Abstract
Copy number variation (CNV) can cause phenotypic changes. However, in contrast to amino acid substitutions and cis-regulatory changes, little is known about the functional categories of genes in which CNV is important for adaptation to novel environments. It is also unclear whether the same genes repeatedly change the copy numbers for adapting to similar environments. Here, we investigate CNV associated with freshwater colonization in fishes, which was observed multiple times across different lineages. Using 48 ray-finned fishes across diverse orders, we identified 23 genes whose copy number increases were associated with freshwater colonization. These genes showed enrichment for peptide receptor activity, hexosyltransferase activity and unsaturated fatty acid metabolism. We further revealed that three of the genes showed copy number increases in freshwater populations compared to marine ancestral populations of the stickleback genus Gasterosteus. These results indicate that copy number increases of genes involved in fatty acid metabolism (FADS2), immune function (PSMB8a) and thyroid hormone metabolism (UGT2) may be important for freshwater colonization at both the inter-order macroevolutionary scale and at the intra-genus microevolutionary scale. Further analysis across diverse taxa will help to understand the role of CNV in the adaptation to novel environments.
This article is part of the theme issue ‘Genetic basis of adaptation and speciation: from loci to causative mutations’.
Keywords: copy number variation, convergent evolution, gene duplication, freshwater colonization
1. Introduction
Parallel and convergent evolution of phenotypic traits in independent lineages that inhabit similar environments is prevalent in nature, indicating the role of natural selection and the repeatability of evolution at the phenotypic level [1–3]. Recent genetic studies have further revealed that mutations in the same genes often occur when organisms are adapted to similar environments, which is called genetic parallelism or genetic convergence [4,5]. There are many striking examples of amino acid substitutions and cis-regulatory changes in the same genes for adapting to similar environments [4–9]. Copy number variation (CNV) is another type of mutation that can lead to phenotypic changes through a variety of mechanisms such as changes in gene expression levels, reorganization of chromatin structure, and subsequent mutations at the cis-regulatory regions and amino acid sequences [10–16]. Several studies have shown that the same genes show repeated increases of copy numbers in organisms treated with antibiotics and pesticides [11] and mammals adapted to starch-rich diets during domestication by the duplication of amylase genes [17,18]. By contrast, there are only a few reports of CNV underlying adaptation in natural systems except for a few cases, such as the adaptation to freshwater habitats by the duplication of fatty acid desaturase genes in fishes [19] and the evolution of floral pigmentation by the duplication of anthocyanin-regulating transcription factors in plants [20]. Therefore, we do not know how prevalent parallel and convergent CNV is in nature.
Fishes provide us an excellent opportunity to test whether any genes exhibit parallel and convergent copy number changes during transitions from marine to freshwater environments, because freshwater colonization occurs repeatedly in multiple independent lineages [21]. Recently, we found that the copy number increases of the fatty acid desaturase gene FADS2, a gene that encodes an enzyme for the biosynthesis of docosahexaenoic acid (DHA), repeatedly underlie freshwater colonization in fishes [19]. Freshwater ecosystems are generally poor in DHA, an omega-3 long-chain polyunsaturated fatty acid (LC-PUFA), which plays crucial roles in growth, survival and reproduction in animals [22]. An increase in the copy number of FADS2 can help fishes overcome DHA deficiency in freshwater habitats. Importantly, FADS2 amplification associated with freshwater colonization was observed not only at the macroevolutionary scale examining 48 species across diverse orders of ray-finned fishes, but also at the microevolutionary scale examining inter-population variations within species. The three-spined stickleback, Gasterosteus aculeatus, is primarily a marine fish species, but repeatedly colonized freshwater habitats on multiple continents [23]. Freshwater populations of the three-spined stickleback were found to have higher FADS2 copy numbers than marine populations across multiple geographical regions. Since marine and freshwater habitats differ not only in fatty acid availability, but also in many other environmental factors, such as salinity, water flow, pathogens and predators, genes other than FADS2 may also show increased copy numbers in freshwater fishes compared to marine fishes.
In the present study, we first tested whether any genes other than FADS2 showed convergent copy number changes in the freshwater fishes than in the marine fishes across orders using whole-genome sequences of 48 ray-finned fishes. Next, we tested whether any of these genes also showed copy number increases associated with freshwater colonization within the stickleback genus Gasterosteus in particular. Previous studies have listed genes showing copy number increases associated with freshwater colonization in the three-spined stickleback [24,25]. However, this work used only North American and European freshwater stickleback populations; thus, no Asian freshwater populations were included in the analyses. Because the genetic basis of freshwater adaptation often differs across geographical regions [26–32], we investigated the CNV among Japanese stickleback populations in the present study and compared our results with previous studies on the North American and European stickleback populations [24,25]. Finally, for the several genes whose copy number increases were associated with freshwater colonization, we investigated whether gene amplification occurred by tandem duplication in a freshwater stickleback population.
2. Methods
(a) . Association between copy number and habitat in the ray-finned fishes
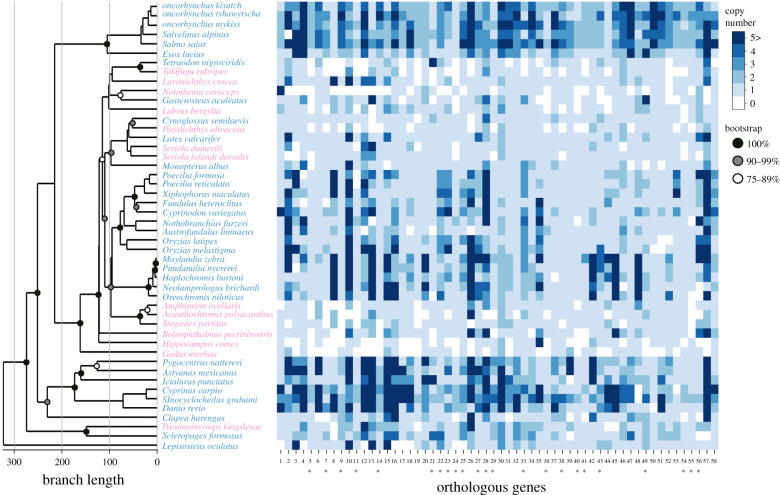
To identify genes that show copy number increases associated with freshwater colonization, we examined the protein sequence data of 48 ray-finned fishes, which were available on Ensembl (https://asia.ensembl.org/index.html) or RefSeq (https://www.ncbi.nlm.nih.gov/refseq/). When the gene ID has several splicing variants, we used a protein sequence from the longest splice variant, ensuring that the dataset contained one peptide sequence for each gene ID. The species were first classified into ‘freshwater species’ that have freshwater populations (34 species) and ‘non-freshwater species' that lack any freshwater populations (14 species) based on Eschmeyer's catalog of fishes [33]. Next, we determined the orthologous relationships of all genes among these species using the SonicParanoid v. 1.2.6 [34]. SonicParanoid was executed with the ‘default’ mode (i.e. the sensitivity parameter for sequence search was set to 4). To identify genes whose copy number increases are strongly associated with freshwater colonization, we selected genes that met the following three criteria: (i) the average copy number of freshwater species is more than twice that of non-freshwater species, (ii) more than 90% of freshwater species have at least one copy of the gene and (iii) more than 70% of non-freshwater species have less than two copies of the gene. Orthologue gene IDs were assigned in the order of copy number ratio between freshwater and non-freshwater species (table 1; electronic supplementary material, table S1; figure 1).
Table 1.
Candidate genes that show increased copy numbers associated with freshwater colonization after phylogenetic correction.
| orthologuos gene ID | gene name | protein name | protein function | references for protein function | mean copy number in freshwater species | mean copy number in non-freshwater species | freshwater/marine CNV ratio | pMCMC | pMCMC (with WGD) |
|---|---|---|---|---|---|---|---|---|---|
| 5 | LTBP1/4 | latent transforming growth factor beta binding protein 1 | pulmonary, gastrointestinal, urinary, musculoskeletal, craniofacial and dermal development | [35] | 1.117647059 | 0.285714286 | 3.911764706 | 0.0136 | 0.0124 |
| 7 | TMEM150A | localization of phosphatidylinositol 4-kinase (PI4K) to the plasma membrane | [36] | 1.352941176 | 0.357142857 | 3.788235294 | 0.0092 | 0.0064 | |
| 9 | YWHAQ | 14-3-3 protein theta | signalling pathways, including metabolism, cell division, stress responses, protein trafficking, and immune responses, insulin sensitivity | [37,38] | 1.647058824 | 0.5 | 3.294117647 | 0.032 | 0.0376 |
| 11 | NFIA | nuclear factor IA | brain maturation, spinal cord development | [39,40] | 1.088235294 | 0.357142857 | 3.047058824 | 0.0128 | 0.0116 |
| 14 | PTH2RA | parathyroid hormone 2 receptor A | calcium and bone homeostasis, corticosterone release, anxiety state, fear response, thermoregulation, prolactin, postnatal pup development | [41,42] | 1.647058824 | 0.571428571 | 2.882352941 | 0.0352 | 0.0272 |
| 21 | HSF5 | heat shock transcription factor 5 | spermatogenesis | [43] | 1.735294118 | 0.714285714 | 2.429411765 | 0.0372 | 0.0348 |
| 22 | FUT9A | fucosyltransferase 9A | tissue development, angiogenesis, fertilization,cell adhesion, inflammation and tumour metastasis | [44] | 2.235294118 | 0.928571429 | 2.407239819 | 0.0048 | 0.0052 |
| 23 | DTX3L | deltex E3 ubiquitin ligase 3L | DNA damage repair, immune system | [45,46] | 2.058823529 | 0.857142857 | 2.401960784 | 0.0368 | 0.0392 |
| 24 | CART1 | cocaine-and amphetamine-regulated transcript protein 1 | feeding behaviour | [47] | 1.029411765 | 0.428571429 | 2.401960784 | 0.034 | 0.0396 |
| 25 | CXCR3 | C-X-C motif chemokine receptor 3 | immune system | [48] | 2.147058824 | 0.928571429 | 2.312217195 | 0.0444 | 0.0368 |
| 27 | PTGR1 | prostaglandin reductase 1-like | inflammation, fatty acid metabolism | [49–51] | 2.411764706 | 1.071428571 | 2.250980392 | 0.004 | 0.0072 |
| 28 | UGT2 | UDP-glucuronosyltransferase 2 | drug metabolism, thyroid homeostasis, glucuronidation | [52–55] | 3.852941176 | 1.714285714 | 2.24754902 | 0.0156 | 0.0124 |
| 29 | CAPN2 | calpain-2 catalytic subunit | early embryonic development | [56,57] | 1.441176471 | 0.642857143 | 2.241830065 | 0.0236 | 0.0268 |
| 33 | FADS2 | fatty acid desaturase 2 | DHA biosynthesis | [19] | 2.205882353 | 1 | 2.205882353 | 0.0032 | 0.0052 |
| 36 | PATJ | PALS1-associated TJ protein | tight junction formation, cell polarization | [58,59] | 1.088235294 | 0.5 | 2.176470588 | 0.0056 | 0.0072 |
| 38 | NAV2A | neuron navigator 2A | neurogenesis | [60] | 1.529411765 | 0.714285714 | 2.141176471 | 0.0472 | 0.082 |
| 40 | si:dkey-22o22.2 | si:dkey-22o22.2 | [61] | 1.058823529 | 0.5 | 2.117647059 | 0.0284 | 0.0328 | |
| 41 | CXorf40A | CXorf40A | inflammatory responses | [62] | 1.205882353 | 0.571428571 | 2.110294118 | 0.0184 | 0.0212 |
| 43 | CD4 | T-cell surface glycoprotein CD4 | immune system | [63,64] | 1.794117647 | 0.857142857 | 2.093137255 | 0.0276 | 0.034 |
| 49 | PSMB8a | proteasome 20S subunit beta 8a | immune system | [65] | 2.029411765 | 1 | 2.029411765 | 0.0488 | 0.0684 |
| 54 | LAMC3 | laminin, gamma 3 | neurogenesis | [66] | 1.147058824 | 0.571428571 | 2.007352941 | 0.026 | 0.0144 |
| 55 | DOC2B | double C2 domain beta | neurotransmission, insulin secretion, insulin sensitivity | [67,68] | 1.147058824 | 0.571428571 | 2.007352941 | 0.0348 | 0.0328 |
| 56 | USP47 | ubiquitin carboxyl-terminal hydrolase 47-like | synapse development, behaviour, immune system | [69–72] | 2.294117647 | 1.142857143 | 2.007352941 | 0.0412 | 0.0548 |
Figure 1.
Genes showing increased copy numbers in freshwater ray-finned fishes. The left panel indicates the phylogenetic tree of ray-finned fishes, which is primarily based on Betancur-R. [73] (see Methods). The light blue and pink letters indicate species with freshwater populations and those that lack freshwater populations, respectively. Circles at nodes indicate bootstrap support values in Betancur-R. [73]. The right panel shows a heat map of the gene copy numbers. The light and deep blue squares indicate lower and higher copy numbers, respectively. The orthologous gene IDs at the bottom correspond to the orthologous gene IDs in table 1 and electronic supplementary material, table S1. The orthologue gene IDs were assigned in the order of copy number ratio between freshwater and non-freshwater species. The asterisks indicate significant increases in copy numbers in freshwater species compared to non-freshwater species after phylogenetic correction (pMCMC < 0.05).
Although the main focus of the present study was to identify candidate freshwater-adaptive genes that increased their copy numbers, we also searched for genes with significantly lower copy numbers in freshwater species than in non-freshwater species. We selected genes that met the following three criteria: (i) the average copy number of non-freshwater species is more than twice that of freshwater species, (ii) more than 90% of non-freshwater species have at least one copy of the gene and (iii) more than 70% of freshwater species have less than two copies of the gene (electronic supplementary material, table S2).
To exclude the possibility that the observed associations between gene copy number and habitat simply reflects their phylogenetic relationships [74], we conducted Bayesian inference for a generalized linear mixed model (GLMM). This accounted for phylogeny as a covariate using the MCMCglmm R package [75] with the published fish phylogenetic tree [73], as described previously [19]. The estimated copy number of each gene was used as a response variable, while the habitat type (freshwater versus non-freshwater species; see above) was used as a predictor.
To investigate what kinds of genes showed CNV, we performed gene ontology (GO) analysis. For the GO term enrichment test of genes that increased in freshwater species, we used the stickleback orthologues, because the sticklebacks contain freshwater populations. By contrast, for the GO term enrichment test of genes that increased in non-freshwater species, we used the spiny choromis (Acanthochromis polyacanthus) orthologues, because the spiny choromis is entirely marine. For genes that had multiple copies with different gene IDs, we randomly selected one gene ID to be conservative. The GO enrichment analysis was performed against all annotated stickleback genes using g: GOSt in g: Profiler (version e104_eg51_p15_3922dba) [76]. We listed GO terms that were significantly enriched (Benjamini–Hochberg FDR p < 0.05) and found in at least two genes in the query.
(b) . Analysis of copy number variation between marine and freshwater sticklebacks
We explored whether the genes whose copy number increases were found to be associated with freshwater colonization at the macroevolutionary scale (see above) showed similar copy number increases in freshwater populations of the three-spined stickleback compared to marine populations. Previous studies have reported a list of genes that show copy number increases associated with freshwater colonization of the three-spined stickleback [24,25]. In these studies, whole-genome sequences of 11 freshwater and 10 marine stickleback populations [77] were analysed [24,25]. Hirase et al. [25] reported 19 genes, but Lowe et al. [24], having developed a novel method for detecting CNV, reported 138 genes, including all 19 genes identified in Hirase et al. [25]. Thus, we investigated whether any of these 138 genes overlapped with genes for which we had found increased copy numbers in freshwater fishes at the macroevolutionary scale.
Because previous studies on sticklebacks did not include freshwater populations from Asia [24,25], we investigated whether Japanese freshwater populations shared copy number increases with the North American and European freshwater populations. Furthermore, we analysed multiple individuals per population, which extends our analytical scope relative to previous studies that sequenced only one individual per population [24,25]. We investigated the copy number of candidate genes using our previously reported whole-genome sequences (WGS) of 75 individuals [19]: two marine populations (five females and five males of Japan Sea stickleback from Akkeshi, six females and four males of Pacific Ocean marine population of the three-spined stickleback from Akkeshi) and four freshwater populations (five females and three males from Nishitappu, six females and two males from Chimikeppu, three females and eight males from Gifu, three females and six males from Ono). Genomic DNA was isolated using the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA). The libraries were constructed using the NEBNext Ultra DNA Library Prep Kit (NEB, Ipswitch, MA, USA) and run in a 150 bp-paired end mode of HiSeqX. Sequence data are available from DDBJ (DRA007515 and DRA001136). The obtained sequence reads were trimmed and mapped to the BROADS1 reference sequence and cDNA coding sequence of each candidate gene, which spanned from the first ATG to the stop codon without introns, using the CLC Genomics Workbench 10.1.1 as described previously [19].
To test whether similar copy number increases occurred in the Japanese freshwater populations compared to the North American and European freshwater populations, we focused on five genes, FADS2 (ENSGACG00000005442), PSMB8a (ENSGACG00000000146), UGT2 (ENSGACG00000013874), PTGR1 (ENSGACG00000019130) and USP47 (ENSGACG00000019203), which possess two or more copies in the BROADS1 stickleback reference genome, which is the sequence of an Alaskan freshwater population belonging to the same Pacific stickleback lineage [28]. Since each candidate gene has multiple cDNA sequences, we conducted a phylogenetic analysis of cDNA sequences using the CLC Genomics Workbench and used the cDNA sequence located at the basal position of the phylogenetic tree for mapping. To calculate the copy number, the coverage of the cDNA was divided by the genome-wide coverage for each individual. All statistical analyses were conducted using the R v. 3.6.2 [78]. To test whether marine and freshwater populations possess different copy numbers for each gene, we used GLMM with a gamma distribution with the glmer function of the lme4 package in R [79], that account for the relative copy number of each gene per individual as the dependent variable, the ecotype (marine or freshwater) and the sex as the independent variables, and the population as the random effect.
(c) . Local genomic similarity analyses around candidate freshwater-adaptive genes in a freshwater stickleback population
To investigate whether duplicated copies were tandemly arrayed, we analysed raw reads of previously generated PacBio long-read sequencing data of a Gifu freshwater population (available at DRA007518) [19]. We conducted BLAST searches to find sequence reads containing FADS2, PSMB8a and UGT2, using the CLC Genomics Workbench. The cDNA sequences used in our search query were as follows: FADS2, ENSGACG00000005442; PSMB8a, ENSGACG00000000146; UGT2, ENSGACG00000013874. The following parameters were used: number of threads = 1, expect = 10, word size = 11, match = 2, mismatch = −2, and gap cost = Existence 5, Extension 2. Local pairwise alignment was conducted using the YASS [80]. Repetitive sequences were identified by searching against a database of known repetitive sequences using the GIRI Repbase in CENSOR software (http://www.girinst.org/censor/index.php) [81].
3. Results
(a) . Genes that show copy number differences between freshwater and non-freshwater fishes
We first screened for genes that showed increased copy numbers in freshwater species compared to non-freshwater species. We identified 58 candidate genes that showed increased copy numbers associated with freshwater colonization (figure 1; electronic supplementary material, table S1). GO analysis indicated enrichment of genes related to G protein-coupled receptor activity, immune receptor activity, cytokine receptor activity, chemotaxis and unsaturated fatty acid metabolism (electronic supplementary material, table S3). Of the 58 genes, 23 genes showed statistical significance even after phylogenetic correction (pMCMC < 0.05; table 1). The FADS2 gene, which we previously found to increase in copy number in freshwater fishes [19], was included in this list. GO analysis of these 23 genes showed enrichment of genes involved in peptide receptor activity, hexosyltransferase activity and unsaturated fatty acid metabolism (electronic supplementary material, table S4).
We identified 18 genes that showed higher copy number in non-freshwater species compared to freshwater species (electronic supplementary material, table S2). GO analysis of these 18 genes showed enrichment of genes involved in carboxylic ester hydrolase activity, catalytic activity, endopeptidase activity and methyltranferase activity (electronic supplementary material, table S5). Of these 18 genes, 12 genes exhibited statistical significance even after phylogenetic correction (pMCMC < 0.05). GO analysis of these 12 genes revealed enrichment of genes involved in carboxylic ester hydrolase activity (electronic supplementary material, table S6).
(b) . Copy number increase in freshwater sticklebacks
Among the 58 candidate freshwater-adaptive genes described above (electronic supplementary material, table S1), three genes, FADS2, GVINP1 and CXCR1, overlapped with those that were previously reported to show higher copy numbers in North American and European three-spined stickleback freshwater populations compared to marine populations (electronic supplementary material, table S1) [24]. Only the FADS2 gene was included in the 23 genes showing a significant difference after phylogenetic correction.
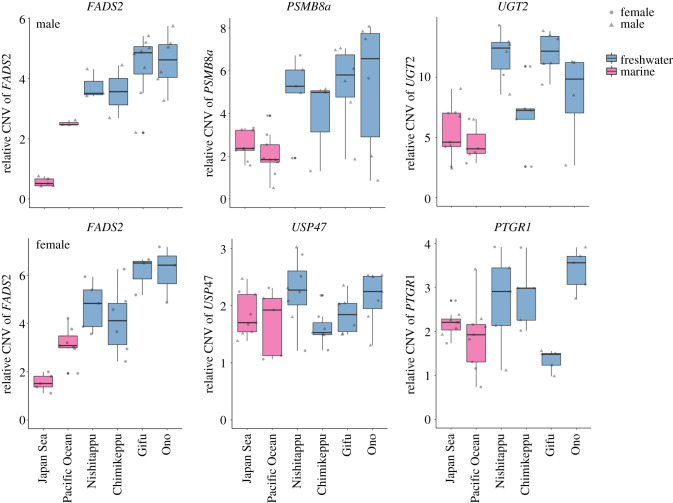
To test whether the copy number increases occurred in the Japanese freshwater populations, we focused on five genes, FADS2, PSMB8a, UGT2, PTGR1 and USP4, which possess two or more copies in the Alaskan freshwater population belonging to the same Pacific stickleback lineage [28]. In the Japanese freshwater three-spined stickleback populations, FADS2, PSMB8a and UGT2 showed significantly higher copy numbers compared to marine populations (GLMM: p = 0.026, p = 0.012 and p = 0.0023, respectively; figure 2; electronic supplementary material, table S7). USP47 and PTGR1 did not show significant differences between habitats (GLMM: p = 0.37 and p = 0.35, respectively), although there are inter-population variations such that Pacific Ocean marine and Gifu freshwater populations possessed significantly lower copy numbers of USP47 and PTGR1, respectively, than other populations (GLM: p = 0.022 and p = 0.0021, respectively).
Figure 2.
Estimated copy number of the FADS2, PSMB8a, UGT2, USP47 and PTGR1 genes in marine and freshwater sticklebacks. The pink and light blue boxes indicate marine and freshwater sticklebacks, respectively. Each dot indicates a single individual. Since at least one copy of FADS2 gene is located on the X chromosome, the relative copy numbers of FADS2 are shown separately for each sex.
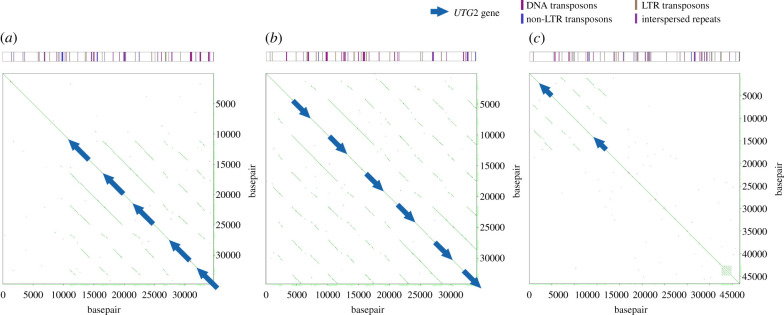
To investigate how the copy number increases have occurred in the freshwater stickleback populations, we analysed the long-read sequencing data of a Japanese freshwater stickleback (Gifu population). We had already found that the FADS2 gene is tandemly duplicated with many transposons in Japanese and North American populations [19]. We also found tandem duplication of UGT2 genes (figure 3). Local genomic similarity analyses revealed that at least six copies of the UGT2 genes were tandemly duplicated. The similarity analyses also detected accumulation of transposons around the tandemly duplicated copies (figure 3; electronic supplementary material, table S8).
Figure 3.
Local pairwise alignment of genomic regions around the UGT2 gene against themselves. The three longest PacBio sequence fragments containing the UGT2 gene are shown (a–c). The green dots represent forward alignments, and the red dots represent reverse alignments; however, the red dots are so rare at these regions that they cannot be seen. The blue arrows indicate the UGT2 genes. The upper bar indicates the positions of repetitive elements identified in Repbase by CENSOR. The red-purple, yellowish-brown, blue and purple bars indicate DNA transposons, long terminal repeat (LTR) transposons, non-LTR transposons and interspersed repeats, respectively. The list of transposons is presented in electronic supplementary material, table S8.
4. Discussion
(a) . Gene amplification associated with freshwater colonization across orders of ray-finned fishes
Our screening for genes whose copy number amplification was associated with freshwater colonization across ray-finned fish orders identified 58 candidate genes. Twenty-three genes remained significant even after phylogenetic correction. Importantly, FADS2, a previously identified gene that shows a copy number increase in freshwater fishes [19], was included in these 23 candidate genes, suggesting that our screening worked well. By contrast, 18 genes showed higher copy number in non-freshwater species, and 12 genes remained significant after phylogenetic correction. Therefore, the number of genes that increased the copy numbers was almost twice as much as that of those that decreased in freshwater species.
GO analysis showed that genes involved in unsaturated fatty acid metabolism and immune functions were enriched in genes that showed copy number increases in freshwater fishes. This is consistent with the hypothesis that increased expression is often beneficial for proteins involved in interaction with the environment, such as stress response and metabolism [14]. Because there is variation in the availability of omega-3 and omega-6 LC-PUFAs between marine and freshwater ecosystems [82], increased copy numbers of PTGR1, which is involved in arachidonic acid metabolism, may be advantageous in freshwater species, which is also the case for the FADS2 gene. Furthermore, freshwater fish species may be exposed to strong selective pressures due to diverse and abundant parasites and pathogens relative to marine fishes [83,84]. Therefore, gene duplication of immune system-related genes may serve as a reservoir from which new genes constantly arise to protect against diverse pathogens [85]. Gene copy number increases concerned with fatty acid metabolism and immune functions may facilitate freshwater colonization and adaptation in ray-finned fishes.
By contrast, genes involved in carboxylic ester hydrolase activity were enriched in genes that showed higher copy number in non-freshwater species. The carboxylic ester hydrolase, also called bile salt-dependent lipase, can hydrolyse wax esters [86,87]. Wax esters are abundant in marine zooplankton, especially the dominating copepods, but generally more resistant to hydrolysis by pancreatic lipase, which can hydrolyse triacylglycerol substrates [87]. Since marine fishes feed mainly on zooplankton including calanoid copepods enriched with wax esters, copy number increases of genes related to carboxylic ester hydrolase activity may help them to use wax esters in marine environments.
(b) . Gene amplification associated with freshwater colonization within the genus Gasterosteus
Three genes (FADS2, GVINP1 and CXCR1) showed copy number increases in North American and European freshwater stickleback populations, whereas FADS2 and two other genes (PSMB8 and UGT2) showed copy number increases in Japanese freshwater stickleback populations. Among these, three genes, PSMB8, GVINP1 and CXCR1, were related to immune functions. PSMB8 encodes a catalytic subunit of the immunoproteasome responsible for generating peptides presented by major histocompatibility complex (MHC) class I molecules [65]. PSMB8 knockout mice show reduced expression of MHC class I molecules on cell surfaces [86]. GVINP1 encodes an interferon-induced very large GTPase 1 [87]. GVINP is known to play a role in immune reactions against pathogens [88]. In Atlantic salmon, the GVINP1 gene is located in a QTL region that explains over 20% of the genetic variance in resistance to amoebic gill disease and is differentially expressed between resistant and susceptible individuals [89]. The CXCR1 gene encodes a C-X-C motif chemokine receptor 1, which is a G protein-coupled receptor for the CLC chemokine interleukin-8 (IL-8), a major mediator of immune and inflammatory responses [90]. The CXCR1 gene plays a crucial role in the IL-8 signal transduction pathway of neutrophils [91–93]. A genome-wide association study revealed that the CXCR1 gene is located at a genomic region explaining the number of piglets born alive, which is potentially affected by CXCR1 immune function [94]. Therefore, high copy numbers of PSMB8a, GVINP1 and CXCR1 genes may contribute to adaptation to the diverse pathogens in freshwater environments.
UGT2 encodes UDP-glucuronosyltransferase 2, which metabolizes and inactivates triiodothyronine (T3), an active form of thyroid hormone found in the liver [55,95,96]. Thyroid hormones play key roles in the regulation of many physiological and behavioural processes, such as metabolism, ion homeostasis, basal activity, growth and development. Importantly, previous studies demonstrated that freshwater sticklebacks possess lower plasma concentrations of T3 and T4 than marine sticklebacks at both juvenile [97] and adult stages [30]. Lower levels of thyroid hormone are correlated with a lower metabolic rate which is likely adaptive in oxygen- and nutrient-limited freshwater environments [30]. Different expression levels of thyroid stimulating hormone ß2 (TSHß2), which may stimulate the synthesis and secretion of thyroid hormones, and iodothyronine deiodinase genes, DIO2 and DIO3, which convert T4 to T3 and T3 to T4, respectively, may underlie this divergence in thyroid hormone levels [30,98]. The higher copy numbers of UGT2 may also contribute to the lower thyroid hormone levels in freshwater sticklebacks. Even in teleosts other than sticklebacks, there are several reports on inter-population and geographical variation in thyroid hormones at adult stages [98]. Thyroid hormones are also deposited in egg of teleosts [99,100]. The eggs of freshwater fishes also contain lower concentrations of T3 than those of non-freshwater fishes [101]. Because freshwater fish larvae have lower metabolic requirements [102], the higher copy numbers of UGT2 in freshwater fishes may be responsible for the lower yolk thyroid hormone levels and lower metabolic activity at the larval stage.
The long-read genome sequencing data revealed tandem duplication of the UGT2 genes with surrounding transposons. It is similar to the FADS2 gene in freshwater sticklebacks, which is tandemly duplicated with many transposons [19]. Because transposons can induce tandem sequence duplications in plants [103], transposons near the UGT2 gene might have facilitated these tandem duplications. Because we did not conduct polishing of PacBio sequences by Illumina short reads, the currently available sequences are not of high enough quality for pinpointing the exact breakpoints of the UGT2 duplication. Precise identification of the boundaries of the duplicated regions with the UGT2 gene clusters may help to understand how adaptive tandem duplication occurs.
(c) . Genetic parallelism versus genetic non-parallelism with regard to copy number variation
Three genes (FADS2, PSMB8 and UGT2) showed a convergent increase in copy number during freshwater colonization, both at the macroevolutionary scale across multiple orders and at the within-genus microevolutionary scale. However, there were many genes that did not overlap between them. Furthermore, even among the stickleback lineages, different geographical populations showed different patterns. For example, PSMB8 and UGT2 showed increased copy numbers only in the Japanese freshwater stickleback populations, not in the North American and European freshwater stickleback populations. These results suggest that the genetic basis for freshwater adaption may differ between lineages and geographic regions.
Consistent with our findings, previous studies have reported that the genetic basis of freshwater adaptation often differs across geographical regions in sticklebacks [26–32]. Furthermore, a meta-analysis indicated that the probability of using the same genes for parallel and convergent phenotypic evolution declines with genetic divergence [104]. Therefore, it is not surprising that different fish lineages use different CNV to adapt to similar environments. Further research regarding CNV underlying repeated adaptation in natural populations will shed light on the generality and lineage specificity in recurrent gene copy number increases.
5. Conclusions
We have identified several genes that show convergent increases in copy numbers during freshwater colonization by analysing macroevolutionary patterns of gene copy numbers across orders of ray-finned fishes. Among freshwater fishes, candidate genes showing increased copy numbers are involved in fatty acid metabolism and immune function. Some of the CNV were also observed even within a genus of Gasterosteus. Currently, little is known about how the identified CNV impact phenotypic traits that help fishes colonize freshwater environments. Further studies on the functional roles of increased copy number of these candidate genes will help to understand the genetic mechanisms of freshwater colonization that occurred repeatedly in fishes.
Acknowledgments
We thank Kitano laboratory members for technical assistance.
Ethics
All animal experiments were approved by the institutional animal care and use committee of the National Institute of Genetics (R2-18). Before dissection, all fish were euthanized with 500 mg l−1 MS-222.
Data accessibility
The whole-genome sequence data are available from DDBJ DRA007515, DRA001136 and DRA007518. The datasets supporting this article have been uploaded as part of the electronic supplementary material [105]. Electronic supplementary material, table S1: candidate genes that show increased copy numbers associated with freshwater colonization. Electronic supplementary material, table S2: candidate genes that show increased copy numbers in non-freshwater fish species. Electronic supplementary material, table S3: GO enrichment analysis for 58 candidate genes that show increased copy numbers associated with freshwater colonization. Electronic supplementary material, table S4: GO enrichment analysis for 23 candidate genes that remained significant even after phylogenetic correction. Electronic supplementary material, table S5: GO enrichment analysis for 18 candidate genes that show increased copy numbers in non-freshwater species. Electronic supplementary material, table S6: GO enrichment analysis for 12 candidate genes that show increased copy numbers in non-freshwater species and remained significant even after phylogenetic correction. Electronic supplementary material, table S7: candidate genes that show convergent increases of copy number in Japanese freshwater sticklebacks. Electronic supplementary material, table S8: transposons near the UGT2 gene in a freshwater stickleback. Electronic supplementary material, figure S1: High-resolution image of figure 3.
Authors' contributions
A.I.: conceptualization, data curation, formal analysis, funding acquisition, investigation, validation, visualization, writing—original draft; S.Y.: data curation, formal analysis, visualization, writing—review and editing; W.I.: funding acquisition, writing—review and editing; J.K.: conceptualization, funding acquisition, project administration, supervision, writing—original draft.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This project was supported by JSPS KAKENHI grant nos. 19H03277, 19KK0187 and 20H04873, the Inamori Research Grants to A.I., JST CREST JPMJCR19S2 to W.I. and JSPS KAKENHI grant no. 19H01003 to J.K.
References
- 1.Schluter D. 2000. The ecology of adaptive radiation. Oxford, UK: Oxford University Press. [Google Scholar]
- 2.Losos JB. 2011. Convergence, adaptation, and constraint. Evolution 65, 1827-1840. ( 10.1111/j.1558-5646.2011.01289.x) [DOI] [PubMed] [Google Scholar]
- 3.Simpson GG. 1953. The major features of evolution. New York, NY: Columbia University Press. [Google Scholar]
- 4.Martin A, Orgogozo V. 2013. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 67, 1235-1250. ( 10.1111/evo.12081) [DOI] [PubMed] [Google Scholar]
- 5.Courtier-Orgogozo V, Martin A. 2020. The coding loci of evolution and domestication: current knowledge and implications for bio-inspired genome editing. J. Exp. Biol. 223, 208934. ( 10.1242/jeb.208934) [DOI] [PubMed] [Google Scholar]
- 6.Dong K, Du Y, Rinkevich F, Nomura Y, Xu P, Wang L, Silver K, Zhorov BS.. 2014. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem. Mol. Biol. 50, 1-17. ( 10.1016/j.ibmb.2014.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra HE. 2010. Convergence in pigmentation at multiple levels: mutations, genes and function. Phil. Trans. R. Soc. B 365, 2439-2450. ( 10.1098/rstb.2010.0104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marques DA, Taylor JS, Jones FC, Di Palma F, Kingsley DM, Reimchen TE.. 2017. Convergent evolution of SWS2 opsin facilitates adaptive radiation of threespine stickleback into different light environments. PLoS Biol. 15, 1-24. ( 10.1371/journal.pbio.2001627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhen Y, Aardema ML, Medina EM, Schumer M, Andolfatto P. 2012. Parallel molecular evolution in an herbivore community. Science 337, 1634-1637. ( 10.1126/science.1226630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauer S, Gresham D. 2019. An evolving view of copy number variants. Curr. Genet. 65, 1287-1295. ( 10.1007/s00294-019-00980-0) [DOI] [PubMed] [Google Scholar]
- 11.Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. R. Soc. B 279, 5048-5057. ( 10.1098/rspb.2012.1108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohno S. 1970. Evolution by gene duplication. Berlin, Germany: Springer. See https://books.google.co.jp/books?id=sxUDAAAAMAAJ. [Google Scholar]
- 13.Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938-950. ( 10.1038/nrg2482) [DOI] [PubMed] [Google Scholar]
- 14.Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11, 97-108. ( 10.1038/nrg2689) [DOI] [PubMed] [Google Scholar]
- 15.Moore RC, Purugganan MD. 2003. The early stages of duplicate gene evolution. Proc. Natl Acad. Sci. USA 100, 15 682-15 687. ( 10.1073/pnas.2535513100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lye ZN, Purugganan MD. 2019. Copy number variation in domestication. Trends Plant Sci. 24, 352-365. ( 10.1016/j.tplants.2019.01.003) [DOI] [PubMed] [Google Scholar]
- 17.Arendt M, Fall T, Lindblad-Toh K, Axelsson E. 2014. Amylase activity is associated with AMY2B copy numbers in dog: implications for dog domestication, diet and diabetes. Anim. Genet. 45, 716-722. ( 10.1111/age.12179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pajic P, et al. 2019. Independent amylase gene copy number bursts correlate with dietary preferences in mammals. Elife 8, 1-22. ( 10.7554/eLife.44628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa A, et al. 2019. A key metabolic gene for recurrent freshwater colonization and radiation in fishes. Science 364, 886-889. ( 10.1126/science.aau5656) [DOI] [PubMed] [Google Scholar]
- 20.Cooley AM, Modliszewski JL, Rommel ML, Willis JH. 2011. Gene duplication in mimulus underlies parallel floral evolution via independent trans-regulatory changes. Curr. Biol. 21, 700-704. ( 10.1016/j.cub.2011.03.028) [DOI] [PubMed] [Google Scholar]
- 21.Betancur-R R, Ortí G, Pyron RA. 2015. Fossil-based comparative analyses reveal ancient marine ancestry erased by extinction in ray-finned fishes. Ecol. Lett. 18, 441-450. ( 10.1111/ele.12423) [DOI] [PubMed] [Google Scholar]
- 22.Arts MT, Brett MT, Kainz MJ. 2009. Lipids in aquatic ecosystems. New York, NY: Springer. [Google Scholar]
- 23.Bell MA, Foster SA. 1994. The evolutionary biology of the threespine stickleback. Oxford, UK: Oxford University Press. [Google Scholar]
- 24.Lowe CB, Sanchez-Luege N, Howes TR, Brady SD, Daugherty RR, Jones FC, Bell MA, Kingsley DM. 2018. Detecting differential copy number variation between groups of samples. Genome Res. 28, 256-265. ( 10.1101/gr.206938.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirase S, Ozaki H, Iwasaki W. 2014. Parallel selection on gene copy number variations through evolution of three-spined stickleback genomes. BMC Genomics 15, 735. ( 10.1186/1471-2164-15-735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitano J, Ishikawa A, Kusakabe M. 2019. Parallel transcriptome evolution in stream threespine sticklebacks. Dev. Growth Differ. 61, 104-113. ( 10.1111/dgd.12576) [DOI] [PubMed] [Google Scholar]
- 27.Jones FC, et al. 2012. A genome-wide SNP genotyping array reveals patterns of global and repeated species-pair divergence in sticklebacks. Curr. Biol. 22, 83-90. ( 10.1016/j.cub.2011.11.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang B, Merilä J, Ribeiro F, Alexandre CM, Momigliano P. 2018. Worldwide phylogeny of three-spined sticklebacks. Mol. Phylogenet. Evol. 127, 613-625. ( 10.1016/j.ympev.2018.06.008) [DOI] [PubMed] [Google Scholar]
- 29.Yamasaki YY, Mori S, Kokita T, Kitano J. 2019. Armour plate diversity in Japanese freshwater threespine stickleback (Gasterosteus aculeatus). Evol. Ecol. Res. 20, 51-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitano J, Lema SC, Luckenbach JA, Mori S, Kawagishi Y, Kusakabe M, Swanson P, Peichel CL. 2010. Adaptive divergence in the thyroid hormone signaling pathway in the stickleback radiation. Curr. Biol. 20, 2124-2130. ( 10.1016/j.cub.2010.10.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang B, Kemppainen P, Momigliano P, Feng X, Merilä J. 2020. On the causes of geographically heterogeneous parallel evolution in sticklebacks. Nat. Ecol. Evol. 4, 1105-1115. ( 10.1038/s41559-020-1222-6) [DOI] [PubMed] [Google Scholar]
- 32.Magalhaes IS, Whiting JR, D'Agostino D, Hohenlohe PA, Mahmud M, Bell MA, Skúlason S, MacColl ADC. 2021. Intercontinental genomic parallelism in multiple three-spined stickleback adaptive radiations. Nat. Ecol. Evol. 5, 251-261. ( 10.1038/s41559-020-01341-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fricke R, Eschmeyer WN, van der Laan R. 2019. Eschmeyer's catalog of fishes: genera, species, references. See www.calacademy.org/scientists/projects/catalog-of-fishes. [DOI] [PubMed]
- 34.Cosentino S, Iwasaki W. 2019. SonicParanoid: fast, accurate and easy orthology inference. Bioinformatics 35, 149-151. ( 10.1093/bioinformatics/bty631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson IB, Horiguchi M, Zilberberg L, Dabovic B, Hadjiolova K, Rifkin DB. 2015. Latent TGF-β-binding proteins. Matrix Biol. 47, 44-53. ( 10.1016/j.matbio.2015.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung J, Nakatsu F, Baskin JM, De Camilli P. 2015. Plasticity of PI 4 KIII α interactions at the plasma membrane. EMBO Rep. 16, 312-320. ( 10.15252/embr.201439151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, Synowsky S, Tinti M, MacKintosh C. 2011. The capture of phosphoproteins by 14-3-3 proteins mediates actions of insulin. Trends Endocrinol. Metab. 22, 429-436. ( 10.1016/j.tem.2011.07.005) [DOI] [PubMed] [Google Scholar]
- 38.Cao J, Tan X. 2018. Comparative and evolutionary analysis of the 14-3-3 family genes in eleven fishes. Gene 662, 76-82. ( 10.1016/j.gene.2018.04.016) [DOI] [PubMed] [Google Scholar]
- 39.Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. 2006. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52, 953-968. ( 10.1016/j.neuron.2006.11.019) [DOI] [PubMed] [Google Scholar]
- 40.Wong Y, Schulze C, Streichert T, Gronostajski RM, Schachner M, Tilling T. 2007. Gene expression analysis of nuclear factor I-A deficient mice indicates delayed brain maturation. Genome Biol. 8, R72. ( 10.1186/gb-2007-8-5-r72) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato E, Muto J, Zhang LJ, Adase CA, Sanford JA, Takahashi T, Nakatsuji T, Usdin TB, Gallo RL. 2016. The parathyroid hormone second receptor PTH2R and its ligand tuberoinfundibular peptide of 39 residues TIP39 regulate intracellular calcium and influence keratinocyte differentiation. J. Invest. Dermatol. 136, 1449-1459. ( 10.1016/j.jid.2016.02.814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, Won HH, Kim Y, Choi JR, Yu N, Lee KA. 2015. Breakpoint mapping by whole genome sequencing identifies PTH2R gene disruption in a patient with midline craniosynostosis and a de novo balanced chromosomal rearrangement. J. Med. Genet. 52, 706-709. ( 10.1136/jmedgenet-2015-103001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saju JM, Hossain MS, Liew WC, Pradhan A, Thevasagayam NM, Tan LSE, Anand A, Olsson PE, Orbán L. 2018. Heat shock factor 5 is essential for spermatogenesis in zebrafish. Cell Rep. 25, 3252-3261.e4. ( 10.1016/j.celrep.2018.11.090) [DOI] [PubMed] [Google Scholar]
- 44.Ma B, Simala-Grant JL, Taylor DE. 2006. Fucosylation in prokaryotes and eukaryotes. Glycobiology 16, 158R-184R. ( 10.1093/glycob/cwl040) [DOI] [PubMed] [Google Scholar]
- 45.Yang CS, et al. 2017. Ubiquitin modification by the E3 ligase/ADP-ribosyltransferase Dtx3L/Parp9. Mol. Cell 66, 503-516.e5. ( 10.1016/j.molcel.2017.04.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. 2015. PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat. Immunol. 16, 1215-1227. ( 10.1038/ni.3279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahi EP, Brunel M, Tsakoumis E, Schmitz M. 2019. Transcriptional study of appetite regulating genes in the brain of zebrafish (Danio rerio) with impaired leptin signalling. Sci. Rep. 9, 1-14. ( 10.1038/s41598-019-56779-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sommer F, Torraca V, Kamel SM, Lombardi A, Meijer AH. 2020. Antagonism between regular and atypical Cxcr3 receptors regulates macrophage migration during infection and injury in zebrafish. J. Leukoc. Biol. 107, 185-203. ( 10.1002/JLB.2HI0119-006R) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitturi DA, et al. 2013. Modulation of nitro-fatty acid signaling: prostaglandin reductase-1 is a nitroalkene reductase. J. Biol. Chem. 288, 25626-25637. ( 10.1074/jbc.M113.486282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang GJ, Lee HJ, Byun HJ, Kim EJ, Kim HJ, Park MK, Lee CH. 2017. Novel involvement of miR-522-3p in high-mobility group box 1-induced prostaglandin reductase 1 expression and reduction of phagocytosis. Biochim. Biophys. Acta 1864, 625-633. ( 10.1016/j.bbamcr.2017.01.006) [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Yin G, Zhang W, Song K, Zhang L, Guo Z. 2021. Prostaglandin reductase 1 as a potential therapeutic target for cancer therapy. Front. Pharmacol. 12, 1-6. ( 10.3389/fphar.2021.717730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fay MJ, Nguyen MT, Snouwaert JN, Dye R, Grant DJ, Bodnar WM, Koller BH. 2015. Xenobiotic metabolism in mice lacking the UDP-glucuronosyltransferase 2 family. Drug Metab. Dispos. 43, 1838-1846. ( 10.1124/dmd.115.065482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J, Liu Y, Barter RA, Klaassen CD. 1995. Alteration of thyroid homeostasis by UDP-glucuronosyltransferase inducers in rats: a dose-response study. J. Pharmacol. Exp. Ther. 273, 977-985. [PubMed] [Google Scholar]
- 54.Vansell NR, Klaassen CD. 2002. Increase in rat liver UDP-glucuronosyltransferase mRNA by microsomal enzyme inducers that enhance thyroid hormone glucuronidation. Drug Metab. Dispos. 30, 240. ( 10.1124/dmd.30.3.240) [DOI] [PubMed] [Google Scholar]
- 55.Richardson TA, Klaassen CD. 2010. Role of UDP-glucuronosyltransferase (UGT) 2B2 in metabolism of triiodothyronine: effect of microsomal enzyme inducers in Sprague Dawley and UGT2B2-deficient Fischer 344 rats. Toxicol. Sci. 116, 413-421. ( 10.1093/toxsci/kfq125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dutt P, Croall DE, Arthur JSC, De Veyra T, Williams K, Elce JS, Greer PA. 2006. m-Calpain is required for preimplantation embryonic development in mice. BMC Dev. Biol. 6, 1-11. ( 10.1186/1471-213X-6-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takano J, Mihira N, Fujioka R, Hosoki E, Chishti AH, Saido TC. 2011. Vital role of the calpain-calpastatin system for placental-integrity-dependent embryonic survival. Mol. Cell. Biol. 31, 4097-4106. ( 10.1128/mcb.05189-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Storrs CH, Silverstein SJ. 2007. PATJ, a tight junction-associated PDZ protein, is a novel degradation target of high-risk human papillomavirus E6 and the alternatively spliced isoform 18 E6. J. Virol. 81, 4080-4090. ( 10.1128/jvi.02545-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shin K, Straight S, Margolis B. 2005. PATJ regulates tight junction formation and polarity in mammalian epithelial cells. J. Cell Biol. 168, 705-711. ( 10.1083/jcb.200408064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merrill RA, Plum LA, Kaiser ME, Clagett-Dame M. 2002. A mammalian homolog of unc-53 is regulated by all-trans retinoic acid in neuroblastoma cells and embryos. Proc. Natl Acad. Sci. USA 99, 3422-3427. ( 10.1073/pnas.052017399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang D, et al. 2007. Efficient genome-wide mutagenesis of zebrafish genes by retroviral insertions. Proc. Natl Acad. Sci. USA 104, 12428-12433. ( 10.1073/pnas.0705502104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y, Liu H, Chen W, Yang T, Zhang W. 2014. EOLA1 protects lipopolysaccharide induced IL-6 production and apoptosis by regulation of MT2A in human umbilical vein endothelial cells. Mol. Cell. Biochem. 395, 45-51. ( 10.1007/s11010-014-2110-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veillette A, Bookman MA, Horak EM, Bolen JB. 1988. The CD4 and CD8 T cell surface antigens are associated with the internal membrane tyrosine-protein kinase p56lck. Cell 55, 301-308. ( 10.1016/0092-8674(88)90053-0) [DOI] [PubMed] [Google Scholar]
- 64.von Herrath MG, Yokoyama M, Dockter J, Oldstone MB, Whitton JL. 1996. CD4-deficient mice have reduced levels of memory cytotoxic T lymphocytes after immunization and show diminished resistance to subsequent virus challenge. J. Virol. 70, 1072-1079. ( 10.1128/jvi.70.2.1072-1079.1996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsukamoto K, Miura F, Fujito NT, Yoshizaki G, Nonaka M. 2012. Long-lived dichotomous lineages of the proteasome subunit beta type 8 (PSMB8) gene surviving more than 500 million years as alleles or paralogs. Mol. Biol. Evol. 29, 3071-3079. ( 10.1093/molbev/mss113) [DOI] [PubMed] [Google Scholar]
- 66.Eve AMJ, Smith JC. 2017. Knockdown of Laminin gamma-3 (Lamc3) impairs motoneuron guidance in the zebrafish embryo [version 1; referees: 2 approved, 2 approved with reservations]. Wellcome Open Res. 2, 1-25. ( 10.12688/wellcomeopenres.12394.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramalingam L, Oh E, Yoder SM, Brozinick JT, Kalwat MA, Groffen AJ, Verhage M, Thurmond DC. 2012. Doc2b is a key effector of insulin secretion and skeletal muscle insulin sensitivity. Diabetes 61, 2424-2432. ( 10.2337/db11-152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Groffen AJ, Martens S, Díez Arazola R. 2010. Doc2b is a high-affinity Ca2+ sensor for spontaneous neurotransmitter release. Science 327, 1614-1618. ( 10.1126/science.1183765) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hodul M, Dahlberg CL, Juo P. 2017. Function of the deubiquitinating enzyme USP46 in the nervous system and its regulation by WD40-repeat proteins. Front. Synapt. Neurosci. 9, 1-7. ( 10.3389/fnsyn.2017.00016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tomida S, et al. 2009. Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nat. Genet. 41, 688-695. ( 10.1038/ng.344) [DOI] [PubMed] [Google Scholar]
- 71.Lei H, et al. 2021. Targeting USP47 overcomes tyrosine kinase inhibitor resistance and eradicates leukemia stem/progenitor cells in chronic myelogenous leukemia. Nat. Commun. 12, 1-16. ( 10.1038/s41467-020-20259-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palazón‐Riquelme P, Worboys JD, Green J, Valera A, Martín‐Sánchez F, Pellegrini C, Brough D, López‐Castejón G. 2018. USP7 and USP47 deubiquitinases regulate NLRP3 inflammasome activation. EMBO Rep. 19, 1-17. ( 10.15252/embr.201744766) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Betancur-R R, Wiley EO, Arratia G, Acero A, Baily N, Miya M, Lecointre G, Orti G. 2017. Phylogenetic classification of bony fishes. BMC Evol. Biol. 17, 162. ( 10.1186/s12862-017-0958-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Felsenstein J. 1985. Phylogenies and the comparative method. Am. Soc. Nat. 125, 1-15. ( 10.1086/284325) [DOI] [Google Scholar]
- 75.Hadfield JD. 2010. MCMCglmm: MCMC methods for multi-response GLMMs in R. J. Stat. Softw. 33, 1-22. ( 10.18637/jss.v033.i02)20808728 [DOI] [Google Scholar]
- 76.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. 2019. G:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191-W198. ( 10.1093/nar/gkz369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jones FC, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55-61. ( 10.1038/nature10944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.R Core Team. 2019. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 79.Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1-48. ( 10.18637/jss.v067.i01) [DOI] [Google Scholar]
- 80.Noé L, Kucherov G. 2005. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 33, 540-543. ( 10.1093/nar/gki478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kohany O, Gentles AJ, Hankus L, Jurka J. 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinf. 7, 1-7. ( 10.1186/1471-2105-7-474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Twining C, et al. 2021. The evolutionary ecology of fatty-acid variation: implications for consumer adaptation and diversification. Ecol. Lett. 24, 1709-1731. ( 10.1111/ele.13771) [DOI] [PubMed] [Google Scholar]
- 83.Ishikawa A, Kusakabe M, Yoshida K, Ravinet M, Makino T, Toyoda A, Fujiyama A, Kitano J. 2017. Different contributions of local- and distant-regulatory changes to transcriptome divergence between stickleback ecotypes. Evolution 71, 565-581. ( 10.1111/evo.13175) [DOI] [PubMed] [Google Scholar]
- 84.Poulin R. 2016. Greater diversification of freshwater than marine parasites of fish. Int. J. Parasitol. 46, 275-279. ( 10.1016/j.ijpara.2015.12.002) [DOI] [PubMed] [Google Scholar]
- 85.Han K, Lou DI, Sawyer SL. 2011. Identification of a genomic reservoir for new trim genes in primate genomes. PLoS Genet. 7, e1002388. ( 10.1371/journal.pgen.1002388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fehling HJ, Swat W, Laplace C, Kühn R, Rajewsky K, Müller U, Von Boehmer H.. 1994. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science 265, 1234-1237. ( 10.1126/science.8066463) [DOI] [PubMed] [Google Scholar]
- 87.Klamp T, Boehm U, Schenk D, Pfeffer K, Howard JC. 2003. A giant GTPase, very large inducible GTPase-1, is inducible by IFNs. J. Immunol. 171, 1255-1265. ( 10.4049/jimmunol.171.3.1255) [DOI] [PubMed] [Google Scholar]
- 88.Kim B-H, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. 2012. IFN-inducible GTPases in host defense. Cell Host Microbe 12, 432-444. ( 10.1016/j.chom.2012.09.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robledo D, Hamilton A, Gutiérrez AP, Bron JE, Houston RD. 2020. Characterising the mechanisms underlying genetic resistance to amoebic gill disease in Atlantic salmon using RNA sequencing. BMC Genomics 21, 1-11. ( 10.1186/s12864-020-6694-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park SH, et al. 2012. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature 491, 779-783. ( 10.1038/nature11580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu Y, Yang S, Lin AA, Cavalli-Sforza LL, Su B. 2005. Molecular evolution of CXCR1, a G protein-coupled receptor involved in signal transduction of neutrophils. J. Mol. Evol. 61, 691-696. ( 10.1007/s00239-005-0039-x) [DOI] [PubMed] [Google Scholar]
- 92.Godaly G, Hang L, Frendéus B, Svanborg C. 2000. Transepithelial neutrophil migration is CXCR1 dependent in vitro and is defective in IL-8 receptor knockout mice. J. Immunol. 165, 5287-5294. ( 10.4049/jimmunol.165.9.5287) [DOI] [PubMed] [Google Scholar]
- 93.Capucetti A, Albano F, Bonecchi R. 2020. Multiple roles for chemokines in neutrophil biology. Front. Immunol. 11, 1-9. ( 10.3389/fimmu.2020.01259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stafuzza NB, Silva RMDO, Fragomeni BDO, Masuda Y, Huang Y, Gray K, Lourenco DAL. 2019. A genome-wide single nucleotide polymorphism and copy number variation analysis for number of piglets born alive. BMC Genomics 20, 1-11. ( 10.1186/s12864-019-5687-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu J, Liu Y, Barter RA, Klaassen CD. 1995. Alteration of thyroid homeostasis by UDP-glucuronosyltransferase inducers in rats: a dose-response study. J. Pharmacol. Exp. Ther. 273, 977-985. [PubMed] [Google Scholar]
- 96.Vansell NR, Klaassen CD. 2002. Increase in rat liver UDP-glucuronosyltransferase mRNA by microsomal enzyme inducers that enhance thyroid hormone glucuronidation. Drug Metab. Dispos. 30, 240-246. ( 10.1124/dmd.30.3.240) [DOI] [PubMed] [Google Scholar]
- 97.Kitano J, Lema SC. 2013. Divergence in thyroid hormone concentrations between juveniles of marine and stream ecotypes of the threespine stickleback (Gasterosteus aculeatus). Evol. Ecol. Res. 15, 143-153. [Google Scholar]
- 98.Ishikawa A, Kitano J. 2012. Ecological genetics of thyroid hormone physiology in humans and wild animals. In Thyroid hormone (ed. Agrawal NK), pp. 37-50. London. UK: IntechOpen. [Google Scholar]
- 99.Brown DD. 1997. The role of thyroid hormone in zebrafish and axolotl development. Dev. Biol. 94, 13 011-13 016. ( 10.1073/pnas.94.24.13011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tagawa M, Hirano T. 1987. Presence of thyroxine in eggs and changes in its content during early development of chum salmon, Oncorhynchus keta. Gen. Comp. Endocrinol. 68, 129-135. ( 10.1016/0016-6480(87)90068-2) [DOI] [PubMed] [Google Scholar]
- 101.Tagawa M, Tanaka M, Matsumoto S, Hirano T. 1990. Thyroid hormones in eggs of various freshwater, marine and diadromous teleosts and their changes during egg development. Fish Physiol. Biochem. 8, 515-520. ( 10.1007/BF00003409) [DOI] [PubMed] [Google Scholar]
- 102.Houde ED. 1992. Are marine and freshwater fish larvae different? Int. Counc. Explor. Sea (C.M. 1992/L:26), 13. [Google Scholar]
- 103.Zhang J, Zuo T, Peterson T. 2013. Generation of tandem direct duplications by reversed-ends transposition of maize ac elements. PLoS Genet. 9, e1003691. ( 10.1371/journal.pgen.1003691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Conte GL, Arnegard ME, Peichel CL, Schluter D. 2012. The probability of genetic parallelism and convergence in natural populations. Proc. R. Soc. B 279, 5039-5047. ( 10.1098/rspb.2012.2146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ishikawa A, Yamanouchi S, Iwasaki W, Kitano J. 2022. Convergent copy number increase of genes associated with freshwater colonization in fishes. Figshare. ( 10.6084/m9.figshare.c.5958820) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Ishikawa A, Yamanouchi S, Iwasaki W, Kitano J. 2022. Convergent copy number increase of genes associated with freshwater colonization in fishes. Figshare. ( 10.6084/m9.figshare.c.5958820) [DOI] [PMC free article] [PubMed]
Data Availability Statement
The whole-genome sequence data are available from DDBJ DRA007515, DRA001136 and DRA007518. The datasets supporting this article have been uploaded as part of the electronic supplementary material [105]. Electronic supplementary material, table S1: candidate genes that show increased copy numbers associated with freshwater colonization. Electronic supplementary material, table S2: candidate genes that show increased copy numbers in non-freshwater fish species. Electronic supplementary material, table S3: GO enrichment analysis for 58 candidate genes that show increased copy numbers associated with freshwater colonization. Electronic supplementary material, table S4: GO enrichment analysis for 23 candidate genes that remained significant even after phylogenetic correction. Electronic supplementary material, table S5: GO enrichment analysis for 18 candidate genes that show increased copy numbers in non-freshwater species. Electronic supplementary material, table S6: GO enrichment analysis for 12 candidate genes that show increased copy numbers in non-freshwater species and remained significant even after phylogenetic correction. Electronic supplementary material, table S7: candidate genes that show convergent increases of copy number in Japanese freshwater sticklebacks. Electronic supplementary material, table S8: transposons near the UGT2 gene in a freshwater stickleback. Electronic supplementary material, figure S1: High-resolution image of figure 3.