Abstract
BACKGROUND
Fusobacterium nucleatum (F. nucleatum) has long been known to cause opportunistic infections and has recently been implicated in colorectal cancer (CRC), which has attracted broad attention. However, the mechanism by which it is involved in CRC development is not fully understood.
AIM
To explore its potential causative role in CRC development, we evaluated the colon pathology, mucosa barrier, colon microbiota and host transcriptome profile after F. nucleatum infection in an azoxymethane/dextran sulfate sodium salt (AOM/DSS) mouse model.
METHODS
Three groups of mice were compared to reveal the differences, i.e., the control, AOM/DSS-induced CRC and AOM/DSS-FUSO infection groups.
RESULTS
Both the AOM/DSS and AOM/DSS-FUSO groups exhibited a significantly reduced body weight and increased tumor numbers than the control group, and AOM/DSS mice with F. nucleatum infection showed the highest tumor formation ratio among the three groups. Moreover, the colon pathology was the most serious in the AOM/DSS-FUSO group. We found that the structure of the colon microbiota changed considerably after F. nucleatum infection; striking differences in mucosal microbial population patterns were observed between the AOM/DSS-FUSO and AOM/DSS groups, and inflammation-inducing bacteria were enriched in the mucosal microbiota in the AOM/DSS-FUSO group. By comparing intestinal transcriptomics data from AOM vs AOM/DSS-FUSO mice, we showed that transcriptional activity was strongly affected by dysbiosis of the gut microbiota. The most microbiota-sensitive genes were oncogenes in the intestine, and the cyclic adenosine monophosphate signaling pathway, neuroactive ligand–receptor interaction, PPAR signaling pathway, retinol metabolism, mineral absorption and drug metabolism were highly enriched in the AOM/DSS-FUSO group. Additionally, we showed that microbial dysbiosis driven by F. nucleatum infection enriched eight taxa belonging to Proteobacteria, which correlates with increased expression of oncogenic genes.
CONCLUSION
Our study demonstrated that F. nucleatum infection altered the colon mucosal microbiota by enriching pathogens related to the development of CRC, providing new insights into the role of F. nucleatum in the oncogenic microbial environment of the colon.
Keywords: Fusobacterium nucleatum, Mucosal microbiota, Transcriptome, Colorectal cancer, Inflammation-inducing bacteria
Core Tip: Fusobacterium nucleatum (F. nucleatum) has long been known to cause opportunistic infections and has recently been implicated in colorectal cancer (CRC). However, the mechanism by which it is involved in CRC development is not fully understood. Here, striking differences in mucosal microbial population patterns were observed and inflammation-inducing bacteria were enriched in the mucosal microbiota in the azoxymethane/dextran sulfate sodium salt-FUSO group. We showed that transcriptional activity was strongly affected by dysbiosis of the gut microbiota. Additionally, we showed that microbial dysbiosis driven by F. nucleatum infection enriched eight taxa belonging to Proteobacteria, which correlates with increased expression of oncogenic genes.
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer and the fourth leading cause of cancer death worldwide[1]. The incidence of CRC in China has increased dramatically during the past decade[2].
Fusobacterium nucleatum (F. nucleatum), a gram-negative anaerobe, was recently identified as an emerging pathogen that has been frequently found in CRC[3-5]. Importantly, overload of F. nucleatum was found to be associated with an advanced tumor stage and a poor prognosis in CRC[6]. Accumulating evidence has revealed that F. nucleatum influences different stages of CRC progression. F. nucleatum can promote the initiation and development of CRC by adhering to epithelial cells through its unique FadA protein[7], which can bind vascular endothelial cadherin and alters endothelial integrity[8]. The activation of oncogenic miR21 under F. nucleatum stimulation results in the overexpression of TLR-4/nuclear factor-kappaB, increasing the proliferation of CRC cells and promoting tumor development[9]. Once a colon tumor has developed, F. nucleatum can adhere to Gal-GalNAc-expressing tumor cells by binding to the Fap2 Lectin, leading to enrichment of F. nucleatum[10]. F. nucleatum infection was also proven to increase the expression levels of inflammatory genes, transcription factors, and oncogenes[8,11]. These studies enhance our knowledge of the mechanisms by which F. nucleatum shapes the tumor microenvironment.
Evidences have revealed that the local microbiota is also an important part of the tumor microenvironment. Gut microbiota dysbiosis can reshape the tumor microenvironment and make it favorable for tumor growth[12]. The mechanisms involved include increasing mutagenesis, regulation of oncogenic pathways, and modulation of host immune system[13]. These oncogenic functions of gut microbiota are probably due to the microbial products or metabolites they produce when the community structure balance is disrupted and a resulting overgrowth of specific bacteria. For example, the overload of colibactin-producing Escherichia coli and other Enterobacteriaceae can induce DNA damage of host cells and exert tumorigenic effect[14]. These facts suggest a close relationship between gut microbiota, the tumor microenvironment, and CRC. However, the regulatory role of F. nucleatum in the “oncogenic microbial environment” of the colon remains mostly unknown. Revealing the effects of F. nucleatum infection on the changes of mucosal microbiota will help to provide further insight into the mechanisms of the tumorigenic effect of F. nucleatum.
In the present study, we explored the role of F. nucleatum in the development of CRC and in altering the mucosal microbial community structure and investigated the association between microbial dysbiosis and the expression of oncogenic signaling pathways. F. nucleatum strain ATCC 25586 was gavaged into azoxymethane/dextran sulfate sodium salt (AOM/DSS) mice to explore the role of Fusobacterium overload in the progression of intestinal adenoma against a background of environmental inflammatory factors. RNA-seq transcriptomic analysis was used to identify the differentially expressed gene (DEG) signatures driven by F. nucleatum infection, and integrated network analysis was conducted to reveal the relationship between microbial dysbiosis and the DEGs. Our study lays a foundation for understanding the regulatory role of F. nucleatum in the oncogenic microbial environment of the colon.
MATERIALS AND METHODS
Bacterial strain and culture conditions
The F. nucleatum strain ATCC 25586 was purchased from American Type Culture Collection (ATCC) and was cultured as described previously[3]. In brief, F. nucleatum was grown in Columbia blood agar supplemented with 5 mg/mL haemin, 5% defibrinated sheep blood, and 1 mg/mL Vitamin K1 in an anaerobic condition with 85% N2, 10% H2, and 5% CO2 at 37 °C. And 109 colony-forming units per mL (CFU/mL) were prepared for F. nucleatum colonization.
Animal model
Thirty-six conventional C57BL/6J wild-type male mice (4 wk of age) were obtained from the Beijing Animal Study Centre and maintained in the Laboratory Animal Services Centre at the Chinese Center for Disease Control and Prevention. The Institutional Animal Care Committee of the Peking University People’s Hospital approved all animal experimentation.
To create the mouse model of colon neoplasia induced by AOM/DSS-induced colon neoplasia[15], 24 mice at 6 wk of age were injected intraperitoneally with a single dose of 10 mg/kg AOM (Sigma), designated week 0. DSS (MP Biochemicals; 36000-50000 mw) was dissolved in filtered drinking water and administered to the mice for 7 d during the following weeks: Weeks 1 (1% DSS), 3 (1% DSS), and 5 (1% DSS). After 1 wk of AOM/DSS treatment, the 24 mice were divided into 2 groups, each with 12 mice. At week 7 they were orally gavaged daily with 109 CFU/mL (in 100 μL volume for each mouse) F. nucleatum (AOM/DSS-FUSO group) or phosphate-buffered saline (AOM/DSS group) for two weeks. The 12 mice in the control group were sibling littermates and received no treatment.
After 2 wk after Fusobacterium inoculation as previous study described[16], the mice were sacrificed, colon mucosal contents were collected for microbiota profiling, the colons were dissected, and the polyps were counted. For a subset of the samples, the distal portion of the colon was fixed in 4% paraformaldehyde, paraffin-embedded, and sectioned for hematoxylin and eosin staining. The remaining colon tissue was then prepared for RNA sequencing.
The body weight, colon length, colorectal tumor formation ratio, immune cell patterns, gut microbiota structure and intestine transcriptomics were assessed to explore the potential role of ATCC 25586 in the carcinogenesis/development of CRC.
Histological evaluation
The colon segments were submitted for histological processing. All slides were stained with hematoxylin and eosin. Dysplasia was assessed by the presence of hyperchromasia, nuclear pleomorphism, and increased nuclear-to-cytoplasmic ratios.
Immunohistochemical staining
The expression of E-cadherin and Ki-67 was determined by immunohistochemistry staining. Serial 3 mm sections of the formalin-fixed paraffin-embedded colon tissues on the slides were heated in a microwave with sodium citrate for antigen repair, followed by 2% goat serum for blocking. The slides were then treated with mouse anti-E-cadherin (BD Biosciences, catalog 610181) and anti-Ki-67 (Cell Signaling Technology, catalog 9192) overnight at 4 °C, visualized with diaminobenzidine, counterstained with hematoxylin, dehydrated and mounted. Staining index of Ki-67 immunostaining was determined by percentage of positive cells having brown nuclear staining from three randomly selected fields at 400 × magnification.
Microbial DNA extraction and 16S rRNA gene sequencing
Microbiota DNA from the colon mucosal contents was extracted using the QIAamp DNA Stool Mini Kit (Qiagen, catalog 51604). For all samples, polymerase chain reaction (PCR) amplification of the 16S rRNA gene with the V3-V4 variable regions was performed using the primers 341F (5’-CCTAYGGGRBGCASCAG-3’) and 806R (5’-GGACTACNNGGGTATCTAAT-3’)[17]. The pooled library was sequenced on the Ion S5TM XL System (Thermo Fisher Scientific). Single-end reads were assigned to the samples based on their unique barcodes and trimmed by removing the barcodes and primer sequences. Raw fastq files were quality-filtered using fastp[18]. Chimeric sequences were detected and removed by vsearch software[19]. Operational taxonomic units (OTUs) were clustered with 97% identity using UPARSE v7.0.1001[20]. The most frequent sequences were selected as the representative sequences for each OTU. Representative sequences for each OTU were annotated using Mothur software at Threshold: 0.8-1 against the SILVA V132 database (http://www.arb-silva.de/) at each taxonomic rank (kingdom, phylum, class, order, family, genus and species)[21]. OTU abundance information was normalized using a standard sequence number corresponding to the sample with the fewest sequences. To avoid biases, the normalized data were subjected to alpha diversity and beta diversity analyses. Alpha diversity indices (Chao1, ACE, Shannon index, Simpson) and UniFrac distance were calculated by QIIME v1.9.1[22,23]. Beta diversities were visualized by principal coordinate analysis (PCoA) and nonmetric multidimensional (NMDS) based on Bray–Curtis dissimilarity.
Quantitative real-time PCR for F. nucleatum
The Q-PCR assay was performed to determine the colonization of F. nucleatum. The 16S primers for F. nucleatum quantification were F-TGCGATAAGCCTAGATAAGTTGCA and R-CTTAATAGATTGCTCCATTCGGAAA. Primers for total bacterial DNA were F-ACTCCTACGGGAGGCAGCAGT and R-GTATTACCGCGGCTGCTGGCAC[16]. The level of F. nucleatum was evaluated by the following formula: The relative value (RV) of F. nucleatum was determined by the 2-Δct method using total bacterial level as a normalizer. The relative abundance (RA) of F. nucleatum was scaled to a reasonable range with the following equation: RA = log2 (RV × 10000 + 1).
RNA extraction and RNA sequencing
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s recommendations. A total of 3 μg RNA per sample was used as the input material for the RNA sample preparations. Sequencing libraries were generated using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, United States) following the manufacturer’s recommendations. The library preparations were sequenced on an Illumina platform, and 150 bp paired-end reads were generated.
The raw reads were first processed through in-house Perl scripts. Adaptor sequences, duplicated sequences, poly-N reads containing more than 10% “N”, and low-quality reads containing more than 50% bases with Q ≤ 5 were removed to obtain high-quality clean reads. Clean reads were aligned to the reference genome using Hisat2 v2.0.5[24]. FeatureCounts v1.5.0-p3 was used to count the read numbers mapped to each gene[25]. Then, the FPKM of each gene was calculated based on the length of the gene and the read count mapped to this gene. Prior to the differential gene expression analysis, for each sequenced library, the read counts were adjusted by the edgeR package. Differential gene expression analysis of the two conditions was performed using the edgeR package v3.18.1. The P values were adjusted using the Benjamini & Hochberg method[26]. A corrected P value of 0.05 and absolute fold change of 2 were set as the thresholds for significantly DEGs. Gene Ontology (GO) enrichment analysis of the DEGs was implemented by the clusterProfiler R package. GO terms with corrected P values less than 0.05 were considered significantly enriched by the DEGs. We used the clusterProfiler R package to test the statistical enrichment of the DEGs in the KEGG pathways[27].
Statistical analysis and visualization
The Kruskal–Wallis test was performed when comparing samples from more than two groups. The difference in ratio of diarrhea and bloody was assessed using Chi-square measure. Permutational multivariate analysis was applied with a parameter of 999 permutations in R[28]. Linear discriminant analysis effect size (LEfSe) was used to estimate which microbiome attributes differed significantly by treatments[29]. The relationship between the gut microbiota and DEGs was determined by calculating Spearman’s correlations, and then network visualization was performed using Cytoscape v3.6.1[30]. Heatmaps were generated using the R package pheatmap (https://github.com/raivokolde/pheatmap). The relationship between the different gut microbiota and the pathways was determined by calculating Spearman’s correlations. The R package ggplot2 was used for the other data visualization in the figures in the present study.
Ethics statement
All animal protocols were approved by the Animal Experimentation Ethics Committee of the Peking University People’s Hospital in China. The experiments were conducted in accordance with the Animal Research: Reporting of in Vivo Experiments (ARRIVE) guidelines.
RESULTS
F. nucleatum infection promotes the development of CRC
A flow chart illustrating the F. nucleatum infection strategy combined with AOM/DSS treatment is shown in Figure 1A. The colonization of F. nucleatum was confirmed by 16S-sequencing and Q-PCR (Supplementary Figure 1). The body weight of the mice in each group increased slowly (Figure 1B), and it differed significantly between the control mice and the AOM/DSS models (two-way ANOVA, P < 0.05). After AOM/DSS treatment for six weeks, four mice in the AOM/DSS group and six mice in the AOM/DSS-FUSO group developed diarrhea and bloody stools (Chi-square, P > 0.05). No bloody stool was observed in the control group. Over the eight-week treatment, only three mice in the AOM/DSS group and five mice in the AOM/DSS-FUSO group survived.
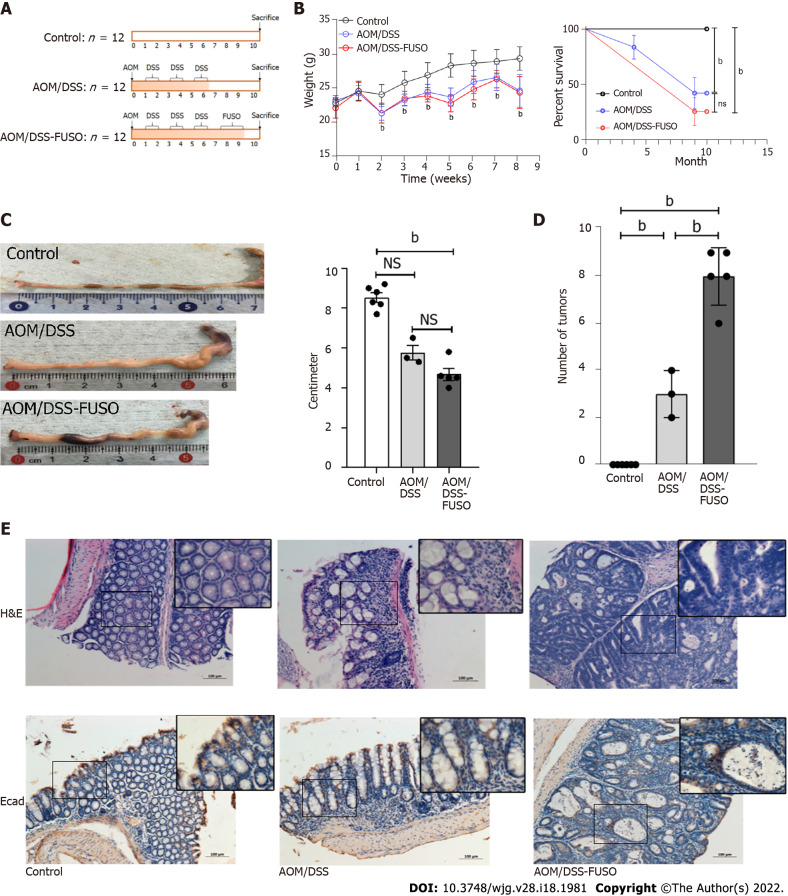
Figure 1.
Effect of F. nucleatum infection on colon carcinogenesis in the azoxymethane/dextran sulfate sodium-induced model. A: Experimental design illustrating the F. nucleatum infection strategy combined with azoxymethane/dextran sulfate sodium salt treatment; B: The body weight and survival curve of the three groups during the experiment was evaluated with two-way ANOVA; C: The colon lengths of the three groups were compared using a t-test; D: The numbers of tumors in the colon of the three groups were compared using a t-test; E: Hematoxylin-eosin staining and immunohistochemistry of E-cadherin-positive cells in the colon of the three groups. aP < 0.05; bP < 0.01. NS: No signification; AOM: Azoxymethane; DSS: Dextran sulfate sodium salt.
The AOM/DSS-FUSO group had a trend but not significantly reduced colon length than the AOM/DSS group (Figure 1C), and AOM/DSS-FUSO had the highest tumor formation ratio among the three groups (Figure 1D). The AOM/DSS mice exhibited histological inflammation. In addition, the colon pathology was the most serious in the AOM/DSS-FUSO group, which was confirmed by hematoxylin-eosin (H&E) staining (Figure 1E). The presence of hyperchromasia and nuclear pleomorphism were observed in the AOM/DSS-FUSO group. We also found increased epithelial cell proliferation in the colons of AOM/DSS-FUSO mice, as indicated by a higher expression of Ki-67 than in the other two groups (Supplementary Figure 2). The abnormal expression of E-cadherin in the AOM/DSS-FUSO mice reflected the impaired function of the intestinal barrier.
Aberrant activation of epithelial-mesenchymal transition (EMT) is considered an important trigger of cancer invasion and metastasis[31]. The AOM/DSS-FUSO colon tissues showed morphological features of partial EMT, which was not observed in the AOM/DSS model (Figure 1E). In addition to the morphology-based analysis of colon tissue by H&E staining, the assessment of E-cadherin expression by immunohistochemistry can support the evaluation of partial EMT[31]. The expression of the barrier protein E-cadherin was modestly reduced in the AOM/DSS-FUSO group. Loss of the prototypical epithelial barrier protein E-cadherin is an additional sign of partial EMT[32]. The abnormal histology changes and the reduction of E-cadherin in AOM/DSS-FUSO mouse colons indicated that the AOM/DSS mice with F. nucleatum infection had undergone partial EMT, which is critical for CRC progression.
F. nucleatum infection changes the mucosal microbiota community structure
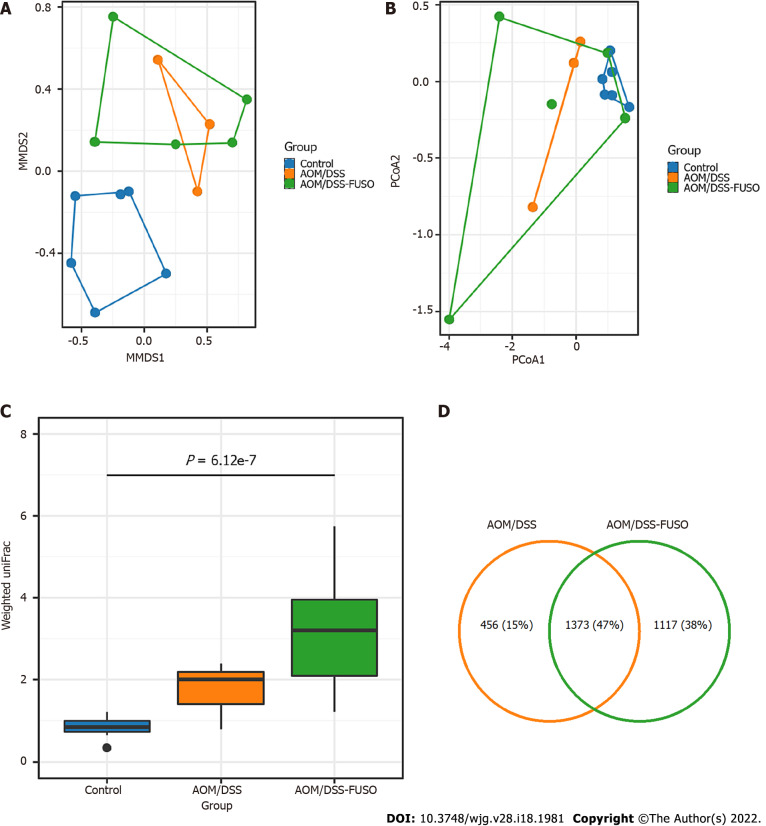
To investigate the effect of F. nucleatum infection on the mucosal microbiota, we compared the mucosal microbial composition of the three groups. The β diversity was very different among the three groups. The control group was clearly separated from the AOM/DSS-FUSO and AOM/DSS groups, as shown in the NMDS analysis (Figure 2A). This trend was also found in the PCoA analysis and was confirmed by the weighted UniFrac distance (Figure 2B and C). Around 12.8% of the bacterial composition variance could be explained by the addition of F. nucleatum (Permutation based analysis of variance). In AOM/DSS models, AOM/DSS-FUSO and AOM/DSS mice developed different mucosal microbiota with F. nucleatum infection as a significant factor in driving the differences. We then assessed the shared and unique OTUs in the AOM/DSS and AOM/DSS-FUSO groups to detect the effect of F. nucleatum infection. We observed that the AOM/DSS-FUSO group had more unique OTUs, approximately 44.9% (1117/2490), than the AOM/DSS group, 24.9% (456/1829), signifying that F. nucleatum-infected mice largely harbor unique inhabitant niches (Figure 2D). The Shannon, Simpson, observed species, and Chao1 indices for alpha diversity showed no significant differences among the three groups (Supplementary Figure 3).
Figure 2.
Diversity and shared operational taxonomic units of the gut microbiota in different groups. A: Nonmetric multidimensional plot of the cecal microbiota from the 14 samples based on Bray-Curtis similarities; B: Principal coordinate analysis plot of the cecal microbiota from the 14 samples based on Bray-Curtis similarities; C: Weighted UniFrac distance of the samples from different groups. Kruskal-Wallis test: P = 6.12 × 10-7; D: Number of shared operational taxonomic units between the samples from the azoxymethane-dextran sulfate sodium salt (AOM/DSS)-FUSO group and the AOM/DSS group. AOM: Azoxymethane; DSS: Dextran sulfate sodium salt.
Differentially abundant mucosal microbial taxa driven by F. nucleatum infection
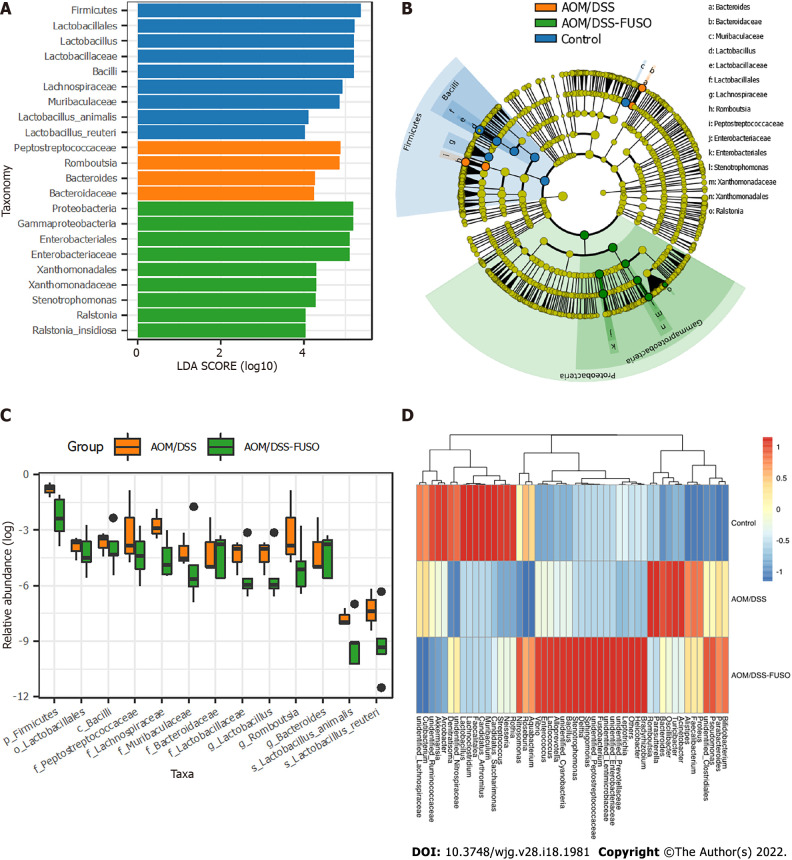
We then explored the specific microbial members related to F. nucleatum infection. First, we compared the bacterial RA between groups at different taxa levels. We showed that the phylum Proteobacteria and eight other potential pathogens belonging to Proteobacteria (LEfSe, Figure 3A and B) were the dominant taxa in AOM/DSS-FUSO mice. Meanwhile, the phylum Bacteroides and the family Bacteroidaceae were enriched in AOM/DSS mice, which were evident CRC signature taxa[33]. The enrichment of potential pathogenic taxa and depletion of probiotic bacteria such as Lactobacillales were features of the mucosal microbiota caused by F. nucleatum infection (Figure 3C). Comparing the two CRC model groups, the mucosal microbiota in the control mice was dominated by the phylum Firmicutes and the probiotic taxa (Figure 3A and B).
Figure 3.
The differentially abundant taxa among the three groups. A: Histogram of the linear discriminant analysis scores computed for taxa that were differentially abundant among the three groups; B: Taxonomic representation of statistically and biologically consistent differences among the three groups. The differences are represented in the color of the most abundant class; C: Relative abundance of the taxa in azoxymethane/dextran sulfate sodium salt (AOM/DSS)-FUSO and AOM/DSS; D: Patterns of the abundance of the top 50 operational taxonomic units in all samples. The colors indicate the Z scores of the mean abundance in each group. AOM: Azoxymethane; DSS: Dextran sulfate sodium salt.
We further compared the distribution of the top 50 most abundant taxa among the three groups. We found that the mucosal microbial population patterns were different in the three groups; the structure of the colon mucosal microbiota changed considerably after F. nucleatum infection in the AOM/DSS-FUSO mice (Figure 3D). Inflammation-inducing bacteria were significantly enriched in the mucosa microbiota of AOM/DSS-FUSO group compared with AOM/DSS group, such as Denitratisom, Stenotrophomonas and unidentified Enterobacteriaceae (P = 0.036, 0.018, and 0.048, respectively, wilcon rank-sum test, one-sided), while the potential probiotic genera were remarkably less abundant in AOM/DSS-FUSO group, such as unidentified Ruminococcaceae, unidentified Lachnospiraceae, and Akkermansia (P = 0.036, 0.036, and 0.071, respectively, wilcon rank-sum test, one-sided). Taken together, these results suggested that F. nucleatum infection further worsens the dysbiosis of the colon mucosal microbiota by increasing the abundance of pathogenic bacteria while reducing probiotics.
Altered host signaling pathways in colon tissue under F. nucleatum infection
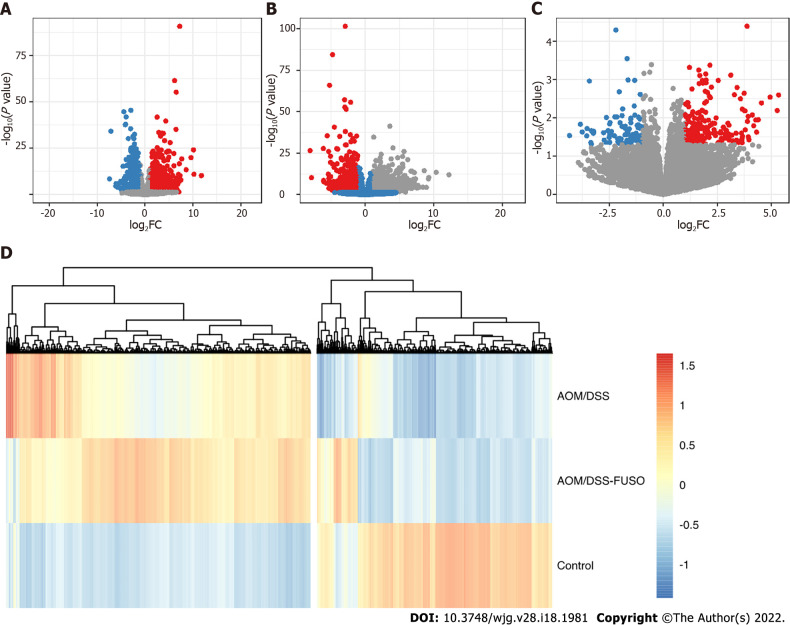
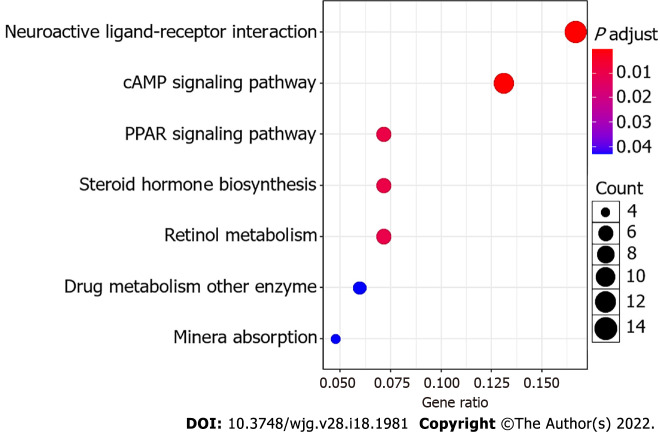
To gain insights into the molecular basis underlying the protumorigenic effect of Fusobacterium infection, RNA-seq was performed to reveal the transcriptomic changes driven by F. nucleatum. We observed different number of DEGs in the three comparions, i.e., AOM/DSS vs Control, AOM/DSS-FUSO vs Control, and AOM/DSS-FUSO vs AOM/DSS (Figure 4A-C), and compared with the AOM/DSS mice, there were 231 upregulated genes and 92 downregulated genes in the AOM/DSS-FUSO mice (Figure 4C). The heatmap of DEGs showed the distinct gene expression profile between AOM/DSS-induced CRC models with or without overloaded F. nucleatum treatment (Figure 4D). These upregulated genes included FABP1 (fatty acid binding protein 1)[34], sucrase isomaltase, defensin, ATP-binding cassette[35], homeobox C6[36] and tolloid-like 2 (NETO2)[37], all of which were overexpressed more than 3-fold and were correlated with a poor prognosis of colorectal carcinoma in previous studies (Supplementary Table 1). KEGG pathway enrichment analysis suggested that the microbiota-transcriptome interaction significantly upregulated neuroactive ligand–receptor interaction, cyclic adenosine monophosphate (cAMP) signaling pathway, PPAR signaling pathway, steroid hormone biosynthesis, retinol metabolism, mineral absorption and drug metabolism in the AOM/DSS-FUSO group (Figure 5), and 36 DEGs were included in these KEGG pathways (Supplementary Figure 4). Among them, several CRC-related DEGs, PPARα[38], PCK1[39,40], GCG[41], VAV2[42], and ANGPTL4[43] were also revealed to be correlated with the CRC prognosis in previous studies. These results suggested that dramatic changes occurred in the host signaling pathways that are related to the tumorigenesis of CRC after F. nucleatum infection and the resulting disturbance of the mucosal microbiota.
Figure 4.
The differentially expressed genes of the mucosa. A: Volcano plot showing the differentially expressed genes between the azoxymethane/dextran sulfate sodium salt (AOM/DSS) group and the control group; B: Volcano plot showing the differentially expressed genes between the AOM/DSS-FUSO group and the control group; C: Volcano plot showing the differentially expressed genes between the AOM/DSS-FUSO group and the AOM/DSS group. Significantly upregulated genes are shown in red (P < 0.05 and log2FC ≥ 1). Significantly downregulated genes are shown in blue (P < 0.05 and log2FC ≤ -1). The numbers of significantly upregulated genes were 1374 for (A), 1939 for (B) and 231 for (C). The numbers of significantly downregulated genes were 1050 for (A), 1104 for (B) and 92 for (C); D: Patterns of the abundance of all of the differentially expressed genes in the upper panels. The colors indicate the Z scores of the mean abundance in each group. AOM: Azoxymethane; DSS: Dextran sulfate sodium salt.
Figure 5.
Bubble plot of the KEGG pathways. The size of the bubble indicates the relative number of genes in the set. The color of the bubble indicates the adjusted P value.
Hub bacteria and key genes involved in the carcinogenesis/development of CRC
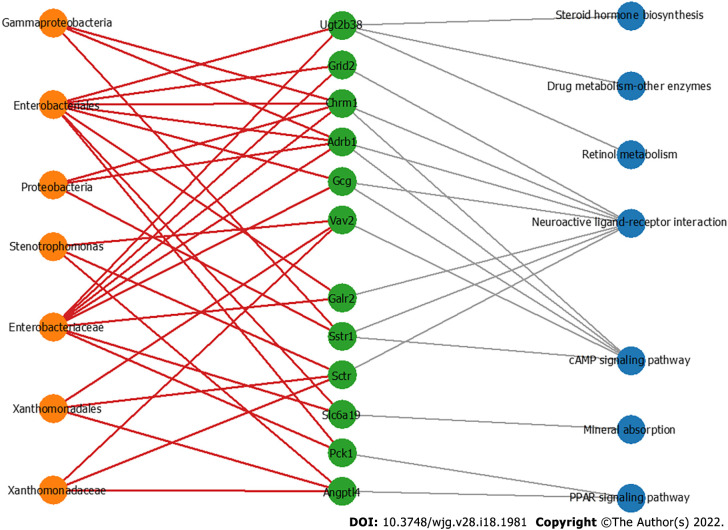
To further explore the hub bacteria and the hub DEGs, MCODE plugged in Cytoscape was used to explore the key modules involved in CRC development (Figure 6). The differential bacterial taxa identified by LEfSe analysis and the genes involved in the top seven significantly differential KEGG pathways were processed. The results showed that seven potentially harmful hub taxa were closely related to the overexpression of 12 hub genes. These taxa, Stenotrophomonas[44], Xanthomonadaceae, Xanthomonadales, Enterobacteriaceae, Enterobacteriales, Gammaproteobacteria, and Proteobacteria, have been shown to exhibit proinflammatory and procarcinogenic properties. This key module with the seven hub microbial taxa and 12 hub genes may regulate the occurrence and development of CRC through CRC-related pathways such as the cAMP and PPAR signaling pathways. Enterobacteriaceae correlated with eight oncogenes, i.e., PCK1, GalR2, GCG, SLC6A19, CHRM1, ADRB1, UGT2B38, and GRID2, and three taxa (Xanthomonadales, Xanthomonadaceae and Stenotrophomonas) were positively correlated with VAV2, SCTR and ANGPTL4. These results suggested that the dysbiosis of gut microbes driven by F. nucleatum infection may have the potential to affect the transcriptional activity of tumor-related metabolic pathways, especially those containing oncogenes.
Figure 6.
The co-occurrence network shows the correlations of the colon mucosa microbiota with the differentially expressed genes. Edges: Correlations with P values < 0.01; red edges: Positive correlations; gray edges: Relationship between the genes and the pathways; green nodes: Genes involved in the top seven significantly differential KEGG pathways in the azoxymethane/dextran sulfate sodium salt (AOM/DSS)-FUSO group compared with the AOM/DSS group; orange nodes: The differential bacterial taxa identified by LEfSe analysis; blue nodes: Related pathways of defferentially expressed genes.
DISCUSSION
F. nucleatum has long been known to cause opportunistic infections and has recently been implicated in CRC, which has attracted broad attention. Previously, we found that the RA of F. nucleatum in the fecal microbiota was significantly increased in CRC patients[33]. However, the effect of F. nucleatum infection on the mucosal microbiota and the resulting relationship between gut microbiome alterations and CRC development are still open questions. AOM/DSS mice can spontaneously develop multiple intestinal adenomas and have been used extensively as a model of colorectal carcinogenesis inflammation-associated tumors[45]. We showed that AOM/DSS mice with F. nucleatum infection have a higher burden of intestinal adenoma and worse colon histology. At the same time, an impaired mucosal barrier as shown by E-cadherin expression and increased epithelial cell proliferation by Ki-67 in the AOM/DSS-FUSO group were observed.
After the introduction of F. nucleatum, we propose that F. nucleatum needs to adapt to and change the symbiotic microbial environment, which could activate and promote the progression of colorectal carcinogenesis. In this study, three increased inflammation-inducing bacteria and three decreased probiotic genera in the top 50 differentially expressed genera were recognized as dysbiotic mucosal taxa after F. nucleatum stimulation. Furthermore, changes in the structure of the mucosal microbiota among the three groups were identified. Proteobacteria and eight other pathogenic bacterial taxa (Figure 3A) were the dominant taxa in the AOM/DSS-FUSO mice compared with the AOM/DSS group. In agreement with previous studies, gut microbial comparisons of CRC patients and non-CRC subjects are characterized by a high prevalence of pro-inflammatory bacteria (mostly Proteobacteria) and a decrease in commensal beneficial bacteria (mostly Firmicutes)[33,46,47]. Notably, the transfer of two Enterobacteriaceae species (Klebsiella pneumoniae and Proteus mirabilis) was sufficient to provoke colitis in the recipient mice[48]. The overload of Proteobacteria promoted intestinal inflammation in a T-bet-/- Rag2-/- mouse model of colitis[48-50]. Similar to studies in mice, microbiota studies in humans have shown that the gut microbial structure of inflammatory bowel disease patients is characterized by an expansion of Proteobacteria, particularly Enterobacteriaceae. Consistent with previous studies, we observed that the RA of Enterobacteriaceae in the mucosal microbiota was positively correlated with the expression of key oncogenes. Moreover, three other taxa, Stenotrophomonas, Xanthomonadales, and Xanthomonadaceae, were positively correlated with ATGPTL4 (angiopoietin-like 4), the VAV2 oncogene, and SCTR (secretin receptor), as shown in Figure 6. Stenotrophomonas was proven to be significantly increased during the exacerbated phase of UC compared to the remission phase[51]. Based on the above evidence, we propose that abnormal expansion of Proteobacteria, especially Enterobacteriaceae and Stenotrophomonas, is the signature of microbial dysbiosis driven by F. nucleatum.
To further reveal the effects of F. nucleatum infection and microbial dysbiosis on the transcriptome of AOM/DSS mice, RNA-seq was performed, and 323 DEGs were identified that were influenced by the oncogenic microbial environment. Interestingly, we found that the F. nucleatum-driven DEGs were significantly enriched in cancer-related KEGG signaling pathways. The neuroactive ligand–receptor interaction, cAMP signaling pathway, and PPAR signaling pathway were significantly overexpressed in the AOM/DSS-FUSO group. Among them, ANGPTL4, VAV2, and PCK1 were significantly highly expressed in the AOM/DSS-FUSO groups and positively correlated with the Proteobacteria taxa. In CRC cell lines, Zheng et al[52] found that F. nucleatum infection provoked glucose uptake and subsequently promoted glycolysis activity of CRC cells via ATGPTL4, which could in turn facilitate its own colonization in CRC cells[52]. F. nucleatum-mediated glycolysis activation via ATGPTL4 plays important roles in the pathogenesis of CRC and subsequently provides a beneficial environment for the outgrowth of CRC-related taxa.
Consistent with our results, other recent studies have demonstrated that F. nucleatum provokes glycolysis by upregulating lncRNA ENO-1[53], and an increase in cellular metabolic activity provides ample cellular energy for EMT-mediated metastasis[54]. Another oncogene, VAV2, related to three Proteobacteria taxa, is a member of the VAV guanine nucleotide exchange factor family of oncogenes and is one of the five combined prognostic genes that was identified to form a prognostic mutation panel for predicting cancer recurrence in stage II and III CRC[55]. Loss of VAV2, VAV3 and TIAM1 is sufficient to strongly suppress the cancer phenotypes and tumorigenesis induced by APC loss[56].
It is important to note that the eight upregulated hub genes shown in the microbiota-transcriptome network were positively correlated with high levels of Enterobacteriaceae, e.g., PCK1, GalR2, GCG, SLC6A19, CHRM1, ADRB1, UGT2B38, and GRID2. Among them, PCK1 enhanced liver metastatic growth by driving pyrimidine nucleotide biosynthesis under hypoxia[40]. G-protein-coupled receptor galanin receptor 1 (GalR1) was identified as a novel regulator of drug resistance[57]. Notably, silencing either GalR1 or its ligand, galanin, induced apoptosis in both drug-sensitive and drug-resistant cell lines and synergistically enhanced the effects of chemotherapy. Mechanistically, GalR1/galanin silencing resulted in downregulation of the endogenous caspase-8 inhibitor FLIP(L), resulting in induction of caspase-8-dependent apoptosis. Galanin mRNA was found to be overexpressed in colorectal tumors, and importantly, high galanin expression was correlated with poor disease-free survival of patients with early-stage CRC. In the future, additional studies need to be performed to explore the effect of F. nucleatum on Proteobacteria-related oncogenes to clarify the roles of F. nucleatum in CRC diseases.
Therefore, our data present the key microbial signature after F. nucleatum infection, which may provide new insights into the role of dysbiosis in the mucosal microbiota driven by F. nucleatum in promoting CRC. We provide direct evidence that Proteobacteria expansion, in particular Enterobacteriaceae and Stenotrophomonas, occurs during F. nucleatum infection, leading to the differential expression of oncogene transcriptomes in the AOM/DSS model. We provide evidence that F. nucleatum interacts with other species, how it remodels the microbiota, and how it alters the tumor microenvironment. Furthermore, we revealed the dysbiosis signature of the colon mucosa microbiota driven by Fusobacterium and the interaction between the hub microbial taxa and the host transcriptome. A deeper understanding of CRC etiology will also help identify new targets for prevention and treatment.
CONCLUSION
In conclusion, these data extend our previous work to provide evidence for microbial dysbiosis in CRC and suggest the existence of a defined set of microbiota whose composition is essential. Our study is also distinct in taking a holistic approach, given the observation of important bacterial-host networks and evidence for an oncogenic microbial environment of the colon promoting the formation of CRC. Our study is limited by: (1) The lack of time-course experiments to validate the effects of F. nucleatum on the symbiotic microenvironment; and (2) The limited sample size due to the deaths of mice when establishing the disease model. Therefore, further studies using more mice to establish the disease model are required to confirm the findings here and to disentangle the defined role of the Proteobacteria-related oncogenes driven by F. nucleatum during the progression of colorectal adenomas.
ARTICLE HIGHLIGHTS
Research background
Fusobacterium nucleatum (F. nucleatum) has long been known to cause opportunistic infections and has recently been implicated in colorectal cancer (CRC), which has attracted broad attention. Exploring the roles and underlying mechanism of F. nucleatum has important theoretical and practical significance for diagnosis and targeted therapy of CRC.
Research motivation
The aim of this study is to reveal the effect of F. nucleatum infection on the changes of mucosal microbiota, and investigate the regulatory role of F. nucleatum in the CRC progression.
Research objectives
The azoxymethane/dextran sulfate sodium salt (AOM/DSS)-induced mice were used to investigate the role of F. nucleatum in the “oncogenic microbial environment” of the colon.
Research methods
The mucosal microbial composition and RNA-seq transcriptomic analysis were used to identify the differentially expressed gene (DEG) signatures driven by F. nucleatum infection, and integrated network analysis was conducted to reveal the relationship between microbial dysbiosis and the DEGs.
Research results
We provide direct evidence that F. nucleatum interacts with other species, remodels the microbiota, and alters the tumor microenvironment. In addition, we revealed the dysbiosis signature of the colon mucosa microbiota driven by F. nucleatum and the interaction between the hub microbial taxa and the host transcriptome.
Research conclusions
Dysbiosis in the mucosal microbiota driven by F. nucleatum, in particular Enterobacteriaceae and Stenotrophomonas, occurs during F. nucleatum infection, leading to the differential expression of oncogene transcriptomes in the AOM/DSS model.
Research perspectives
Our study provides a deeper insight into CRC etiology driven by F. nucleatum infection and the mechanisms of the tumorigenic effect of F. nucleatum. Targeting F. nucleatum infection is a potential treatment for CRC.
Footnotes
Institutional animal care and use committee statement: The study was reviewed and approved by the Institutional Animal Care Committee of the Peking University People’s Hospital. All animal experiments conformed to the internationally accepted principles for the care and use of laboratory animals.
Conflict-of-interest statement: The authors declare no competing interests.
ARRIVE guidelines statement: The authors have read the ARRIVE guidelines, and the manuscript was prepared and revised according to the ARRIVE guidelines.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: January 30, 2022
First decision: February 24, 2022
Article in press: March 26, 2022
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Batyrbekov K, Kazakhstan; Díez M, Germany S-Editor: Fan JR L-Editor: A P-Editor: Yu HG
Contributor Information
Na Wu, Department of Central Laboratory & Institute of Clinical Molecular Biology, Peking University People’s Hospital, Beijing 100044, China.
Yu-Qing Feng, State Key Laboratory of Animal Nutrition, College of Animal Science and Technology, China Agricultural University, Beijing 100193, China.
Na Lyu, CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China.
Di Wang, Department of Central Laboratory & Institute of Clinical Molecular Biology, Peking University People’s Hospital, Beijing 100044, China.
Wei-Dong Yu, Department of Central Laboratory & Institute of Clinical Molecular Biology, Peking University People’s Hospital, Beijing 100044, China.
Yong-Fei Hu, State Key Laboratory of Animal Nutrition, College of Animal Science and Technology, China Agricultural University, Beijing 100193, China. huyongfei@cau.edu.cn.
Data sharing statement
The raw 16S amplicon sequencing and transcriptome data in this study have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA767246.
References
- 1.Song M, Garrett WS, Chan AT. Nutrients, foods, and colorectal cancer prevention. Gastroenterology. 2015;148:1244–60.e16. doi: 10.1053/j.gastro.2014.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B, Wan YJ, Nie YQ. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016;22:3227–3233. doi: 10.3748/wjg.v22.i11.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, Kostic AD, Giannakis M, Bullman S, Milner DA, Baba H, Giovannucci EL, Garraway LA, Freeman GJ, Dranoff G, Garrett WS, Huttenhower C, Meyerson M, Meyerhardt JA, Chan AT, Fuchs CS, Ogino S. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J Biol Chem. 2007;282:25000–25009. doi: 10.1074/jbc.M611567200. [DOI] [PubMed] [Google Scholar]
- 8.Fardini Y, Wang X, Témoin S, Nithianantham S, Lee D, Shoham M, Han YW. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol. 2011;82:1468–1480. doi: 10.1111/j.1365-2958.2011.07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, Li H, Guo B, Zhu Q, Wei Q, Moyer MP, Wang P, Cai S, Goel A, Qin H, Ma Y. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating Toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology. 2017;152:851–866.e24. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang W, Sun T, Tang D, Zhou J, Feng Q. Time-course transcriptome analysis of gingiva-derived mesenchymal stem cells reveals that Fusobacterium nucleatum triggers oncogene expression in the process of cell differentiation. Front Cell Dev Biol. 2019;7:359. doi: 10.3389/fcell.2019.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Peng F, Peng C, Du JR. Gut microbiota in tumor microenvironment: A critical regulator in cancer initiation and development as potential targets for Chinese medicine. Am J Chin Med. 2021;49:609–626. doi: 10.1142/S0192415X21500270. [DOI] [PubMed] [Google Scholar]
- 13.Wong-Rolle A, Wei HK, Zhao C, Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell. 2021;12:426–435. doi: 10.1007/s13238-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrett WS. Cancer and the microbiota. Science. 2015;348:80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doulberis M, Angelopoulou K, Kaldrymidou E, Tsingotjidou A, Abas Z, Erdman SE, Poutahidis T. Cholera-toxin suppresses carcinogenesis in a mouse model of inflammation-driven sporadic colon cancer. Carcinogenesis. 2015;36:280–290. doi: 10.1093/carcin/bgu325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Liang L, Liang H, Wang M, Lu B, Xue M, Deng J, Chen Y. Fusobacterium nucleatum aggravates the progression of colitis by regulating M1 macrophage polarization via AKT2 pathway. Front Immunol. 2019;10:1324. doi: 10.3389/fimmu.2019.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y, Lee C, Kim J, Hwang S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng. 2005;89:670–679. doi: 10.1002/bit.20347. [DOI] [PubMed] [Google Scholar]
- 18.Chen S, Zhou Y, Chen Y, Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 21.Schloss PD. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS One. 2009;4:e8230. doi: 10.1371/journal.pone.0008230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 26.Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, Karp M, Benjamini Y, Hochberg Y, Laron Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med Hypotheses. 1995;45:486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 27.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (N Y) 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zapala MA, Schork NJ. Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. Proc Natl Acad Sci U S A. 2006;103:19430–19435. doi: 10.1073/pnas.0609333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neufert C, Heichler C, Brabletz T, Scheibe K, Boonsanay V, Greten FR, Neurath MF. Inducible mouse models of colon cancer for the analysis of sporadic and inflammation-driven tumor progression and lymph node metastasis. Nat Protoc. 2021;16:61–85. doi: 10.1038/s41596-020-00412-1. [DOI] [PubMed] [Google Scholar]
- 32.Stemmler MP, Eccles RL, Brabletz S, Brabletz T. Non-redundant functions of EMT transcription factors. Nat Cell Biol. 2019;21:102–112. doi: 10.1038/s41556-018-0196-y. [DOI] [PubMed] [Google Scholar]
- 33.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, Chen Y, Yang F, Lu N, Wang Z, Luan C, Liu Y, Wang B, Xiang C, Wang Y, Zhao F, Gao GF, Wang S, Li L, Zhang H, Zhu B. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Zhang W, Xia M, Xie Z, An F, Zhan Q, Tian W, Zhu T. High expression of FABP4 in colorectal cancer and its clinical significance. J Zhejiang Univ Sci B. 2021;22:136–145. doi: 10.1631/jzus.B2000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q, Zhao H, Wu D, Cao D, Ma W. A comprehensive analysis of the microbiota composition and gene expression in colorectal cancer. BMC Microbiol. 2020;20:308. doi: 10.1186/s12866-020-01938-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji M, Feng Q, He G, Yang L, Tang W, Lao X, Zhu D, Lin Q, Xu P, Wei Y, Xu J. Silencing homeobox C6 inhibits colorectal cancer cell proliferation. Oncotarget. 2016;7:29216–29227. doi: 10.18632/oncotarget.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu L, Chen HY, Cai J, Yang GZ, Feng D, Zhai YX, Gong H, Qi CY, Zhang Y, Fu H, Cai QP, Gao CF. Upregulation of NETO2 expression correlates with tumor progression and poor prognosis in colorectal carcinoma. BMC Cancer. 2015;15:1006. doi: 10.1186/s12885-015-2018-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang H, Zhao F, Xie X, Liao Y, Song Y, Liu C, Wu Y, Wang Y, Liu D, Zou J, Qi Z. PPARα suppresses Th17 cell differentiation through IL-6/STAT3/RORγt pathway in experimental autoimmune myocarditis. Exp Cell Res. 2019;375:22–30. doi: 10.1016/j.yexcr.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 39.Yang J, Gao S, Qiu M, Kan S. Integrated Analysis of Gene Expression and metabolite data reveals candidate molecular markers in colorectal carcinoma. Cancer Biother Radiopharm. 2020 doi: 10.1089/cbr.2020.3980. [DOI] [PubMed] [Google Scholar]
- 40.Yamaguchi N, Weinberg EM, Nguyen A, Liberti MV, Goodarzi H, Janjigian YY, Paty PB, Saltz LB, Kingham TP, Loo JM, de Stanchina E, Tavazoie SF. PCK1 and DHODH drive colorectal cancer liver metastatic colonization and hypoxic growth by promoting nucleotide synthesis. Elife. 2019;8:e52135. doi: 10.7554/eLife.52135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bian Q, Chen J, Qiu W, Peng C, Song M, Sun X, Liu Y, Ding F, Zhang L. Four targeted genes for predicting the prognosis of colorectal cancer: A bioinformatics analysis case. Oncol Lett. 2019;18:5043–5054. doi: 10.3892/ol.2019.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hebeis B, Vigorito E, Kovesdi D, Turner M. Vav proteins are required for B-lymphocyte responses to LPS. Blood. 2005;106:635–640. doi: 10.1182/blood-2004-10-3919. [DOI] [PubMed] [Google Scholar]
- 43.Wen L, Zhang Y, Yang B, Han F, Ebadi AG, Toughani M. Knockdown of Angiopoietin-like protein 4 suppresses the development of colorectal cancer. Cell Mol Biol (Noisy-le-grand) 2020;66:117–124. [PubMed] [Google Scholar]
- 44.Merlo LMF, DuHadaway JB, Montgomery JD, Peng WD, Murray PJ, Prendergast GC, Caton AJ, Muller AJ, Mandik-Nayak L. Differential roles of IDO1 and IDO2 in T and B cell inflammatory immune responses. Front Immunol. 2020;11:1861. doi: 10.3389/fimmu.2020.01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat Protoc. 2007;2:1998–2004. doi: 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- 46.Chamorro N, Montero DA, Gallardo P, Farfán M, Contreras M, De la Fuente M, Dubois K, Hermoso MA, Quera R, Pizarro-Guajardo M, Paredes-Sabja D, Ginard D, Rosselló-Móra R, Vidal R. Landscapes and bacterial signatures of mucosa-associated intestinal microbiota in Chilean and Spanish patients with inflammatory bowel disease. Microb Cell. 2021;8:223–238. doi: 10.15698/mic2021.09.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wassenaar TM. E. coli and colorectal cancer: A complex relationship that deserves a critical mindset. Crit Rev Microbiol. 2018;44:619–632. doi: 10.1080/1040841X.2018.1481013. [DOI] [PubMed] [Google Scholar]
- 48.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 50.Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M, Gallini CA, Beal C, van Hylckama-Vlieg JE, Ballal SA, Morgan XC, Glickman JN, Gevers D, Huttenhower C, Garrett WS. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walujkar SA, Kumbhare SV, Marathe NP, Patangia DV, Lawate PS, Bharadwaj RS, Shouche YS. Molecular profiling of mucosal tissue associated microbiota in patients manifesting acute exacerbations and remission stage of ulcerative colitis. World J Microbiol Biotechnol. 2018;34:76. doi: 10.1007/s11274-018-2449-0. [DOI] [PubMed] [Google Scholar]
- 52.Zheng X, Liu R, Zhou C, Yu H, Luo W, Zhu J, Liu J, Zhang Z, Xie N, Peng X, Xu X, Cheng L, Yuan Q, Huang C, Zhou X. ANGPTL4-mediated promotion of glycolysis facilitates the colonization of Fusobacterium nucleatum in colorectal cancer. Cancer Res. 2021;81:6157–6170. doi: 10.1158/0008-5472.CAN-21-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong J, Guo F, Lu SY, Shen C, Ma D, Zhang X, Xie Y, Yan T, Yu T, Sun T, Qian Y, Zhong M, Chen J, Peng Y, Wang C, Zhou X, Liu J, Liu Q, Ma X, Chen YX, Chen H, Fang JY. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. 2021;70:2123–2137. doi: 10.1136/gutjnl-2020-322780. [DOI] [PubMed] [Google Scholar]
- 54.Teo Z, Sng MK, Chan JSK, Lim MMK, Li Y, Li L, Phua T, Lee JYH, Tan ZW, Zhu P, Tan NS. Elevation of adenylate energy charge by angiopoietin-like 4 enhances epithelial-mesenchymal transition by inducing 14-3-3γ expression. Oncogene. 2017;36:6408–6419. doi: 10.1038/onc.2017.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sho S, Court CM, Winograd P, Russell MM, Tomlinson JS. A prognostic mutation panel for predicting cancer recurrence in stages II and III colorectal cancer. J Surg Oncol. 2017;116:996–1004. doi: 10.1002/jso.24781. [DOI] [PubMed] [Google Scholar]
- 56.Pickering KA, Gilroy K, Cassidy JW, Fey SK, Najumudeen AK, Zeiger LB, Vincent DF, Gay DM, Johansson J, Fordham RP, Miller B, Clark W, Hedley A, Unal EB, Kiel C, McGhee E, Machesky LM, Nixon C, Johnsson AE, Bain M, Strathdee D, van Hoof SR, Medema JP, Anderson KI, Brachmann SM, Stucke VM, Malliri A, Drysdale M, Turner M, Serrano L, Myant K, Campbell AD, Sansom OJ. A RAC-GEF network critical for early intestinal tumourigenesis. Nat Commun. 2021;12:56. doi: 10.1038/s41467-020-20255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stevenson L, Allen WL, Turkington R, Jithesh PV, Proutski I, Stewart G, Lenz HJ, Van Schaeybroeck S, Longley DB, Johnston PG. Identification of galanin and its receptor GalR1 as novel determinants of resistance to chemotherapy and potential biomarkers in colorectal cancer. Clin Cancer Res. 2012;18:5412–5426. doi: 10.1158/1078-0432.CCR-12-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw 16S amplicon sequencing and transcriptome data in this study have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA767246.