between the selfing C. rubella and the outcrossing C. grandiflora, interspecific crosses between two species yield fertile descendants, making QTL mapping possible by using recombinant inbred lines (RIL) or F2 individuals. Two studies independently identified QTLs for the traits of the selfing syndrome [46,47]. Sicard et al. [46] identified several QTLs involved in such traits, including petal size, flower-opening angle and the distance between stigma and anthers, and found that some of the QTLs had major effects on phenotypic variation [46]. Slotte et al. [47] identified a few loci for selfing syndrome traits and revealed that the additive effects of major QTLs for floral size traits explain a considerable proportion of variance within F2 generations as well as divergence between species (32% of variance within F2; 26% of interspecific divergence for petal width). This suggests that changes at a few genomic regions are responsible for the drastic reduction in petal size in C. rubella [47].
These QTL mapping analyses paved the way for the further fine-mapping of the selfing syndrome traits in Capsella. Sicard et al. [18] fine-mapped the QTL of petal size, demonstrating that variation in the intron of a general growth regulator affects an organ-specific enhancer regulating the level of the STERILE APETALA (SAP) protein in petals [18]. Fujikura et al. [19] provided another functional follow-up of QTL mapping in Capsella, focusing on the QTL peak for petal size on chromosome 6, which explains about 10% of the variation between the parental C. rubella and C. grandiflora [19]. They demonstrated that allelic variation in the gene encoding the brassinosteroid (BR)-biosynthesis enzyme CYP724A1 was responsible for the petal size difference between these two species [19]. This allelic variation led to higher BR levels in plants bearing the C. rubella allele, and increased amounts of BR inhibit cell proliferation, resulting in smaller petals. Increased CYP724A1 activity was shown to result from more efficient splicing of the C. rubella allele. Sas et al. [20] addressed the genetic basis of reduced floral scent associated with the transition to selfing in C. rubella [20]. The outcrossing C. grandiflora plants emit a strong, marzipan-like scent. The major constituent of this floral scent is benzaldehyde, which has been lost in the selfing C. rubella. QTL mapping identified a strong peak at chromosome 2 and fine-mapping of the peak revealed that the cinnamate : CoA ligase-like protein (CNL1) underlies the variation in benzaldehyde emission.
In the genus Capsella, predominant selfing evolved independently in C. rubella and C. orientalis, and both share almost identical flower characteristics of the typical selfing syndrome. Woźniak et al. [48] performed QTL mapping and transcriptome analyses, demonstrating that low-pleiotropic and organ-specific gene regulatory networks are involved in the evolution of the selfing syndrome, and that petal size reduction at least has a similar genetic basis to C. rubella [48]. By contrast, in the case of the loss of benzaldehyde emission, an inactivating mutation in CNL1 was not responsible in C. orientalis, unlike in C. rubella [49].
(b) . Arabidopsis
Arabidopsis thaliana has been the most intensively studied species as a model of the evolution of selfing. Unlike its close relatives—such as A. lyrata, A. arenosa or A. halleri—A. thaliana is a self-compatible and predominantly selfing species [14,50–52]. The loss of SI had a major impact on the evolution of selfing, and independent gene-disruptive mutations at the specificity-determining genes for SI were shown to be responsible for the loss of SI [53–61]. Based on the non-synonymous/synonymous substitution ratio, Bechsgaard et al. estimated that the A. thaliana lineage has retained SI for at least 91.7% of the time since its split from A. lyrata [62]. This timing corresponds to 0.413 Ma, given that the divergence time of the two species is estimated to be 5 Ma [63]. The alternative estimate of the evolution of selfing based on the genome-wide pattern of linkage disequilibrium (LD) was of the order of 1 Ma [58].
Arabidopsis thaliana shows typical floral phenotypes considered as the selfing syndrome, such as small petals or reduced pollen number [14] (figure 1). There are several studies on the genetic and molecular basis of the reproductive traits associated with the evolution of selfing in this species. While most molecular and functional studies of the selfing syndrome in Capsella stem from QTL mapping using outcrossing and selfing individuals as parents, since the species split of A. thaliana with the outcrossing A. lyrata is relatively ancient (approx. 5 Ma) and chromosome numbers are different, the crosses between them do not yield fertile F2 individuals. Therefore, studies in A. thaliana usually exploit alternative approaches, such as comparative transcriptome or genome-wide association studies (GWAS).
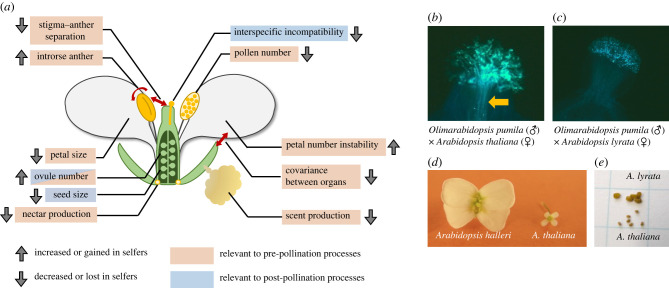
Figure 1.
Representative traits of the selfing syndrome. (a) A schematic figure of the traits related to the selfing syndrome. Arrows indicate directions in change through the evolution of selfing. Traits relevant to pre- and post-pollination processes are shown in orange and blue boxes, respectively. Ovule number could be related to both processes. (b,c) Interspecific crosses observed using fluorescence microscopy. Crosses between Olimarabidopsis pumila × Arabidopsis thaliana (a selfer) (b) and O. pumila × Arabidopsis lyrata (an outcrosser) (c). For both crosses, O. pumila was used as a pollen donor. In (b) the arrow indicates a bundle of pollen tubes stained with aniline blue. (d) Flowers of Arabidopsis halleri (an outcrosser) and A. thaliana (a selfer). (e) Seeds of A. lyrata and Arabidopsis thaliana.
Using GWAS in A. thaliana, Tsuchimatsu et al. [21] investigated the genetic basis of quantitative natural variation in pollen number, a representative trait involved in the selfing syndrome [21]. The mean pollen number per flower in an outcrossing population of A. lyrata is approximately 18 000 [64], which is several times higher than that of A. thaliana (approx. 2000–8000). GWAS identified a strong peak that explains approximately 20% of the total phenotypic variance between accessions [21]. The peak included the gene Reduced Pollen Number1 (RDP1), which encodes a ribosome-biogenesis factor, and a quantitative complementation test using CRISPR/Cas9-generated mutants revealed that natural variants confer variation in pollen number without detectable pleiotropy.
Comparative transcriptomics provides an alternative approach to identify candidate genes when forward genetic approaches are not applicable. Although attempts to compare floral differences directly between selfing A. thaliana and outcrossing A. lyrata are still limited, a comparative transcriptomics analysis using Capsella and Arabidopsis suggested the presence of parallel floral transcriptome changes in C. rubella and A. thaliana compared with C. grandiflora and A. lyrata, respectively: 373 genes were expressed more strongly in C. rubella compared with C. grandiflora, and 75 orthologues were also expressed more strongly in A. thaliana than in A. lyrata [65]. Further functional analyses of these genes may provide insights into the extent of the shared genetic basis involved in the selfing syndrome in selfing lineages that evolved in parallel.
(c) . Solanum
Studies of the molecular basis of the selfing syndrome were first pioneered in tomato (Solanum sp.). Tomato species include a diverse range of mating systems, from obligatory outcrossing with self-incompatibility, to facultative outcrossing, to predominantly selfing [66,67]. The major determinant of the selfing rate is the degree of stigma exsertion: obligate outcrossers bear flowers with highly exserted stigmas [67]. The major QTL for style length variation, which influences the stigma exsertion, was cloned and named Style2.1 [24]. The gene encodes a basic helix–loop–helix-related protein that regulates cell elongation in developing styles. Downregulation of Style2.1 owing to a 450 bp deletion in the promoter region was responsible for reduced anther–stigma separation in domesticated tomatoes. Recently, another stigma exsertion gene (SE3.1) was also identified through a GWAS in a population of 277 tomato accessions [25]. A loss-of-function mutation in the SE3.1 gene was shown to create an inserted stigma, having a major impact on the selfing rate. Population genetic data combined with functional analysis suggested the two-step transition from exserted to inserted stigmas: loss of function of Style2.1 contributed to the transition from exserted to flush stigmas, and that of SE3.1 contributed to the transition from flush to inserted stigmas.
3. Selfing and post-pollination reproductive barriers
(a) . Interspecific pollen transfer, selfing and interspecific incompatibility
While the previous section mainly focused on traits involved in pre-pollination processes, here we highlight post-pollination reproductive barriers and discuss how the evolution of selfing could be related to interspecific pollen transfer (IPT).
Interspecific incompatibility is a plant trait that serves to reject heterospecific pollen carried to the pistils via IPT. IPT results from two or more sympatric plant species sharing pollinators. IPT can cause the loss of female and male resources that could otherwise be used for intraspecific sexual reproduction [68]. IPT is now accepted to occur more often than previously thought, as most pollinators are generalists [69–71]. This review focuses on the interrelation between IPT and post-pollination reproductive barriers and is not intended to cover IPT research comprehensively. Readers who are interested in the recent advances in the IPT research itself should consult other sources [68,72,73]. When restricted to female fitness costs resulting from IPT, heterospecific pollen deposition on stigmas can cause stigma clogging, stigma closure, allelopathic inhibition of conspecific pollen, or ovule usurpation by interspecific fertilization [68]. IPT often leads to reproductive interference between one or more sympatric species, which might even endanger the survival of vulnerable species in some cases [74,75].
Recent works have studied the ecological impacts of IPT, and much has been revealed about its fitness costs, and also about how plants cope with it [69,73,76–81]. In a recent review, selfing and interspecific incompatibility were listed among the evolutionary pathways for plants to tolerate IPT-related fitness decline [72]. Thus, the evolutionary trajectories of these two plant traits might be associated when related to IPT. However, because of the lack of identified molecular factors, less has been discussed about the evolutionary relationships of IPT, interspecific incompatibility and selfing. Here, we re-review these traits and try to sort out the order of the evolutionary pathways by considering the genes involved in expressing interspecific incompatibility and selfing.
(b) . Selfing and interspecific incompatibility: two physiological strategies to tolerate heterospecific pollen transfer
The ecological effect of selfing as a means to tolerate IPT is mostly supported by field studies, investigating self-incompatible and self-compatible congenic species. Prior autonomous selfing was found to reduce the negative effect of heterospecific pollen transfer in two Commelina species that overlap in their habitats [76]. Earlier selfing was predicted to have evolved in a population of Collinsia rattanii as a response to selection against heterospecific pollen transport from its sympatric sister species Collinsia linearis [77]. These and other studies found that timing of selfing, especially prior selfing (in some studies referred to as ‘pre-emptive selfing’) before heterospecific pollen has arrived, seemed to be the effective mechanism against the fitness cost of IPT [72,79]. However, not all studies have supported the role of selfing in facilitating the coexistence of two different species [82,83]. A geographical study failed to find a correlation between mating system (i.e. selfing or outcrossing) and range overlaps of the sister species pairs [82]. Rather, it was suggested that other mechanisms such as pollinator shifts or post-pollination interspecific incompatibilities might also contribute to such coexistence [82].
From ecological studies, the negative effect of IPT could also be alleviated by pistil-associated interspecific incompatibility. Geographically sympatric populations of the two species of Costus were found to be pollen–pistil incompatible when distant populations were prone to create interspecific hybrids [84]. A study of Clarkia species showed that a population with a history of exposure to heterospecific pollen was more tolerant against the negative fitness effects of IPT [80]. Thus, IPT could be an important driver for the evolution of interspecific incompatibility. The advantage of this over selfing could be that it is capable of reconciling tolerance against IPT and reproductive assurance, with outcrossing being preserved.
Interspecific incompatibility might result from two mechanisms [85]. One is incongruity, the passive loss of gene interactions resulting from evolutionary divergence of male and female components of each species, which may cause the failure of interspecific fertilization. The other is the active incompatibility function which serves to reject an undesired male partner. Some case studies have reported the acquisition of interspecific pollen–pistil incompatibility functions. A unilateral incompatibility relationship between species, known as the ‘SI × SC rule’, is one of the most prominent examples [85,86]. Self-compatible (SC) species are prone to accept the pollen tubes of self-incompatible species, whereas pollen is rejected in reciprocal crosses. In some cases, factors involved in the SI system are directly involved in interspecific incompatibility. In many angiosperm species, including the Solanaceae family, a cytotoxic pistil-side protein S-RNase and a pollen-side detoxification protein S-locus F-box (SLF), determine the non-self-recognition-based SI mechanism [87]. In this system, the male determinant for SI is composed of multiple SLF proteins, and these SLFs can collaboratively detoxify the S-RNase in the non-self pistils (see [87] for review). In some cases, the absence of pollen SLF complex could explain the unilateral interspecific incompatibility in Solanaceae species [88,89]. The Cullin component of the SLFcomplex, or the one of the SLF-encoding gene were found to be the cause of the unilateral incompatibility relationships in tomato [88,89]. There are also numbers of pollen and pistil factors that are not linked to the SI-determining locus (see [90] for review), suggesting that interspecific incompatibility could be far more complex than the molecular mechanisms that could simply be suspected from the term ‘SI × SC rule’. More recently, a molecular factor controlling the stigmatic ability to reject heterospecific pollen was identified from the model species A. thaliana. The gene, Stigmatic Privacy 1 (SPRI1), was found to be responsible for rejecting pollen grains from many other Brassicaceae species in the stigma [22]. SPRI1 was found through GWAS using the variation in the interspecific incompatibility phenotype in A. thaliana. SPRI1 was found to encode a 221 amino acid-long plasma-membrane- localized protein with four transmembrane domains. Multiple and parallel loss-of-function alleles of the SPRI1 gene were found in A. thaliana. Because all known A. thaliana strains are self-compatible, SPRI1 and the SI system were considered to be functionally distinct. Indeed, the reconstituted SI system in A. thaliana could operate even when the SPRI1 gene was knocked out [22], suggesting that the two systems are not interdependent.
So, how are selfing and interspecific incompatibility related to each other evolutionarily? As discussed above, interspecific incompatibility and the SI system could be functionally separated in some cases. Thus, to fully explain the evolutionary relationships of selfing and interspecific incompatibility, we need to compare the demographic conditions of outcrossers and selfers, other than that they differ most of the time by being self-incompatible and self-compatible. In theory, paternal competition and/or parental conflicts in outcrossing species are high compared with selfing species. Compared with selfers which receive one type of pollen, outcrossers typically receive pollen from diverse origins, including both conspecific and heterospecific. The ‘weak inbreeder/strong outbreeder’ (WISO) hypothesis postulates that outcrossing parents ‘overpower’ selfing parents because of higher parental conflicts, and unilateral incompatibility between outbreeders and selfers is formed [27]. We suggest that the SI × SC rule could be transformed into a special case of the WISO hypothesis.
To untangle the relationship between selfing and interspecific incompatibility, tracking the evolutionary decay of the SPRI1 gene in A. thaliana could be helpful. First, the ancestral outcrossing A. thaliana species carried a functional SPRI1 similar to its relative A. lyrata, and when IPT occurred on the pistils, they were able to reject heterospecific pollen efficiently. The evolution of selfing and the reduction in pollinators might have released this species from the selective pressure of IPT. The SPRI1 locus became selectively neutral in A. thaliana, and loss-of-function mutations were not disadvantageous. This explains why at least 24% of the A. thaliana strains investigated have lost SPRI1 function by missense codon substitutions [22]. Thus, it could be speculated that interspecific incompatibility conferred by SPRI1 was acquired in the outcrossers, and that the evolutionary pressure to maintain this function was weakened in the selfers. This was also supported by the experiment that showed that the reconstructed ancestral form of the SPRI1 protein was preserved for its function to reject heterospecific pollen [22].
Rejection mechanisms of heterospecific pollen similar to SPRI1 could exist in the reproductive systems of other plant families. There are a few non-SI molecular mechanisms that confer interpopulational incompatibilities in the Poaceae [91] and Brassicaceae [92]. A pectin methyl esterase protein was found responsible for the pistil-barrier between sympatric populations of domesticated maize and its wild ancestor teosinte [91]. A duplicated copy of the self-incompatibility locus was found to cause the ligand-receptor mediated unilateral pollen rejection between Turkish and Japanese Brassica rapa strains [92]. A mechanism analogous to these might also cause interspecific incompatibility. In summary, although more needs to be understood on the distribution of these SI-independent pistil-based incompatibility mechanisms not controlled by the S-locus, molecular studies are starting to find that the SI × SC rule does not always accurately explain the interspecific incompatibility. These factors may rather fit the evolutionary pattern expected from the WISO hypothesis, which implies that selfers tend to lose the barrier functions via the loss of parental conflict [27].
4. Other notable traits associated with the evolution of selfing
While most studies have focused on a priori traits that are already well known to be associated with the evolution of selfing, we expect that even more traits might be found to be associated, by compiling the knowledge of independently evolved selfing species. Here we feature notable traits that have been found to be associated with the evolution of selfing. A caveat is that these associations may not directly reflect the effect of selfing, because of possible confounding factors such as polyploidization, which is often associated with the evolution of selfing [93].
(a) . Seed size and paternal–maternal conflict
De Jong et al. [94] proposed a mathematical model predicting that seed size variations between species would be influenced by the evolution of selfing [94]. This is based on the kinship theory of genetic imprinting [95], which postulates that the optimal seed mass for mothers differs from that for offspring, considering the conflict of interests between maternally and paternally derived alleles. Given the weaker conflict in selfing species, De Jong et al. [94] predicted that evolutionarily stable seed size would be smaller in selfers than in outcrossers [94]. There is some empirical support for this prediction. First, Mazer et al. [96] investigated the effects on seed size of both mating system and climate in Clarkia (Onagraceae), using seeds from three pairs of sister taxa, with each pair including a predominantly outcrossing and a facultatively selfing taxon [96]. Mazer et al. [96] found that the selfing taxon had smaller seeds than outcrossers in each taxon pair, while the local climate was also associated with seed size [96]. Second, a large-scale statistical analysis using 642 species from three plant families revealed that selfing species generally have smaller seed mass compared with outcrossing congeners, by controlling for possible confounding factors such as phylogeny and growth forms [97]. These results are consistent with the parental conflict hypothesis, but there are also non-mutually exclusive alternative hypotheses, such as colonization ability, which should be associated with both selfing and small seeds, and the prediction of sex allocation theory, in which the P/O ratio should increase linearly with increasing seed mass among seeding plants [98]. Nonetheless, there are multiple reports supporting the relationship between parental conflict, seed size, and endosperm development in A. lyrata and Dalechampia scandens [99–101]. Furthermore, the maternally imprinted gene MEDEA, which is involved in parent–offspring conflict, shows reduced selection signatures in selfing A. thaliana compared with outcrossing A. lyrata [102], consistent with smaller seed size in the former. These findings support the notion that small seeds are also a feature of selfing species, and that paternal–maternal conflict might partly explain this variation across species.
(b) . Covariance between floral organs
Selfers are predicted to show weaker covariance between floral organs than outcrossers with their specialized pollinators [103,104]. This is because floral traits should be coordinated in efficient pollen transfer in outcrossers—phenotypic integration [105]—which are no longer subjected to correlational selection in selfers. This prediction is indeed supported by a comparison between self-compatible and self-incompatible populations of Leavenworthia alabamica [106] (but see also another unsupported observation in A. lyrata [107]). In predominantly selfing Cardamine hirsuta, the petal number is unstable within individuals, possibly because of the released selection pressure. This instability was shown to be caused by epistatic interactions between QTLs and the floral regulator gene APETALA1 [23]. In addition to such covariance between traits, the evolution of selfing might also increase the variance of each floral trait once released from stabilizing selection. GWAS methods to detect loci controlling the covariance and variance of each trait have now been developed [108,109], so the genetic basis of correlated selection associated with mating system could be elucidated.
5. Emerging patterns in the evolution of the selfing syndrome
(a) . Is the selfing syndrome adaptive?
(i) . Selection versus neutral degradation
There are two alternative explanations for why the selfing syndrome is observed so widely in independently evolved selfing lineages: (1) the traits are adaptive in selfing populations; or (2) the traits were adaptive in outcrossing populations but became neutral and were released from selection pressure in selfing populations. In a general context, the selfing syndrome would represent a model of relaxed selection, an elimination of a source of selection that was formerly important for maintaining a trait [110,111]. The decay of nonfunctional traits could be due to direct or indirect selection for the trait as well as an accumulation of mutations [110,111].
There is support for the adaptive significance of the selfing syndrome, particularly in those traits promoting efficient auto-pollination. Sicard et al. [46] generated Capsella grandiflora-like but self-compatible plants through repeated introgression of the S-locus of Capsella rubella, which mimics the initial state of the evolution of selfing, just after the loss of SI [46]. The efficiency of auto-pollination was significantly higher in the C. rubella individuals than in the C. grandiflora-like SC plants. Petal opening angle was most strongly associated with selfing efficiency in the RIL population, suggesting that reduction in the opening of the petals and the flower as a whole might have been selected directly via efficient auto-pollination [46]. In C. rubella, Slotte et al. [47] also suggested directional selection on the selfing syndrome, based on the directionality of QTL effects and population genetic patterns of polymorphism and divergence at QTL [47]. A field study using F2 populations of Arabis alpina showed that a combination of introrse anthers and reduced anther–stigma distance was favoured at a site where pollinator activity was low, because of their effects on assuring self-pollination [112].
The evidence of adaptation for reduced allocation to outcrossing-related traits or male function is rather limited. Emission of benzaldehyde was lost in the selfing C. rubella, but the reason for its loss remains elusive [20]. Sas et al. [20] proposed three hypotheses [20]. First, benzaldehyde could attract both pollinators and herbivores; thus fitness cost imposed by herbivores would select for reduced scent emission in selfing species. Second, the loss of benzaldehyde emission might result in a metabolic benefit. Third, this could reflect a release from selective constraint. These hypotheses could be tested by measuring the fitness under an environment with relevant pollinators and herbivores. One of the few examples detecting selection would be the evolution of pollen number in A. thaliana. Selection scans revealed that genomic windows including pollen number-associated loci were significantly enriched in extreme tails of long-range haplotype-based selection statistics, supporting polygenic selection on a considerable number of loci associated with pollen numbers throughout the genome [21]. The RDP1 locus also appeared to be under selection; importantly, accessions with the long-haplotype variants produced lower pollen numbers than those with alternative haplotype variants, suggesting selection toward reduced pollen numbers. This finding is consistent with the sex allocation theory, which predicts that reduced investment for male gametes should be advantageous in selfing species because potential fitness gain through male function decreases in highly selfing populations where the proportion of female gametes available for outcrossing becomes low [98,113]. However, the fitness gain with reduced pollen number has not yet been measured directly, such as by increased seed set. We note that, although the trade-off between different functions such as male and female functions is always assumed as a basis of sex allocation theory, this often is not supported or is difficult to detect, and even positive correlations between male and female allocations tend to be reported (e.g. [114]).
Rather, it is possible that repeatedly observed traits in selfing species might simply reflect relaxed selection for traits that had important functions in the ancestral outcrossing populations. The loss of interspecific incompatibility in A. thaliana by multiple loss-of-function mutations in SPRI1 would be a good example of reduced constraint in selfing species [22]. Similarly, reduced selection signatures in the maternally imprinted gene MEDEA in A. thaliana would also represent a release from selection pressure in terms of parent–offspring conflict [102]. Consistently, in outcrossing A. lyrata, selection on MEDEA has been supported in population genetic studies [102,115,116]. Weaker covariance between floral organs in selfing populations would also represent neutral degradation, although the genetic basis is still unclear [106]. In summary, the relative contributions of adaptation and neutral degradation seem to differ among traits, while those for promoting efficient self-pollination show clear evidence of adaptation. Nonetheless, there are still too few examples to establish a general picture. Direct and precise measurement of the fitness consequences of causative mutations in field conditions would be important for clarifying the role of selection in evolution of the selfing syndrome.
(ii) . Traits segregating in a population or species
It is also important to note that the traits involved in the selfing syndrome are often segregating in a population or species, and the evolution may be even on-going. In the case of pollen number, in addition to fixed differences from outcrossing A. lyrata, there is still considerable standing genetic variation in predominantly selfing A. thaliana [21,117]. The long-range haplotypes at the pollen-associated loci would suggest that selection for pollen number is ongoing in current populations. Luo & Widmer [37] also found variation in herkogamy in A. thaliana, and a field experiment demonstrated that the distance between stigma and anthers was positively correlated with the outcrossing rate [37]. Bomblies et al. [118] showed that, among local sites of A. thaliana, there was considerable heterogeneity in outcrossing rates, with rural stands of the plant exhibiting greater heterozygosity than urban stands. Therefore, it is possible that the variation in the selfing syndrome traits might be associated with variation in local-scale outcrossing rates [118]. Other selfing syndrome traits, such as ovule number, were also found to be variable in current A. thaliana populations and thus might be under recent or ongoing selection [21,119].
(b) . Nature of mutations for the selfing syndrome
Several genes and mutations have been identified successfully as involved in the selfing syndrome. One obvious finding from these reports is that, although the selfing syndrome evolves as a set of traits, there are no ‘master switch’ genes governing all these traits, but rather each trait has an independent genetic basis, albeit some overlapping QTLs possibly because of linkage or pleiotropy [47].
Studies have revealed that the nature of mutations involved in the selfing syndrome differs among traits. For example, in the case of the SAP gene involved in the specific reduction of petal size in C. rubella, there were several polymorphisms associated with petal size variation in the extant outcrossing C. grandiflora populations. Species-specific polymorphisms were absent at the causal region in the selfing C. rubella, suggesting that the allele for small petals was captured from existing genetic variation in the ancestral outcrossing population. In contrast, the CYP724A1 gene involved in petal size reduction in C. rubella harboured two mutations, and population genetic analysis from 182 C. grandiflora individuals revealed that the more efficiently spliced C. rubella allele, which confers reduced petal size, most likely arose by two de novo mutations in the C. rubella lineage after its divergence from C. grandiflora [19]. The CNL1, gene involved in reduced scent emission, was inactivated twice independently in C. rubella by different de novo mutations in its coding sequence [20,49].
It appears to be relatively common that multiple independent mutations at the same genes are involved for the same trait even within a species [19,20,22]. Such patterns might be observed particularly when the selfing syndrome phenotype arises by gene-disruptive mutations (e.g. SPRI1 in Arabidopsis, SE3.1 in Solanum, and CNL1 in Capsella). This would be expected in the case that the loss-of-function mutations are causative for the phenotype because any nonsense mutations in the gene should have the same phenotypic consequences. Such observation would be analogous to the studies of the loss of sporophytic self-incompatibility in the Brassicaceae, in which multiple gene-disruptive mutations were often found to be causative [14,57,58,61,120]. It is important to note that the loss of function at the gene level does not necessarily indicate the neutral degradation at the phenotypic level as loss-of-function mutations can often provide substantial fitness benefits [121–123].
6. Perspectives
Except for pioneering studies featured in this review, the molecular basis of the parallel evolution of the selfing syndrome remains largely unknown. Information on the nature of causative mutations is essential for general understanding and quantitative predictions of evolution. Given that the evolution of selfing is generally recent [9,10], the traits of the selfing syndrome should evolve relatively rapidly. However, it is still unclear how rapid this is and how the speed differs between traits. For example, in recently evolved selfing populations of A. lyrata, the extent of change from outcrossing populations differs among traits [107], suggesting that the speed of evolution may be variable between traits. Whether and how quickly a trait will decay when a source of selection is eliminated is a major question of relaxed selection [110]. The molecular basis of each trait should help us in charting the detailed evolutionary history of coordinated sets of traits, and its knowledge in multiple selfing species will be essential for generalization. Species that harbour mating system variations would be of particular interest (e.g. A. lyrata, A. alpina and Petunia [64,107,124,125]), because it might be possible to trace quite recent or ongoing evolutionary processes of the selfing syndrome.
Acknowledgements
We thank Hiroyuki Kakui, Kazuho Isono and two anonymous reviewers for their helpful comments on the manuscript.
Contributor Information
Takashi Tsuchimatsu, Email: tsuchimatsu@bs.s.u-tokyo.ac.jp.
Sota Fujii, Email: a-fujii@g.ecc.u-tokyo.ac.jp.
Data accessibility
This article has no additional data.
Authors' contributions
T.T.: conceptualization, funding acquisition, investigation, visualization, writing—original draft, writing—review and editing; S.F.: conceptualization, funding acquisition, investigation, visualization, writing—original draft, writing—review and editing.
Both authors gave final approval for publication and agreed to be held accountable for the work performed herein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This work was supported in part by Grants-in-Aid for Scientific Research (19H04851 and 19H03271 to T.T., 18H02456 to S.F.) and a Grant-in-Aid for Challenging Exploratory Research (20K21417 to S.F.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). This work was also supported by the Suntory Rising Stars Encouragement Program in Life Sciences (SunRiSE; to S.F.).
References
- 1.Fisher RA. 1930. The genetical theory of natural selection. Oxford, UK: Clarendon Press. [Google Scholar]
- 2.Orr HA. 2005. The genetic theory of adaptation: a brief history. Nat. Rev. Genet. 6, 119-127. ( 10.1038/nrg1523) [DOI] [PubMed] [Google Scholar]
- 3.Hoekstra HE, Coyne JA. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61, 995-1016. ( 10.1111/j.1558-5646.2007.00105.x) [DOI] [PubMed] [Google Scholar]
- 4.Stern DL. 2013. The genetic causes of convergent evolution. Nat. Rev. Genet. 14, 751-764. ( 10.1038/nrg3483) [DOI] [PubMed] [Google Scholar]
- 5.Tsuchimatsu T, Shimizu KK. 2013. Effects of pollen availability and the mutation bias on the fixation of mutations disabling the male specificity of self-incompatibility. J. Evol. Biol. 26, 2221-2232. ( 10.1111/jeb.12219) [DOI] [PubMed] [Google Scholar]
- 6.Barrett SCH. 2002. The evolution of plant sexual diversity. Nat. Rev. Genet. 3, 274-284. ( 10.1038/nrg776) [DOI] [PubMed] [Google Scholar]
- 7.Barrett SCH. 2003. Mating strategies in flowering plants: the outcrossing–selfing paradigm and beyond. Phil. Trans. R. Soc. Lond. B 358, 991-1004. ( 10.1098/rstb.2003.1301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stebbins GL. 1974. Flowering plants: evolution above the species level. Cambridge, MA: Belknap Press of Harvard University Press. [Google Scholar]
- 9.Igic B, Bohs L, Kohn JR. 2006. Ancient polymorphism reveals unidirectional breeding system shifts. Proc. Natl Acad. Sci. USA 103, 1359-1363. ( 10.1073/pnas.0506283103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldberg EE, Kohn JR, Lande R, Robertson KA, Smith SA, Igić B. 2010. Species selection maintains self-incompatibility. Science 330, 493-495. ( 10.1126/science.1194513) [DOI] [PubMed] [Google Scholar]
- 11.Fisher RA. 1941. Average excess and average effect of a gene substitution. Ann. Eugen. 11, 53-63. ( 10.1111/j.1469-1809.1941.tb02272.x) [DOI] [Google Scholar]
- 12.Darwin C. 1876. The effects of cross and self fertilisation in the vegetable kingdom. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 13.Sicard A, Lenhard M. 2011. The selfing syndrome: a model for studying the genetic and evolutionary basis of morphological adaptation in plants. Ann. Bot. 107, 1433-1443. ( 10.1093/aob/mcr023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu KK, Tsuchimatsu T. 2015. Evolution of selfing: recurrent patterns in molecular adaptation. Annu. Rev. Ecol. Evol. Syst. 46, 593-622. ( 10.1146/annurev-ecolsys-112414-054249) [DOI] [Google Scholar]
- 15.Weigel D, Nordborg M. 2015. Population genomics for understanding adaptation in wild plant species. Annu. Rev. Genet. 49, 315-338. ( 10.1146/annurev-genet-120213-092110) [DOI] [PubMed] [Google Scholar]
- 16.Bamba M, Kawaguchi YW, Tsuchimatsu T. 2019. Plant adaptation and speciation studied by population genomic approaches. Dev. Growth Differ. 61, 12-24. ( 10.1111/dgd.12578) [DOI] [PubMed] [Google Scholar]
- 17.Ellegren H. 2014. Genome sequencing and population genomics in non-model organisms. Trends Ecol. Evol. 29, 51-63. ( 10.1016/j.tree.2013.09.008) [DOI] [PubMed] [Google Scholar]
- 18.Sicard A, Kappel C, Lee YW, Woźniak NJ, Marona C, Stinchcombe JR, Wright SI, Lenhard M. 2016. Standing genetic variation in a tissue-specific enhancer underlies selfing-syndrome evolution in Capsella. Proc. Natl Acad. Sci. USA 113, 13 911-13 916. ( 10.1073/pnas.1613394113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujikura U, Jing R, Hanada A, Takebayashi Y, Sakakibara H, Yamaguchi S, Kappel C, Lenhard M. 2018. Variation in splicing efficiency underlies morphological evolution in Capsella. Dev. Cell. 44, 192-203.e5. ( 10.1016/j.devcel.2017.11.022) [DOI] [PubMed] [Google Scholar]
- 20.Sas C, Müller F, Kappel C, Kent TV, Wright SI, Hilker M, Lenhard M. 2016. Repeated inactivation of the first committed enzyme underlies the loss of benzaldehyde emission after the selfing transition in Capsella. Curr. Biol. 26, 3313-3319. ( 10.1016/j.cub.2016.10.026) [DOI] [PubMed] [Google Scholar]
- 21.Tsuchimatsu T, et al. 2020. Adaptive reduction of male gamete number in the selfing plant Arabidopsis thaliana. Nat. Commun. 11, 2885. ( 10.1038/s41467-020-16679-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii S, et al. 2019. A stigmatic gene confers interspecies incompatibility in the Brassicaceae. Nat. Plants 5, 731-741. ( 10.1038/s41477-019-0444-6) [DOI] [PubMed] [Google Scholar]
- 23.Monniaux M, Pieper B, McKim SM, Routier-Kierzkowska A-L, Kierzkowski D, Smith RS, Hay A. 2018. The role of APETALA1 in petal number robustness. eLife 7, e39399. ( 10.7554/eLife.39399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen K-Y, Cong B, Wing R, Vrebalov J, Tanksley SD. 2007. Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science 318, 643-645. ( 10.1126/science.1148428) [DOI] [PubMed] [Google Scholar]
- 25.Shang L, et al. 2021. A mutation in a C2H2-type zinc finger transcription factor contributed to the transition towards self-pollination in cultivated tomato. Plant Cell 33, 3293-3308. ( 10.1093/plcell/koab201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cutter AD. 2008. Reproductive evolution: symptom of a selfing syndrome. Curr. Biol. 18, R1056-R1058. ( 10.1016/j.cub.2008.09.008) [DOI] [PubMed] [Google Scholar]
- 27.Brandvain Y, Haig D. 2005. Divergent mating systems and parental conflict as a barrier to hybridization in flowering plants. Am. Nat. 166, 330-338. ( 10.1086/432036) [DOI] [PubMed] [Google Scholar]
- 28.Mattila TM, Laenen B, Slotte T. 2020. Population genomics of transitions to selfing in Brassicaceae model systems. In Statistical population genomics (ed. Dutheil JY), pp. 269-287. New York, NY: Springer. [DOI] [PubMed] [Google Scholar]
- 29.Glémin S, François CM, Galtier N. 2019. Genome evolution in outcrossing vs. selfing vs. asexual species. In Evolutionary genomics: statistical and computational methods (ed. Anisimova M), pp. 331-369. New York, NY: Springer. [DOI] [PubMed] [Google Scholar]
- 30.Barrett SCH, Arunkumar R, Wright SI. 2014. The demography and population genomics of evolutionary transitions to self-fertilization in plants. Phil. Trans. R. Soc. B 369, 20130344. ( 10.1098/rstb.2013.0344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cutter AD. 2019. Reproductive transitions in plants and animals: selfing syndrome, sexual selection and speciation. New Phytol. 224, 1080-1094. ( 10.1111/nph.16075) [DOI] [PubMed] [Google Scholar]
- 32.Wang X-J, Barrett SCH, Zhong L, Wu Z-K, Li D-Z, Wang H, Zhou W. 2021. The genomic selfing syndrome accompanies the evolutionary breakdown of heterostyly. Mol. Biol. Evol. 38, 168-180. ( 10.1093/molbev/msaa199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright SI, Kalisz S, Slotte T. 2013. Evolutionary consequences of self-fertilization in plants. Proc. R. Soc. B 280, 20130133. ( 10.1098/rspb.2013.0133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fishman L, Kelly AJ, Willis JH. 2002. Minor quantitative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution 56, 2138-2155. ( 10.1111/j.0014-3820.2002.tb00139.x) [DOI] [PubMed] [Google Scholar]
- 35.Goodwillie C, Ritland C, Ritland K. 2006. The genetic basis of floral traits associated with mating system evolution in Leptosiphon (Polemoniaceae): an analysis of quantitative trait loci. Evolution 60, 491-504. ( 10.1111/j.0014-3820.2006.tb01131.x) [DOI] [PubMed] [Google Scholar]
- 36.Georgiady MS, Whitkus RW, Lord EM. 2002. Genetic analysis of traits distinguishing outcrossing and self-pollinating forms of currant tomato, Lycopersicon pimpinellifolium (Jusl.) Mill. Genetics 161, 333-344. ( 10.1093/genetics/161.1.333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo Y, Widmer A. 2013. Herkogamy and its effects on mating patterns in Arabidopsis thaliana. PLoS ONE 8, e57902. ( 10.1371/journal.pone.0057902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duncan TM, Rausher MD. 2013. Evolution of the selfing syndrome in Ipomoea. Front. Plant Sci. 4, 301. ( 10.3389/fpls.2013.00301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frazee LJ, Rifkin J, Maheepala DC, Grant A-G, Wright S, Kalisz S, Litt A, Spigler R. 2021. New genomic resources and comparative analyses reveal differences in floral gene expression in selfing and outcrossing Collinsia sister species. G3 Genes Genomes Genet. 11, jkab177. ( 10.1093/g3journal/jkab177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sicard A, Lenhard M. 2018. Capsella. Curr. Biol. 28, R920-R921. ( 10.1016/j.cub.2018.06.033) [DOI] [PubMed] [Google Scholar]
- 41.Paetsch M, Mayland-Quellhorst S, Neuffer B. 2006. Evolution of the self-incompatibility system in the Brassicaceae: identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity 97, 283-290. ( 10.1038/sj.hdy.6800854) [DOI] [PubMed] [Google Scholar]
- 42.Guo Y-L, Bechsgaard JS, Slotte T, Neuffer B, Lascoux M, Weigel D, Schierup MH. 2009. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc. Natl Acad. Sci. USA 106, 5246-5251. ( 10.1073/pnas.0808012106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachmann JA, et al. 2019. Genetic basis and timing of a major mating system shift in Capsella. New Phytol. 224, 505-517. ( 10.1111/nph.16035) [DOI] [PubMed] [Google Scholar]
- 44.Foxe JP, Slotte T, Stahl EA, Neuffer B, Hurka H, Wright SI. 2009. Recent speciation associated with the evolution of selfing in Capsella. Proc. Natl Acad. Sci. USA 106, 5241-5245. ( 10.1073/pnas.0807679106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brandvain Y, Slotte T, Hazzouri KM, Wright SI, Coop G. 2013. Genomic identification of founding haplotypes reveals the history of the selfing species Capsella rubella. PLoS Genet. 9, e1003754. ( 10.1371/journal.pgen.1003754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sicard A, Stacey N, Hermann K, Dessoly J, Neuffer B, Bäurle I, Lenhard M. 2011. Genetics, evolution, and adaptive significance of the selfing syndrome in the genus Capsella. Plant Cell 23, 3156-3171. ( 10.1105/tpc.111.088237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slotte T, Hazzouri KM, Stern D, Andolfatto P, Wright SI. 2012. Genetic architecture and adaptive significance of the selfing syndrome in Capsella. Evolution 66, 1360-1374. ( 10.1111/j.1558-5646.2011.01540.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woźniak NJ, Kappel C, Marona C, Altschmied L, Neuffer B, Sicard A. 2020. A similar genetic architecture underlies the convergent evolution of the selfing syndrome in Capsella. Plant Cell 32, 935-949. ( 10.1105/tpc.19.00551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jantzen F, et al. 2019. Retracing the molecular basis and evolutionary history of the loss of benzaldehyde emission in the genus Capsella. New Phytol. 224, 1349-1360. ( 10.1111/nph.16103) [DOI] [PubMed] [Google Scholar]
- 50.Novikova PY, et al. 2016. Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat. Genet. 48, 1077-1082. ( 10.1038/ng.3617) [DOI] [PubMed] [Google Scholar]
- 51.Bomblies K, Weigel D. 2007. Arabidopsis—a model genus for speciation. Curr. Opin. Genet. Dev. 17, 500-504. ( 10.1016/j.gde.2007.09.006) [DOI] [PubMed] [Google Scholar]
- 52.Mitchell-Olds T. 2001. Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends Ecol. Evol. 16, 693-700. ( 10.1016/S0169-5347(01)02291-1) [DOI] [Google Scholar]
- 53.Dwyer KG, et al. 2013. Molecular characterization and evolution of self-incompatibility genes in Arabidopsis thaliana: the case of the Sc haplotype. Genetics. 193, 985-994. ( 10.1534/genetics.112.146787) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kusaba M, Dwyer K, Hendershot J, Vrebalov J, Nasrallah JB, Nasrallah ME. 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13, 627-643. ( 10.1105/tpc.13.3.627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nasrallah ME, Liu P, Nasrallah JB. 2002. Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297, 247-249. ( 10.1126/science.1072205) [DOI] [PubMed] [Google Scholar]
- 56.Sherman-Broyles S, Boggs N, Farkas A, Liu P, Vrebalov J, Nasrallah ME, Nasrallah JB. 2007. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell 19, 94-106. ( 10.1105/tpc.106.048199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimizu KK, Shimizu-Inatsugi R, Tsuchimatsu T, Purugganan MD. 2007. Independent origins of self-compatibility in Arabidopsis thaliana. Mol. Ecol. 17, 704-714. ( 10.1111/j.1365-294X.2007.03605.x) [DOI] [PubMed] [Google Scholar]
- 58.Tang C, et al. 2007. The evolution of selfing in Arabidopsis thaliana. Science 317, 1070-1072. ( 10.1126/science.1143153) [DOI] [PubMed] [Google Scholar]
- 59.Tsuchimatsu T, et al. 2010. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464, 1342-1346. ( 10.1038/nature08927) [DOI] [PubMed] [Google Scholar]
- 60.Tsuchimatsu T, et al. 2017. Patterns of polymorphism at the self-incompatibility locus in 1,083 Arabidopsis thaliana genomes. Mol. Biol. Evol. 34, 1878-1889. ( 10.1093/molbev/msx122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Durvasula A, et al. 2017. African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 114, 5213-5218. ( 10.1073/pnas.1616736114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bechsgaard JS, Castric V, Charlesworth D, Vekemans X, Schierup MH. 2006. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 23, 1741-1750. ( 10.1093/molbev/msl042) [DOI] [PubMed] [Google Scholar]
- 63.Koch MA, Haubold B, Mitchell-Olds T. 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17, 1483-1498. ( 10.1093/oxfordjournals.molbev.a026248) [DOI] [PubMed] [Google Scholar]
- 64.Willi Y. 2013. Mutational meltdown in selfing Arabidopsis lyrata. Evolution 67, 806-815. ( 10.1111/j.1558-5646.2012.01818.x) [DOI] [PubMed] [Google Scholar]
- 65.Slotte T, et al. 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45, 831-835. ( 10.1038/ng.2669) [DOI] [PubMed] [Google Scholar]
- 66.Rick CM. 1988. Evolution of mating systems in cultivated plants. In Plant evolutionary biology (eds Gottlieb LD, Jain SK), pp. 133-147. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 67.Chen K-Y, Tanksley SD. 2004. High-resolution mapping and functional analysis of se2.1: a major stigma exsertion quantitative trait locus associated with the evolution from allogamy to autogamy in the genus Lycopersicon. Genetics 168, 1563-1573. ( 10.1534/genetics.103.022558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morales CL, Traveset A. 2008. Interspecific pollen transfer: magnitude, prevalence and consequences for plant fitness. Crit. Rev. Plant Sci. 27, 221-238. ( 10.1080/07352680802205631) [DOI] [Google Scholar]
- 69.Briggs HM, Anderson LM, Atalla LM, Delva AM, Dobbs EK, Brosi BJ. 2016. Heterospecific pollen deposition in Delphinium barbeyi: linking stigmatic pollen loads to reproductive output in the field. Ann. Bot. 117, 341-347. ( 10.1093/aob/mcv175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arceo-Gómez G, Alonso C, Ashman TL, Parra-Tabla V. 2018. Variation in sampling effort affects the observed richness of plant–plant interactions via heterospecific pollen transfer: implications for interpretation of pollen transfer networks. Am. J. Bot. 105, 1601-1608. ( 10.1002/ajb2.1144) [DOI] [PubMed] [Google Scholar]
- 71.Johnson AL, Ashman TL. 2019. Consequences of invasion for pollen transfer and pollination revealed in a tropical island ecosystem. New Phytol. 221, 142-154. ( 10.1111/nph.15366) [DOI] [PubMed] [Google Scholar]
- 72.Moreira-Hernández JI, Muchhala N. 2019. Importance of pollinator-mediated interspecific pollen transfer for angiosperm evolution. Annu. Rev. Ecol. Evol. Syst. 50, 191-217. ( 10.1146/annurev-ecolsys-110218-024804) [DOI] [Google Scholar]
- 73.Ashman TL, Arceo-Gómez G. 2013. Toward a predictive understanding of the fitness costs of heterospecific pollen receipt and its importance in co-flowering communities. Am. J. Bot. 100, 1061-1070. ( 10.3732/ajb.1200496) [DOI] [PubMed] [Google Scholar]
- 74.Burdfield-Steel ER, Shuker DM. 2011. Reproductive interference. Curr. Biol. 21, R450-R451. ( 10.1016/j.cub.2011.03.063) [DOI] [PubMed] [Google Scholar]
- 75.Nishida S, Kanaoka MM, Hashimoto K, Takakura KI, Nishida T. 2014. Pollen–pistil interactions in reproductive interference: comparisons of heterospecific pollen tube growth from alien species between two native Taraxacum species. Funct. Ecol. 28, 450-457. ( 10.1111/1365-2435.12165) [DOI] [Google Scholar]
- 76.Katsuhara KR, Ushimaru A. 2019. Prior selfing can mitigate the negative effects of mutual reproductive interference between coexisting congeners. Funct. Ecol. 33, 1504-1513. ( 10.1111/1365-2435.13344) [DOI] [Google Scholar]
- 77.Randle AM, Spigler RB, Kalisz S. 2018. Shifts to earlier selfing in sympatry may reduce costs of pollinator sharing. Evolution 72, 1587-1599. ( 10.1111/evo.13522) [DOI] [PubMed] [Google Scholar]
- 78.Lyu N, Du W, Wang XF. 2017. Unique growth paths of heterospecific pollen tubes result in late entry into ovules in the gynoecium of Sagittaria (Alismataceae). Plant Biol. 19, 108-114. ( 10.1111/plb.12508) [DOI] [PubMed] [Google Scholar]
- 79.Brys R, van Cauwenberghe J, Jacquemyn H. 2016. The importance of autonomous selfing in preventing hybridization in three closely related plant species. J. Ecol. 104, 601-610. ( 10.1111/1365-2745.12524) [DOI] [Google Scholar]
- 80.Arceo-Gómez G, Raguso RA, Geber MA. 2016. Can plants evolve tolerance mechanisms to heterospecific pollen effects? An experimental test of the adaptive potential in Clarkia species. Oikos 125, 718-725. ( 10.1111/oik.02594) [DOI] [Google Scholar]
- 81.Arceo-Gómez G, Ashman TL. 2011. Heterospecific pollen deposition: does diversity alter the consequences? New Phytol. 192, 738-746. ( 10.1111/j.1469-8137.2011.03831.x) [DOI] [PubMed] [Google Scholar]
- 82.Grossenbacher D, Briscoe Runquist RD, Goldberg EE, Brandvain Y. 2016. No association between plant mating system and geographic range overlap. Am. J. Bot. 103, 110-117. ( 10.3732/ajb.1500078) [DOI] [PubMed] [Google Scholar]
- 83.Matallana G, Godinho MAS, Guilherme FAG, Belisario M, Coser TS, Wendt T. 2010. Breeding systems of Bromeliaceae species: evolution of selfing in the context of sympatric occurrence. Plant Syst. Evol. 289, 57-65. ( 10.1007/s00606-010-0332-z) [DOI] [Google Scholar]
- 84.Kay KM, Schemske DW. 2008. Natural selection reinforces speciation in a radiation of neotropical rainforest plants. Evolution 62, 2628-2642. ( 10.1111/j.1558-5646.2008.00463.x) [DOI] [PubMed] [Google Scholar]
- 85.de Nettancourt D. 2001. Incompatibility and incongruity in wild and cultivated plants. Berlin, Germany: Springer. [Google Scholar]
- 86.Hiscock SJ, Dickinson HG. 1993. Unilateral incompatibility within the Brassicaceae: further evidence for the involvement of the self-incompatibility (S)-locus. Theor. Appl. Genet. 86, 744-753. ( 10.1007/BF00222665) [DOI] [PubMed] [Google Scholar]
- 87.Fujii S, Kubo K, Takayama S. 2016. Non-self- and self-recognition models in plant self-incompatibility. Nat. Plants 2, 16130. ( 10.1038/nplants.2016.130) [DOI] [PubMed] [Google Scholar]
- 88.Li W, Chetelat RT. 2015. Unilateral incompatibility gene ui1.1 encodes an S-locus F-box protein expressed in pollen of Solanum species. Proc. Natl Acad. Sci. USA 112, 4417-4422. ( 10.1073/pnas.1423301112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li W, Chetelat RT. 2010. A pollen factor linking inter- and intraspecific pollen rejection in tomato. Science 330, 1827-1830. ( 10.1126/science.1197908) [DOI] [PubMed] [Google Scholar]
- 90.Broz AK, Bedinger PA. 2021. Pollen-pistil interactions as reproductive barriers. Annu. Rev. Plant Biol. 72, 615-639. ( 10.1146/annurev-arplant-080620-102159) [DOI] [PubMed] [Google Scholar]
- 91.Lu Y, Hokin SA, Kermicle JL, Hartwig T, Evans MMS. 2019. A pistil-expressed pectin methylesterase confers cross-incompatibility between strains of Zea mays. Nat. Commun. 10, 2304. ( 10.1038/s41467-019-10259-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Takada Y, et al. 2017. Duplicated pollen–pistil recognition loci control intraspecific unilateral incompatibility in Brassica rapa. Nat. Plants 3, 17096. ( 10.1038/nplants.2017.96) [DOI] [PubMed] [Google Scholar]
- 93.Barringer BC. 2007. Polyploidy and self-fertilization in flowering plants. Am. J. Bot. 94, 1527-1533. ( 10.3732/ajb.94.9.1527) [DOI] [PubMed] [Google Scholar]
- 94.De Jong TJ, Van Dijk H, Klinkhamer PGL. 2005. Hamilton's rule, imprinting and parent–offspring conflict over seed mass in partially selfing plants. J. Evol. Biol. 18, 676-682. ( 10.1111/j.1420-9101.2004.00856.x) [DOI] [PubMed] [Google Scholar]
- 95.Haig D. 1997. Parental antagonism, relatedness asymmetries, and genomic imprinting. Proc. R. Soc. Lond. B 264, 1657-1662. ( 10.1098/rspb.1997.0230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mazer SJ, Park IM, Kimura M, Maul EM, Yim AM, Peach K. 2020. Mating system and historical climate conditions affect population mean seed mass: evidence for adaptation and a new component of the selfing syndrome in Clarkia. J. Ecol. 108, 1523-1539. ( 10.1111/1365-2745.13338) [DOI] [Google Scholar]
- 97.Tateyama H, Chimura K, Tsuchimatsu T. 2021. Evolution of seed mass associated with mating systems in multiple plant families. J. Evol. Biol. 34, 1931-1937. ( 10.1111/jeb.13949) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Charnov EL. 1982. The theory of Sex allocation. Princeton, NJ: Princeton University Press. [Google Scholar]
- 99.Willi Y. 2013. The battle of the sexes over seed size: support for both kinship genomic imprinting and interlocus contest evolution. Am. Nat. 181, 787-798. ( 10.1086/670196) [DOI] [PubMed] [Google Scholar]
- 100.Raunsgard A, Opedal ØH, Ekrem RK, Wright J, Bolstad GH, Armbruster WS, Pélabon C. 2018. Intersexual conflict over seed size is stronger in more outcrossed populations of a mixed-mating plant. Proc. Natl Acad. Sci. USA 115, 11 561-11 566. ( 10.1073/pnas.1810979115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.İltaş Ö, Svitok M, Cornille A, Schmickl R, Lafon Placette C. 2021. Early evolution of reproductive isolation: a case of weak inbreeder/strong outbreeder leads to an intraspecific hybridization barrier in Arabidopsis lyrata. Evolution 75, 1466-1476. ( 10.1111/evo.14240) [DOI] [PubMed] [Google Scholar]
- 102.Spillane C, et al. 2007. Positive darwinian selection at the imprinted MEDEA locus in plants. Nature 448, 349-352. ( 10.1038/nature05984) [DOI] [PubMed] [Google Scholar]
- 103.Berg RL. 1959. A general evolutionary principle underlying the origin of developmental homeostasis. Am. Nat. 93, 103-105. ( 10.1086/282061) [DOI] [Google Scholar]
- 104.Berg RL. 1960. The ecological significance of correlation pleiades. Evolution 14, 171-180. ( 10.1111/j.1558-5646.1960.tb03076.x) [DOI] [Google Scholar]
- 105.Pigliucci M. 2003. Phenotypic integration: studying the ecology and evolution of complex phenotypes. Ecol. Lett. 6, 265-272. ( 10.1046/j.1461-0248.2003.00428.x) [DOI] [Google Scholar]
- 106.Anderson IA, Busch JW. 2006. Relaxed pollinator-mediated selection weakens floral integration in self-compatible taxa of Leavenworthia (Brassicaceae). Am. J. Bot. 93, 860-867. ( 10.3732/ajb.93.6.860) [DOI] [PubMed] [Google Scholar]
- 107.Carleial S, van Kleunen M, Stift M. 2017. Small reductions in corolla size and pollen:ovule ratio, but no changes in flower shape in selfing populations of the North American Arabidopsis lyrata. Oecologia 183, 401-413. ( 10.1007/s00442-016-3773-4) [DOI] [PubMed] [Google Scholar]
- 108.Korte A, Vilhjálmsson BJ, Segura V, Platt A, Long Q, Nordborg M. 2012. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat. Genet. 44, 1066-1071. ( 10.1038/ng.2376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shen X, Pettersson M, Rönnegård L, Carlborg Ö. 2012. Inheritance beyond plain heritability: variance-controlling genes in Arabidopsis thaliana. PLoS Genet. 8, e1002839. ( 10.1371/journal.pgen.1002839) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lahti DC, Johnson NA, Ajie BC, Otto SP, Hendry AP, Blumstein DT, Coss RG, Donohue K, Foster SA. 2009. Relaxed selection in the wild. Trends Ecol. Evol. 24, 487-496. ( 10.1016/j.tree.2009.03.010) [DOI] [PubMed] [Google Scholar]
- 111.Fong DW, Kane TC, Culver DC. 1995. Vestigialization and loss of nonfunctional characters. Annu. Rev. Ecol. Syst. 26, 249-268. ( 10.1146/annurev.es.26.110195.001341) [DOI] [Google Scholar]
- 112.Toräng P, Vikström L, Wunder J, Wötzel S, Coupland G, Ågren J. 2017. Evolution of the selfing syndrome: anther orientation and herkogamy together determine reproductive assurance in a self-compatible plant. Evolution 71, 2206-2218. ( 10.1111/evo.13308) [DOI] [PubMed] [Google Scholar]
- 113.de Jong T, Klinkhamer P. 2005. Evolutionary ecology of plant reproductive strategies. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 114.Ashman T-L, Majetic CJ. 2006. Genetic constraints on floral evolution: a review and evaluation of patterns. Heredity 96, 343-352. ( 10.1038/sj.hdy.6800815) [DOI] [PubMed] [Google Scholar]
- 115.Kawabe A, Fujimoto R, Charlesworth D. 2007. High diversity due to balancing selection in the promoter region of the Medea gene in Arabidopsis lyrata. Curr. Biol. 17, 1885-1889. ( 10.1016/j.cub.2007.09.051) [DOI] [PubMed] [Google Scholar]
- 116.Miyake T, Takebayashi N, Wolf DE. 2009. Possible diversifying selection in the imprinted gene, MEDEA, in Arabidopsis. Mol. Biol. Evol. 26, 843-857. ( 10.1093/molbev/msp001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Beaudry FEG, Rifkin JL, Barrett SCH, Wright SI. 2020. Evolutionary genomics of plant gametophytic selection. Plant Commun. 1, 100115. ( 10.1016/j.xplc.2020.100115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bomblies K, Yant L, Laitinen RA, Kim S-T, Hollister JD, Warthmann N, Fitz J, Weigel D. 2010. Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet. 6, e1000890. ( 10.1371/journal.pgen.1000890) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yuan J, Kessler SA. 2019. A genome-wide association study reveals a novel regulator of ovule number and fertility in Arabidopsis thaliana. PLoS Genet. 15, e1007934. ( 10.1371/journal.pgen.1007934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tsuchimatsu T, Kaiser P, Yew C-L, Bachelier JB, Shimizu KK. 2012. Recent loss of self-incompatibility by degradation of the male component in allotetraploid Arabidopsis kamchatica. PLoS Genet. 8, e1002838. ( 10.1371/journal.pgen.1002838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hottes AK, Freddolino PL, Khare A, Donnell ZN, Liu JC, Tavazoie S. 2013. Bacterial adaptation through loss of function. PLoS Genet. 9, e1003617. ( 10.1371/journal.pgen.1003617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Monroe JG, Powell T, Price N, Mullen JL, Howard A, Evans K, Lovell JT, Mckay JK. 2018. Drought adaptation in Arabidopsis thaliana by extensive genetic loss-of-function. eLife 7, e41038. ( 10.7554/eLife.41038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Monroe JG, McKay JK, Weigel D, Flood PJ. 2021. The population genomics of adaptive loss of function. Heredity 126, 383-395. ( 10.1038/s41437-021-00403-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tsukamoto T, Ando T, Takahashi K, Omori T, Watanabe H, Kokubun H, Marchesi E, Kao T. 2003. Breakdown of self-incompatibility in a natural population of Petunia axillaris caused by loss of pollen function. Plant Physiol. 131, 1903-1912. ( 10.1104/pp.102.018069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tedder A, Carleial S, Gołębiewska M, Kappel C, Shimizu KK, Stift M. 2015. Evolution of the selfing syndrome in Arabis alpina (Brassicaceae). PLoS ONE 10, e0126618. ( 10.1371/journal.pone.0126618) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.