SUMMARY
We investigated the antibiotic and heavy-metal resistance mechanisms, virulence genes and clonal relationships of macrolide- and/or lincosamide-resistant (M+/−LR) Streptococcus agalactiae (group B Streptococcus, GBS) isolates from pregnant women in La Rioja in Northern Spain, a region with a significant immigrant population. In total 375 GBS isolates were recovered during 2011. About three-quarters of isolates were from European nationals and the remainder distributed among 23 other nationalities. Seventy-five (20%) were classified as M+/−LR strains and 28 (37%) of these were resistant to ⩾3 classes of antibiotics. Capsular serotypes III (29·3%), V (21·3%) and II (12%) were the most frequent. A wide variety of antibiotic resistance genes were detected in M+/−LR strains; notably, 5·3% harboured the lsa(C) gene associated with cross-resistance, and tet(W) was identified in a single strain. We report, for the first time, the detection of cadmium and copper resistance encoded by tcrB + cadA + cadC genes in 20 M+/−LR strains, which raises the possibility of co-selection of antibiotic and heavy-metal resistance disseminated through mobile genetic elements. The M+/−LR strains were highly diverse by DNA macrorestriction profiles (65 patterns) and 16 multilocus sequence types (STs) distributed among six clonal complexes; the most frequent were ST1, ST19, and ST12, and two strains were novel (ST586 and ST601). In conclusion, a wide diversity of genetic lineages of macrolide, lincosamide and heavy-metal- resistant GBS strains was observed in an ethnically diverse maternal population.
Key words: Antibiotic resistance, bacterial typing, streptococcal infections
INTRODUCTION
Streptococcus agalactiae (group B Streptococcus, GBS) is recognized as an important cause of invasive diseases in neonates and pregnant women, as well as infections in diabetic, elderly, and immunocompromised patients. This microorganism is a natural resident of the human gastrointestinal tract, but colonization of the urogenital tract in pregnant women constitutes a risk factor for acquisition by newborns [1]. In many industrialized countries, S. agalactiae is an important cause of sepsis and meningitis during the first week of life. Generally, colonization rates in pregnant women oscillate from 10% to 30%, varying widely among different ethnic groups, geographical areas and age groups [1]. In Spain, maternal vaginal colonization by GBS rates vary between 12% and 20% [2], and similar prevalence levels have been reported in other European countries [3]. Although colonization of the newborn from GBS-positive mothers is high, only 1–2% of those infants will show an early-onset of GBS infection in the absence of prevention measures [1]. In Spain pregnant women are usually screened for colonization at 35–37 weeks of gestation, and if colonized, receive intrapartum antimicrobial prophylaxis. Penicillin is the drug of choice for the treatment of GBS, and macrolide and lincosamide antibiotics are reserved as alternatives for penicillin-allergic patients as recommended by US and Spanish guidelines [1, 2]. However, elevated erythromycin and clindamycin resistance rates have been reported worldwide [4].
Metals such as silver, iron, and copper are widely used for environmental control, water disinfection, or incorporated into medical devices (e.g. intravascular catheters), as well as in fungicides, antiseptics and preservatives in pharmaceuticals and cosmetics [5] (https://www.cdc.gov/hicpac/Disinfection_Sterilization/10_0MiscAgents.html). Additionally, heavy metals (such as cadmium and copper) have been suggested to promote co-selection and spread of antibiotic resistance, due to genetic elements encoding resistance to both groups of compounds as shown for Staphylococcus aureus and Enterococcus spp. [6, 7], but, as yet, not for GBS isolates.
S. agalactiae is genetically diverse and several molecular typing methods including DNA macrorestriction analysis by pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST) and serotype or capsular polysaccharides typing (cps operons), have been used for epidemiological studies of the organism [8]. In recent years, a circulating genetic lineage with enhanced invasive capacity, expressing serotype III and defined by MLST (ST17) has been identified worldwide. This homogeneous epidemic clone has been significantly associated with cases of invasive neonatal disease and appears to be of bovine origin [8–10].
The pathogenesis of S. agalactiae is related not only to particular capsular polysaccharide types, but also to a wide range of cell surface proteins, which may contribute significantly to the invasive potential of a particular lineage [11]. This variety of virulence factors and the multiplicity of circulating genetic lineages in different geographical areas together pose a major obstacle for the development of effective vaccines [12]. To date, there is limited knowledge regarding the epidemiology, antibiotic susceptibility, virulence gene complement, and genetic lineages of GBS in pregnant women in Spain, and specifically in the La Rioja region in the north of the country. As a consequence, this study was undertaken on a large collection of GBS isolates from an unselected population of pregnant women in the region presenting at antenatal clinics during a single year. Strains exhibiting macrolide and/or lincosamide-resistance (M+/−LR) were further characterised for antimicrobial and resistance gene complement, and genetic lineage.
METHODS
Samples and bacterial isolates
S. agalactiae were isolated from vaginal-rectal samples of all pregnant women (35–37 weeks of pregnancy) who attended the main and reference hospital (Hospital San Pedro) in La Rioja region in 2011. Samples were cultured using selective media (Todd-Hewitt broth and Granada agar medium, Becton Dickinson, Germany) and after 18–24 h of incubation, suspected colonies were identified by biochemical tests (API 20 STREP, bioMérieux, France).
Antimicrobial susceptibility testing
All isolates were tested for susceptibility to the following antibiotics (penicillin, erythromycin (ERY), azithromycin, spiramycin, pristinamycin, clindamycin (CLI), lincomycin, tetracycline, levofloxacin, chloramphenicol, vancomycin, teicoplanin, daptomycin, linezolid, quinupristin-dalfopristin, trimethoprim-sulfamethoxazol, high-level resistance to aminoglycosides (streptomycin, gentamicin, kanamycin)] by disc-diffusion method and MicroScan® microdilution system (Beckman Coulter, USA). Susceptibility categorization was carried out according to CLSI recommendations [13], except for spiramycin, pristinamycin, lincomycin, teicoplanin, and high-level resistance to aminoglycosides, for which CA-SFM criteria were used [14]. Inducible CLI resistance was detected by double-disc diffusion method (D test) using CLI and ERY discs [13, 14].The phenotypes of resistance to macrolide-lincosamides (ML) were classified as: constitutive (cML), inducible (iML), phenotype M (ERY-resistant, CLI-susceptible), and the unusual phenotype of resistance to lincosamides (LR) (ERY-susceptible, CLI-resistant) [13, 14].
Antimicrobial resistance genotype
DNA extraction of M+/−LR GBS isolates was carried out by InstaGene Matrix (Bio-Rad, USA). Genes involved in resistance to macrolides and lincosamides [erm(A) and erm(A) [erm(TR) variant], erm(B), erm(C), erm(T), erm(F), mef(A/E), msr(A), lnu(A), lnu(B), lsa(C)] [15–18], to tetracycline [tet(L), tet(K), tet(M), tet(O), tet(S), tet(W)] [19], to chloramphenicol [catA, cat(pC194) cat(pC221), cat(pC223), cfr, fexA] [19, 20], and to aminoglycosides [ant(6)-I, ant(4’)-Ia, aph(3’)-IIIa, aac(6’)-aph(2”)] [20, 21] were detected by polymerase chain reaction (PCR). The tet(W) gene was amplified with primers designed in this study [tet(W)-F: 5’-GGTGCAGTTGGAGGTTGTTT-3’ and tet(W)-R: 5’-TTTTTACCTGGACCGTTTCG-3’]. The gyrA and parC genes were amplified by PCR in levofloxacin-resistant isolates and sequenced on both strands [22], and the sequences were compared to those previously reported for gyrA and parC genes of S. agalactiae NEM316 strain (GenBank accession no. AL732656).
Susceptibility to copper and cadmium
Minimum inhibitory concentrations (MICs) were determined by agar dilution assay using serial double dilutions of cadmium acetate [Cd(CH3COO)2] (0·0625–128 mm) and copper sulphate (CuSO4) (0·0625–64 mm) on Mueller-Hinton II agar plates (Becton Dickinson, France) as described previously [6]. E. faecalis ATCC29212, S. aureus strains C1906, C2940 and C2941 were used as control strains.
The genes implicated in copper resistance (copB, cueO, tcrB, mco) were detected according to published PCRs [7] but cadA and cadC cadmium resistance genes were amplified using primers designed in this study (cadA-F: 5′-AGGGACGACAACCATTGAAG-3′ and cadA-R: 5′-AAGGCACAAGGACAACCAAC-3′; and cadC-F: 5′-TGACGAAGAAAAGGTCAATCG-3′ and cadC-R: 5′-GTTCGCAGGTGATGAGAGGT-3′, respectively).
Molecular typing
M+/−LR strains were typed by PFGE following a modified protocol previously reported [23]. DNA in agarose plugs was digested with 20 U of SmaI (New England Biolabs Inc., UK) according to the manufacturer's instructions. Electrophoresis was performed in a CHEF-DR III (Bio-Rad Laboratories Inc., USA), at 6 V/cm for 22 h at 14 °C, with pulse times ranging from 5 s to 45 s. DNA profiles were interpreted according to the Tenover criteria [24] and van Belkum et al. [25], with BioNumerics software v. 2.0 (Applied Maths, Belgium), using the Dice coefficient with an 80% similarity cut-off to determine strain relatedness.
MLST was performed according to the published scheme [8] and sequence types (STs) were determined by reference to the S. agalactiae MLST database and assigned to clonal complexes (CCs) using the program eBURST (The Department of Infectious Disease Epidemiology Imperial College, London, UK).
Capsular typing
Capsular serotypes were determined by multiplex PCR of cps genes as described previously [26].
Virulence genes
M+/−LR strains were screened by PCR for the following virulence-associated genes: immunoglobulin A binding protein (bac), C5a peptidase precursor (scpB), laminin-binding protein (lmb), surface protein (spb1) and fibrinogen-binding proteins A and B (fbsA and fbsB) [27–29], and additionally, the genes encoding the Alp protein family (named Alpha-C, Rib, Epsilon, Alp2, Alp3, Alp4) were assayed by multiplex PCR [30].
Statistical analysis
Statistical analysis of the data was performed using SPSS v. 20.0 (IBM, UK). Possible associations between current and previous GBS colonization of subjects were tested by χ2 with the level of significance set at P = 0·05.
RESULTS
Epidemiological and clinical data
A total of 2730 vaginal-rectal samples from pregnant women were processed and 375 (13·7%) S. agalactiae isolates were collected; 75 (20%) of the collection exhibited the M+/−LR phenotype. Table 1 lists the age group and country of origin of the 375 pregnant women colonized with S. agalactiae. The average age was 33·5 years (range 18–49), with most falling in the 31–35 years group. Most subjects were Spanish nationals (65·9%) and 74·7% were European. Of the 75 women harbouring M+/−LR strains, 28·6% originated from Asia (three countries), 17·9% from Latin America (12 countries) and 13% from Africa (eight countries).
Table 1.
Characteristics of pregnant women harbouring S. agalactiae during 2011
| No. positive for | No. with M+/−LR | ||
|---|---|---|---|
| GBS (N = 375) | GBS (N = 75) | ||
| Characteristics of subjects | n (%) | n (%) | |
| Age (range of years) | |||
| <25 | 37 (9·9) | 8 (10·7) | |
| 25–30 | 88 (23·5) | 15 (20) | |
| 31–35 | 138 (36·8) | 33 (8·8) | |
| 36–40 | 90 (24) | 16 (21·3) | |
| >40 | 22 (5·9) | 3 (4) | |
| Europe | Spain | 247 (65·9) | 50 (66·7) |
| Romania | 28 (7·5) | 6 (8) | |
| Russia | 2 (0·5) | 1 (1·3) | |
| Other (single isolates)* | 3 (0·8) | 1 (1·3) | |
| Africa | Morocco | 36 (9·6) | 3 (4) |
| Algeria | 4 (1·1) | 1 (1·3) | |
| Other (single isolates)† | 6 (1·6) | 2 (2·7) | |
| Latin America | Colombia | 8 (2·1) | 2 (2·7) |
| Bolivia | 4 (1·1) | 1 (1·3) | |
| Brazil | 4 (1·1) | 0 | |
| Chile | 3 (0·8) | 1 (1·3) | |
| Ecuador | 2 (0·5) | 1 (1·3) | |
| Other (single isolates)‡ | 7 (2·1) | 0 | |
| Asia | Pakistan | 19 (5·1) | 4 (5·3) |
| Other (single isolates)§ | 2 (0·5) | 2 (2·7) | |
| Previous isolation of GBS¶ | 135 (36) | 24 (32) | |
| Weeks of gestation | |||
| <35 | 9 (2·4) | 2 (2·7) | |
| 35–36 | 9 (2·4) | 3 (4) | |
| 37–40 | 182 (48·5) | 38 (50·7) | |
| >40 | 163 (43·5) | 29 (38·7) | |
| No data | 12 (3·2) | 3 (4) | |
| Birth delivery | |||
| Vaginal | 299 (79·8) | 57 (76) | |
| Caesarean | 67 (17·9) | 15 (20) | |
| No data | 9 (2·4) | 3 (4) | |
| No. of pregnancies | |||
| First | 198 (52·8) | 39 (52) | |
| Second | 139 (37·1) | 30 (40) | |
| Third | 27 (7·2) | 4 (5·3) | |
| Fourth | 8 (2·1) | 0 | |
| Fifth | 3 (0·8) | 2 (2·7) | |
| GBS colonization other pregnancies|| | |||
| Yes | 66 (37·3) | 10 (5·6) | |
| No | 69 (39) | ||
| No data | 42 | ||
| Previous miscarriages | 66 (17·5) | 10 (13·3) |
Bulgaria, Italy, and Moldova.
Angola, Equatorial Guinea, Ghana, Guinea, Ivory Coast, and Mali.
Argentina, Cuba, Dominican Republic, Honduras, Paraguay, Peru, and Venezuela.
China, and Iraq.
Colonized or infected prior to 2011.
First-time mothers excluded.
Thirty-six percent of those harbouring GBS had previously been colonized or infected with GBS prior to 2011 at the beginning of the survey. Most (92%) had a gestation of >37 weeks and only 5% women underwent a preterm delivery (<37 weeks). None of the newborns was diagnosed with neonatal sepsis. Almost half (47%) of subjects had previously been pregnant, and 66 (37·6%) of these had prior GBS colonization in earlier pregnancies (χ2 = 0·067, P = 0·796).
Resistance phenotype/genotype of M+/−LR strains
Table 2 shows that 52 (69·3%) of the 75 strains expressed the cML phenotype and 92·3% of these harboured the erm(B) gene; the erm(A) gene [erm(TR) variant] was found in five strains, and one contained erm(B) in addition. The iML phenotype was detected in 16 (21·3%) strains, distributed among four genotype patterns, the most frequent being erm(A) [erm(TR) variant] in 10 strains. Six mef(A/E)-positive strains were of the M phenotype and a single LR phenotype strain harboured the lsa(C) gene. The predominant genes in all strains were erm(B) (68%) and erm(A) [erm(TR) variant] (20%). Six strains harboured two M+/−L resistance genes.
Table 2.
Resistance phenotypes and genotypes of 75 M+/−LR S. agalactiae strains
| Macrolides and lincosamides | Tetracycline, aminoglycosides and chloramphenicol | Heavy metals | No. of strains | |
|---|---|---|---|---|
| Phenotype (no.) | Genotype* | Genotype | Genotype | |
| Constitutive (52) | erm(B) | tet(M) | 9 | |
| erm(B) | tet(M) + cat(pC194) | tcrB + cadA + cadC | 1 | |
| erm(B) | tet(M) + aph(3’)-IIIa + cat(pC194) | tcrB + cadA + cadC | 2 | |
| erm(B) | tet(M) + aph(3’)-IIIa + ant(6)-Ia | 2 | ||
| erm(B) | tet(M) + aph(3’)-IIIa + ant(6)-Ia | tcrB + cadA + cadC | 3 | |
| erm(B) | tet(M) + aph(3’)-IIIa + ant(6)-Ia + cat(pC194) | tcrB + cadA + cadC | 4 | |
| erm(B) | tet(O) | 6 | ||
| erm(B) | tet(O) + cat(pC194) | tcrB + cadA + cadC | 1 | |
| erm(B) | tet(O) + aph(3’)-IIIa + ant(6)-Ia | 6 | ||
| erm(B) | tet(M) + tet(O) | 3 | ||
| erm(B) | aph(3’)-IIIa + cat(pC194) | tcrB + cadA + cadC | 1 | |
| erm(B) | aph(3’)-IIIa + ant(6)-Ia | 1 | ||
| erm(B) | aph(3’)-IIIa + ant(6)-Ia | tcrB + cadA + cadC | 1 | |
| erm(B) | aph(3’)-IIIa + ant(6)-Ia + cat(pC194) | tcrB + cadA + cadC | 2 | |
| erm(B) | tet(W) | 1 | ||
| erm(B) + msr(A) | tet(M) | 1 | ||
| erm(B) + lsa(C) | tet(M) + aph(3’)-IIIa + ant(6)-Ia + cat(pC194) | tcrB + cadA + cadC | 2 | |
| erm(B) + lsa(C) | aph(3’)-IIIa + ant(6)-Ia + cat(pC194) | tcrB + cadA + cadC | 1 | |
| erm(B) + erm(A) [erm(TR) variant] | tet(M) + aph(3’)-IIIa + ant(6)-Ia | 1 | ||
| erm(A) [erm(TR) variant] | tet(M) | 2 | ||
| erm(A) [erm(TR) variant] | tet(O) | 2 | ||
| Inducible (16) | erm(B) | tet(M) + tet(O) | 1 | |
| erm(B) | tet(O) | 1 | ||
| erm(B) + mef(A/E) | tet(M) + aph(3’)-IIIa + ant(6)-Ia | 1 | ||
| erm(A) [erm(TR) variant] | tet(M) | 6 | ||
| erm(A) [erm(TR) variant] | tet(M) | tcrB + cadA + cadC | 2 | |
| erm(A) [erm(TR) variant] | tet(M) + aph(3’)-IIIa + ant(6)-I | 1 | ||
| erm(A) [erm(TR) variant] | tet(O) | 1 | ||
| erm(T) | tet(M) | 3 | ||
| M (6) | mef(A/E) | tet(M) | 5 | |
| mef(A/E) | No genes | 1† | ||
| ERY(S)-CLI(R) (1) | lsa(C) | tet(M) | 1 | |
The erm(A) gene was amplified with erm(TR)-variant specific primers.
Resistant to levofloxacin (amino-acid substitutions Ser81Leu in GyrA and Ser79Phe in ParC).
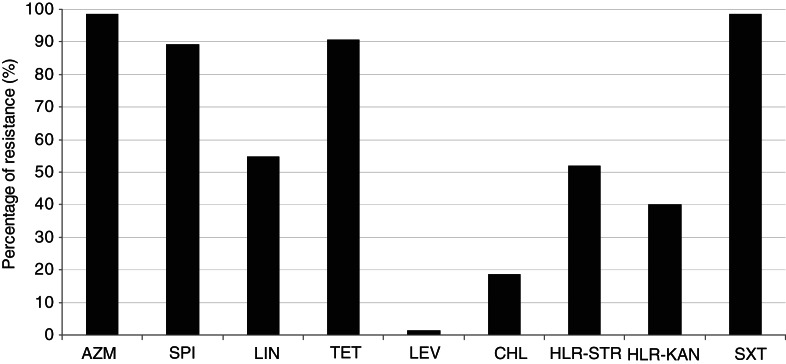
Figure 1 shows that most of M+/−LR strains were resistant to azithromycin (98·7%), trimethoprim-sulfamethoxazole (98·7%), tetracycline (90·7%), and spiramycin (89·3%). About half were resistant to lincomycin (54·7%), or expressed high-level resistance to streptomycin (52%) and kanamycin (40%). By contrast, all but one strains were susceptible to levofloxacin, and other antibiotics tested.
Fig. 1.
Percentages of resistance of 75 M+/−LR GBS strains. AZM, Azithromycin; SPI, spiramycin; LIN, lincomycine; TET, tetracycline; LEV, levofloxacin; CHL, chloramphenicol; HLR-STR, high-level resistance to streptomycin; HLR-KAN, high-level resistance to kanamycin; SXT, trimethoprim-sulfamethoxazole.
Among the 68 tetracyline-resistant strains, the detected encoding genes were tet(M) (67·6%), tet(O) (25%), tet(M) + tet(O) (5·9%), and tet(W) in a single strain (Table 2). Aminoglycoside resistance genes ant(6)-Ia and aph(3')-IIIa accounted, respectively, for the majority of streptomycin- (25/38) and kanamycin- (28/30) resistant strains. All 14 chloramphenicol-resistant strains were positive for the cat(pC194) gene. The amino-acid substitutions Ser81Leu in GyrA and Ser79Phe in ParC were detected in the single levofloxacin-resistant strain. Overall, 28 (37%) strains were resistant to ⩾3 classes of antibiotics and the most frequent multiresistance phenotype/genotype association (14 strains) was resistance to macrolides, tetracycline and aminoglycosides, encoded respectively by genes erm(B)/erm(A) [erm(TR) variant] + tet(M)/tet(O) + aph(3')-IIIa + ant(6)-Ia (Table 2).
Cadmium and copper resistance
The MICs for Cd(CH3COO)2 ranged from ⩽0·0625 to 4 mm, and for CuSO4 from 4 to 8 mm in the 75 strains tested. Twenty strains which were positive for cadA and cadC genes showed the highest cadmium MICs (from 1 to 4 mm), and the copper resistance gene tcrB was only detected in these strains.
Genetic diversity, capsule types and virulence factors
In total, 65 different PFGE patterns were distinguished in the 75 M+/−LR strains. Sixteen different STs were identified in the 65 strains with unique PFGE patterns, with the five most frequent STs being: ST19 (21 strains), ST1 (10), ST12 (9), ST23 (7), and ST17 (4). Six STs, each represented by single strains were assigned to other previously identified types but two strains proved to be novel to the database and were assigned as ST586 and ST601, which are single locus variants of ST22 and ST19, respectively, falling in clonal complexes CC22 and CC19. It is noteworthy that ST1, ST19 and ST23 contained strains of different PFGE patterns (Table 3). The majority of the 65 strains with different PFGE patterns belonged to capsular types III and V (33·8% and 24·1%, respectively), with the remainder distributed among capsular types II, Ia, Ib and IV (13·8, 12·3, 12·3, 3·1%, respectively) (Table 3).
Table 3.
Type characteristics and virulence genes of M+/−LR S. agalactiae strains*
| PFGE (no). | Sequence type [clonal complex] (no.) | Capsular type (no.) | Virulence factors genes | |
|---|---|---|---|---|
| Alp protein family gene (no.) | Other virulence genes (no.)‡ | |||
| P1 (5), P4 (2), P5 (2), | ST1 [CC1] (10) | Ib (1) | alp2/3 (10) | scpB-lmb-fbsB (1) |
| P7–P13† (7) | V (9) | scpB-lmb-fbsA[900]-fbsB (1) | ||
| scpB-lmb-fbsB (6) | ||||
| scpB-lmb (2) | ||||
| P14 (1), P15 (1) | ST2 [CC1] (2) | II (1) | epsilon (2) | scpB-lmb-fbsA[600]-fbsB (1) |
| V (1) | scpB-lmb-fbsB (1) | |||
| P16 (1) | ST8 [CC9] (1) | Ib (1) | alphaC + bac (1) | scpB-lmb-fbsB (1) |
| P17 (1), P18 (1) | ST10 [CC9] (2) | Ib (1) | alphaC (1) | scpB-lmb-fbsA[1300]-fbsB (1) |
| II (1) | alphaC + bac (1) | scpB-lmb-fbsA[600]-fbsB (1) | ||
| P19-P27† (9) | ST12 [CC9] (9) | Ib (5) | alphaC + bac (5) | scpB-lmb-fbsA[1300]-fbsB (4) |
| scpB-lmb (1) | ||||
| II (4) | alphaC (1) | scpB-lmb-fbsA[900]-fbsB (1) | ||
| alphaC + bac (3) | scpB-lmb-fbsA[600]-fbsB (3) | |||
| P28-P31† (4) | ST17 [CC17] (4) | III (4) | rib (4) | scpB-lmb-fbsA[900]- fbsB-spb1 (4) |
| P2 (2), P3 (2), P32-P50† (19) | ST19 [CC19] (21) | III (16) | rib (16) | scpB-lmb-fbsA[1600]-fbsB (16) |
| V (5) | epsilon (4) | scpB-lmb-fbsA[1600]-fbsB (4) | ||
| epsilon + bac (1) | scpB-lmb-fbsA[1600]-fbsB (1) | |||
| P51 (1) | ST22 [CC22] (1) | II (1) | aphaC (1) | scpB-lmb-fbsA[750]-fbsB (1) |
| P6 (3), P52–P57† (6) | ST23 [CC23] (7) | Ia (7) | epsilon (7) | scpB-lmb-fbsB (7) |
| P58 (1) | ST28 [CC19] (1) | II (1) | rib (1) | scpB-lmb-fbsA[600]-fbsB (1) |
| P59 (1) | ST110 [CC19] (1) | V (1) | rib (1) | scpB-lmb-fbsA[600]-fbsB (1) |
| P60 (1), P61 (1) | ST196 [CC1] (2) | IV (2) | epsilon (2) | scpB-lmb-fbsA-fbsB (2) |
| P62 (1) | ST335 [CC19] (1) | III (1) | rib (1) | scpB-lmb-fbsA[600]-fbsB (1) |
| P63 (1) | ST498 [singleton] (1) | Ia (1) | alphaC (1) | scpB-lmb-fbsB (1) |
| P64 (1) | ST586 [CC22] (1) | II (1) | alphaC (1) | scpB-lmb-fbsA[750]-fbsB (1) |
| P65 (1) | ST601 [CC19] (1) | III (1) | rib (1) | fbsA[600]-fbsB (1) |
All 75 M+/−LR S. agalactiae strains typed by PFGE; MLST, capsular type and virulence gene patterns determined for 65 strains with different PFGE patterns.
One isolate for each PFGE pattern.
The number in square brackets shows the PCR amplicon size of fbsA.
The virulence factor related to the Alp protein family most frequently found in the 75 M+/−LR strains was the rib gene (37%); other virulence factor genes occurred at lower frequency: epsilon (25%), alphaC (23%), and alp2/3 (15%) (Table 3). The bac gene was present in 11 strains in association with alphaC + bac (10) or epsilon + bac (1). None of the strains harboured the alp4 gene (Table 3). Some correspondence of capsular types with Alp protein family genes was observed, e.g. capsular types Ib and II and alphaC in seven of eight strains, Ia with epsilon, and all capsular type III strains with rib (Table 3). Regarding other virulence-associated genes, all but one of 65 strains were positive for the scpB and lmb genes. The amplicon size of fbsA gene was variable (from 600 to 1600 bp) and fbsA + fbsB genes were found in 43 strains, and fbsB without fbsA in 19 strains. The spb1 gene was only found in the four strains assigned to ST17, which showed the same virulence pattern (Table 3).
Analysis of sequence types, capsular types and Alp family virulence genes showed that 9 of 10 strains of ST1 were capsular type V, and were also positive for alp2/3, and tet(M) tetracycline-resistance genes. Other notable associations were ST17 and capsular type III, possessing the rib virulence gene as well as erm(B) and tet(O) resistance genes. The erm(B) gene was found in all 21 strains of lineage ST19, and 17 of these strains contained cadmium and copper resistance genes (cadA, cadC, tcrB).
DISCUSSION
S. agalactiae is a frequent colonist of the urogenital tract in pregnant women worldwide and is a recognized major risk factor for neonatal sepsis associated with this organism. The need to monitor the epidemiology of S. agalactiae and their antibiotic resistance is emphasized worldwide for the prevention and control of such infections [1]. This is the first study of the frequency, and characteristics of S. agalactiae isolated from pregnant women in the La Rioja region (Northern Spain). We found a colonization rate of 14% in the study population which is within the range previously described in the country (12–20%) [2]. No cases of neonatal sepsis resulted in the cohort owing to effective maternal antimicrobial prophylaxis. In the study year (2011) 14·3% of the population of La Rioja region were classed as immigrants to Spain (national average 12·2%) from several countries. However, according to maternal nationality, 29·1% of children born in the region in that year were from immigrant mothers, which was marginally higher than the corresponding 19·5% recorded for Spain as a whole (http://www.larioja.org/larioja-client/cm/estadistica/images?idMmedia=621742). Additionally, to our knowledge, this is the first study investigating the epidemiology, antibiotic and heavy-metal susceptibilities, virulence and molecular types of S. agalactiae strains recovered from pregnant women in Spain.
The percentage of ERY- and/or CLI-resistant strains in our study (20%) was similar to that previously published by others [2]. Most of these strains contained erm(B) or erm(A) [erm(TR) variant] genes, associated with cML or iML phenotypes, respectively. The erm(T) gene associated with iML, was rare in the M+/−LR strains studied but may be of significance since it has been described in resistance and virulence plasmids in groups A and B streptococci [31] and also has been shown to be readily transferable among methicillin-resistant S. aureus [6]. Only four of the M+/−LR strains contained the lsa(C) gene, which encodes an ABC transporter, and could be an emerging resistance mechanism among LR S. agalactiae [18].
About 90% of M+/−LR strains showed resistance to tetracycline which might reflect the widespread use of these agents in the community. Indeed, it has been suggested that the extensive use of tetracycline has resulted in the emergence of relatively few resistant virulent clones adapted to survival in the host which have displaced less virulent clones of S. agalactiae [32]. The tet(M) gene was the most prevalent among tetracycline-resistant strains but 5·9% of these strains also harboured a tet(O) gene, which is a high percentage compared to previous studies [33]. Notably, this is the first report of the presence the tet(W) gene in S. agalactiae strains in Spain.
Resistance to metals is a widespread property in many bacterial species but data on heavy-metal resistance phenotypes and genotypes in S. agalactiae have not been previously described. Our work has demonstrated the existence of metal resistance mechanisms in M+/−LR S. agalactiae through the tcrB and cadA genes which codify a cation transport ATPase, and the cadC gene encoding a regulator protein with an arsR-type DNA-binding domain (http://bacmet.biomedicine.gu.se/index.html). Other copper-resistant M+/−LR tcrB-negative S. agalactiae strains could have acquired the copper resistance due to other mechanisms such as membrane reduced permeability, and expressed drug and metal efflux pumps. Therefore the selective pressure exerted by the use of copper and cadmium in feedstuffs or in clinical settings such as antisepsis, disinfection, and anti-infective chemotherapy or by the use of antibiotics in hospitals, farms and agriculture could potentially co-select for resistance to both metals [5–7]. This has been previously described with E. faecium and S. aureus, among others, in which metal resistance is co-selected with resistance to macrolides, glycopeptides, aminoglycosides or tetracycline, suggesting not only a genetic association between antibiotic and metal resistance mechanisms but also their co-dissemination through mobile genetic elements [6, 7].
This study has demonstrated a high degree of genetic diversity in antibiotic-resistant S. agalactiae strains within our patient community. The high frequency of capsular type III, which is associated with serious infections in humans [11], is consistent with other surveys in both European and African countries [34]. However, the emergence of capsular type V strains (24·6%), which has also been described in maternal colonization studies [34] is noteworthy. Although capsular type IV strains were infrequent in this and other European countries [9, 34], this serotype may be an emerging pathogen given reports of its prevalence in the United Arab Emirates, or recently in the United States and Portugal [34–36]. Furthermore, our work and a few other studies have shown an association between capsular type IV and ST196 [35, 37] which has also been also related to erythromycin-resistant strains [38].
As observed by others [37, 39–41], a strong correlation between capsular types and virulence genes was evident, namely, Ia/epsilon, Ib/alphaC, II/alphaC, and III/rib. The most predominant STs identified here (ST1, ST12, ST17, ST19, ST23) are, with the exception of ST12, also the most prevalent worldwide, and the main cause of human infections [8]. The common STs also showed strong correspondence with capsular serotypes, e.g. ST1/V, ST17 and 19/III, and ST23/Ia) as reported by others [8, 9]. It is noteworthy that four of our strains exhibited the characteristic of ST17/III/rib/spb1 which has been recognized as a hypervirulent clone and strongly associated with invasive neonatal infections such as meningitis, due to a novel ST17 specific surface-anchored protein, called hypervirulent GBS adhesin (HvgA), that enhances adherence of the organisms and their translocation across intestinal epithelium and the blood–brain barrier [42]. In addition, the surface protein Spb1 promotes invasion of respiratory and cervical epithelial cells, and contributes to the pathogenesis of neonatal and maternal infection by mediating internalization of virulent serotype III strains [42, 43].
The diversity of amplicon sizes of fbsA gene was expected because FbsA carries a repeated motif of 16 amino acids which varies among isolates in the number and sequence of repeats [28]. Only one strain lacked the scpB-lmb region. This region in S. agalactiae of human origin is surrounded by two ISSag2 elements, but interestingly, in the bovine strains, the scpB-lmb region is not necessary to cause bovine infections. Hence, its absence in our strain, might be due to homologous recombination between the flanking repeated IS sequences or could reveal a bovine origin for the strain [29, 44].
A high percentage of our erythromycin-resistant strains were typed as ST19/III/rib; although some studies have noted a strong association of ST1/V/alp3 with erythromycin resistance [37, 38, 41]. This tendency was expected as strains of CC19 (founder type ST19) have been linked with macrolide resistance in Poland [41].
In conclusion, surveillance of macrolide, lincosamide and heavy-metal resistant S. agalactiae strains is crucial not only to avoid their dissemination, nationally and internationally, but also to further our understanding of the dynamics of circulating genetic lineages within different community populations. The key findings from this study are the extreme clonal diversity of M+/−LR S. agalactiae strains recovered from an ethnically heterogeneous pregnant cohort and the marked associations of capsular serotypes and specific virulence genes. Further research is warranted to explore in depth the mechanisms that mobilize the antibiotic resistance determinants in this group of microorganisms.
ACKNOWLEDGEMENTS
The authors are grateful to Pilar Martínez and Carmen Rojo (Área de Diagnóstico Biomédico, Laboratorio de Microbiología, Hospital San Pedro) for technical support. This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Verani JR, et al. Prevention of perinatal group B streptococcal disease. Revised Guidelines from CDC. Morbidity and Mortality Weekly Report 2010; 59(RR-10): 1–32. [PubMed] [Google Scholar]
- 2.Alós Cortés JI, et al. Prevention of neonatal group B streptococcal infection. Spanish recommendations. Update 2012. SEIMC/SEGO/SEN/SEQ/SEMFYC Consensus Document [in Spanish]. Enfermedades Infecciosas y Microbiología Clínica 2013; 31: 159–172. [DOI] [PubMed] [Google Scholar]
- 3.Lambiase A, et al. In vitro resistance to macrolides and clindamycin by Group B Streptococcus isolated from pregnant and nonpregnant women. Infectious Diseases in Obstetrics and Gynecology 2012; 2012: 913603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betriu C, et al. Erythromycin and clindamycin resistance and telithromycin susceptibility in Streptococcus agalactiae. Antimicrobial Agents and Chemotherapy 2003; 47: 1112–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Gorman J, Humphreys H. Application of copper to prevent and control infection. Where are we now? Journal of Hospital Infection. 2012; 81: 217–223. [DOI] [PubMed] [Google Scholar]
- 6.Gómez-Sanz E, et al. Novel erm(T)-carrying multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST398 that also harbor cadmium and copper resistance determinants. Antimicrobial Agents and Chemotherapy 2013; 57: 3275–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silveira E, et al. Co-transfer of resistance to high concentrations of copper and first-line antibiotics among Enterococcus from different origins (humans, animals, the environment and foods) and clonal lineages. Journal of Antimicrobial Chemotherapy 2014; 69: 899–906. [DOI] [PubMed] [Google Scholar]
- 8.Jones N, et al. Multilocus sequence typing system for group B streptococcus. Journal of Clinical Microbiology 2003; 41: 2530–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bisharat N, et al. Hyperinvasive neonatal group B Streptococcus has arisen from a bovine ancestor. Journal of Clinical Microbiology 2004; 42: 2161–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sørensen UB, et al. Emergence and global dissemination of host-specific Streptococcus agalactiae clones. mBio 2010; 1: e00178–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindahl G, Stålhammar-Carlemalm M, Areschoug T. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clinical Microbiology Reviews 2005; 18: 102–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johri AK, et al. Group B Streptococcus: global incidence and vaccine development. Nature Reviews Microbiology 2006; 4: 932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Second Informational Supplement M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA, 2012.
- 14.CA-SFM. Comite de L'Antibiogramme de la Societe Francaise de Microbiologie. 2011. (http://www.sfm-microbiologie.org/page/page/showpage/page_id/105.html). Accessed 10 April 2016.
- 15.Arana DM, et al. First clinical isolate in Europe of clindamycin-resistant group B Streptococcus mediated by the lnu(B) gene. Revista Española de Quimioterapia 2014; 27: 106–109. [PubMed] [Google Scholar]
- 16.Kataja J, et al. Erythromycin resistance genes in group A streptococci in Finland. The Finnish Study Group for Antimicrobial Resistance. Antimicrobial Agents and Chemotherapy 1999; 43: 48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung WO, et al. Host range of the ermF rRNA methylase gene in bacteria of human and animal origin. Journal of Antimicrobial Chemotherapy 1999; 43: 5–14. [DOI] [PubMed] [Google Scholar]
- 18.Malbruny B, et al. Cross-resistance to lincosamides, streptogramins A, and pleuromutilins due to the lsa(C) gene in Streptococcus agalactiae UCN70. Antimicrobial Agents and Chemotherapy 2011; 55: 1470–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aarestrup FM, et al. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagnostic Microbiology and Infectious Disease 2000; 37: 127–137. [DOI] [PubMed] [Google Scholar]
- 20.Lozano C, et al. Changes in genetic lineages, resistance, and virulence in clinical methicillin-resistant Staphylococcus aureus in a Spanish hospital. Journal of Infection and Chemotherapy 2013; 19: 233–242. [DOI] [PubMed] [Google Scholar]
- 21.Clark NC, et al. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrobial Agents and Chemotherapy 1999; 43: 157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wehbeh W, et al. Fluoroquinolone-resistant Streptococcus agalactiae: epidemiology and mechanism of resistance. Antimicrobial Agents and Chemotherapy 2005; 49: 495–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turabelidze D, et al. Improved pulsed-field gel electrophoresis for typing vancomycin-resistant enterococci. Journal of Clinical Microbiology 2000; 38: 4242–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenover FC, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. Journal of Clinical Microbiology 1995; 33: 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Belkum A, et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clinical Microbiology and Infection 2007; 13 (Suppl. 3): 1–46. [DOI] [PubMed] [Google Scholar]
- 26.Poyart C, et al. Multiplex PCR assay for rapid and accurate capsular typing of group B streptococci. Journal of Clinical Microbiology 2007; 45: 1985–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeland JA, et al. Streptococcus agalactiae beta gene and gene product variations. Journal of Medical Microbiology 1997; 46: 999–1005. [DOI] [PubMed] [Google Scholar]
- 28.Brochet M, et al. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes and Infection 2006; 8: 1227–1243. [DOI] [PubMed] [Google Scholar]
- 29.Rato MG, et al. Antimicrobial resistance and molecular epidemiology of streptococci from bovine mastitis. Veterinary Microbiology 2013; 161: 286–294. [DOI] [PubMed] [Google Scholar]
- 30.Creti R, et al. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. Journal of Clinical Microbiology 2004; 42: 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiPersio LP, et al. Identification and characterization of plasmid-borne erm(T) macrolide resistance in group B and group A Streptococcus. Diagnostic Microbiology and Infectious Disease 2011; 71: 217–223. [DOI] [PubMed] [Google Scholar]
- 32.Da Cunha V, et al. Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nature Communications 2014; 5: 4544. doi: 10.1038/ncomms5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betriu C, et al. In vitro activities of tigecycline against erythromycin-resistant Streptococcus pyogenes and Streptococcus agalactiae: mechanisms of macrolide and tetracycline resistance. Antimicrobial Agents and Chemotherapy 2004; 48: 323–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ippolito DL, et al. Group B Streptococcus serotype prevalence in reproductive-age women at a tertiary care military medical center relative to global serotype distribution. BMC Infectious Diseases 2010; 10: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diedrick MJ, et al. Clonal analysis of colonizing group B Streptococcus, serotype IV, an emerging pathogen in the United States. Journal of Clinical Microbiology 2010; 48: 3100–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Florindo C, et al. Epidemiological surveillance of colonising group B Streptococcus epidemiology in the Lisbon and Tagus Valley regions, Portugal (2005 to 2012): emergence of a new epidemic type IV/clonal complex 17 clone. Eurosurveillance 2014; 19: 20825. [DOI] [PubMed] [Google Scholar]
- 37.Gherardi G, et al. Molecular epidemiology and distribution of serotypes, surface proteins, and antibiotic resistance among group B streptococci in Italy Journal of Clinical Microbiology 2007; 45: 2909–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Francesco MA, et al. Phenotypes, genotypes, serotypes and molecular epidemiology of erythromycin-resistant Streptococcus agalactiae in Italy. European Journal of Clinical Microbiology & Infectious Diseases 2012; 31: 1741–1747. [DOI] [PubMed] [Google Scholar]
- 39.Brimil N, et al. Epidemiology of Streptococcus agalactiae colonization in Germany. International Journal of Medical Microbiology 2006; 296: 39–44. [DOI] [PubMed] [Google Scholar]
- 40.Persson E, et al. Characterisation of invasive group B streptococci based on investigation of surface proteins and genes encoding surface proteins. Clinical Microbiology and Infection 2008; 14: 66–73. [DOI] [PubMed] [Google Scholar]
- 41.Sadowy E, Matynia B, Hryniewicz W. Population structure, virulence factors and resistance determinants of invasive, non-invasive and colonizing Streptococcus agalactiae in Poland. Journal of Antimicrobial Chemotherapy 2010; 65: 1907–1914. [DOI] [PubMed] [Google Scholar]
- 42.Tazi A, et al. The surface protein HvgA mediates group B Streptococcus hypervirulence and meningeal tropism in neonates. Journal of Experimental Medicine 2010; 207: 2313–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adderson EE, et al. Subtractive hybridization identifies a novel predicted protein mediating epithelial cell invasion by virulent serotype III group B Streptococcus agalactiae. Infection and Immunity 2003; 71: 6857–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franken C, et al. Horizontal gene transfer and host specificity of beta-haemolytic streptococci: the role of a putative composite transposon containing scpB and lmb. Molecular Microbiology 2001; 41: 925–935. [DOI] [PubMed] [Google Scholar]