Abstract
The identification of a gene (yiaE) encoding 2-ketoaldonate reductase (2KR) in our previous work led to the hypothesis that Escherichia coli has other ketogluconate reductases including 2,5-diketo-d-gluconate reductase (25DKGR) and to study of the related ketogluconate metabolism. By using the deduced amino acid sequences of 5-diketo-d-gluconate reductase (5KDGR) of Gluconobacter oxydans and 25DKGR of Corynebacterium sp., protein databases were screened to detect homologous proteins. Among the proteins of E. coli, an oxidoreductase encoded by yjgU and having 56% similarity to 5KDGR of G. oxydans and two hypothetical oxidoreductases encoded by yqhE and yafB and having 49.8 and 42% similarity, respectively, to 25DKGR of Corynebacterium sp. were detected. Recently, the yjgU gene was identified as encoding 5KDGR and renamed idnO (C. Bausch, N. Peekhaus, C. Utz, T. Blais, E. Murray, T. Lowary, and T. Conway, J. Bacteriol. 180:3704–3710, 1998). The pathways involved in the metabolism of ketogluconate by E. coli have been predicted by biochemical analysis of purified enzymes and chemical analysis of the pathway intermediates. The gene products of yqhE and yafB were identified as 25DKGR-A, and 25DKGR-B, respectively, catalyzing the reduction of 25KDG to 2-keto-l-gulonate (2KLG). The native 25DKGR-A, 25DKGR-B, and 5KDGR had apparent molecular weights of about 30,000, 30,000, and 54,000, respectively. In sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels, all three enzymes showed protein bands with a molecular weight of about 29,000, which indicated that 25DKGR-A, 25DKGR-B, and 5KDGR may exist as monomeric, monomeric, and dimeric proteins, respectively. The optimum pHs for reduction were 7.5, 7.0, and 8.0, respectively. The 5KDGR was active with NADH, whereas 25DKGR-A and 25DKGR-B were active with NADPH as a preferred electron donor. 25DKG can be converted to 5KDG by 2KR, which is then reduced to d-gluconate by 5KDGR. The pathways were compared with those of Erwinia sp. and Corynebacterium sp. A BLAST search of published and incomplete microbial genome sequences revealed that the ketogluconate reductases and their related metabolism may be widespread in many species.
Incomplete oxidation of glucose to ketogluconates, 2-keto-d-gluconate (2KDG), 5-keto-d-gluconate (5KDG), and 2,5-diketo-d-gluconate (25DKG), in a variety of microorganisms has been shown to proceed via membrane-bound dehydrogenases linked to the electron transport chain (4, 33). Some ketogluconates can be used as the source of carbon and energy for many bacteria. The metabolic pathways involved in the use of such ketogluconates have been studied for Corynebacterium sp. (32) and Erwinia spp. (5, 35). The pathways in the two species are quite similar, except that, in Erwinia sp., 25DKG is converted to 5KDG but in Corynebacterium sp., 25DKG is converted to 2KDG before being converted to d-gluconate. The sequential conversion of ketogluconates to d-gluconate is mediated by soluble NAD(P)H-dependent reductases. In those bacteria, d-gluconate is phosphorylated to 6-phosphogluconate and further metabolized through the pentose phosphate pathway. In our previous work, it was found that the decrease of 2KDG produced from d-gluconate in the cultivation of recombinant Escherichia coli harboring the cloned membrane-bound gluconate dehydrogenase gene (38) was due to 2-ketoaldonate reductase (2KR) as the cytosolic enzyme responsible for conversion of 2KDG to d-gluconate (37). We also identified a gene (yiaE) encoding 2KR and found that 2KR catalyzes the reduction of 25DKG to 5KDG and of 2-keto-l-gulonate (2KLG) to l-idonate (IA), as well as of 2KDG to d-gluconate (37). This result suggested strongly that the other ketogluconate reductases known in other bacteria, 5-keto-d-gluconate reductase (5KDGR) and 2,5-diketo-d-gluconate reductase (25DKGR), and the related ketogluconate metabolism could also exist in E. coli.
Several ketogluconate reductases, 5KDGR, 2KDGR, 2KR, and 25DKGR, have been purified and characterized (1–3, 27, 34, 36) elsewhere, and the genes encoding 5KDGR (21) and 25DKGR (6, 17, 18) have been cloned elsewhere. To find ketogluconate reductases in E. coli, we searched homologous proteins in the protein databases with the amino acid sequences of 5KDGR of Gluconobacter oxydans (21) and 25DKGR of Corynebacterium sp. (6). As a result, our attention was drawn to two hypothetical oxidoreductases, encoded by yqhE and yafB, showing similarity to 25DKGR. A hypothetical oxidoreductase, encoded by yjgU, showing similarity to 5KDGR of G. oxydans was also found; however, the yjgU gene was found to encode 5KDGR and renamed idnO recently (8). 5KDGR of E. coli was identified by sequence analysis of the GntII (subsidiary) system for gluconate metabolism. The GntII system encodes a pathway for catabolism of IA, in which IA is converted to 5KDG by IA dehydrogenase before being reduced to d-gluconate by 5KDGR. d-Gluconate is phosphorylated to 6-phosphogluconate and further metabolized through the Entner-Doudoroff pathway.
In this report, we will show that the proteins encoded by yqhE and yafB are 25DKGR-A and 25DKGR-B, catalyzing the reduction of 25DKG to 2KLG, and the pathways involved in the metabolism of ketogluconates by E. coli are similar to those involved in the metabolism of ketogluconates by Erwinia and Corynebacterium spp. 25DKGR is an important enzyme in the bioconversion of 2KLG, a key intermediate in the biosynthesis of ascorbic acid (vitamin C) by the glucose pathway (17, 27, 31, 32, 34). Besides the application of these 25DKGRs, the understanding of ketogluconate metabolism should lead to further development of an E. coli host strain that lacks the catabolic pathway of ketogluconates produced when a membrane-bound gluconate dehydrogenase (38), a 2KDG dehydrogenase, and a 25DKGR are expressed for the conversion (5, 6, 17) of glucose to 2KLG.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli W3110 [F− IN(rrnD-rrnE)], DH5α [F− endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA relA1 Δ(argF-lac)U169 deoR Φ80lacZΔM15], and ER2566 [F− λ− fhuA2 (lon) ompT lacZ::T7 gene 1 gal sulA11 Δ(mcrC-mrr)114::IS10 R(mcr-73::mini-Tn10)2 R(zgb-210::Tn10)1 (Tets) endA1 (dcm)] (13) were used as host strains. All strains were grown in Luria-Bertani (LB) medium (1% Bacto Tryptone, 0.5% yeast extract, 0.5% NaCl) or M9 minimal medium (26) with 2KDG, 5KDG, 25DKG, or 2KLG as the carbon source. For antibiotic selection, the concentration of 100 μg/ml (ampicillin) was used.
Preparation of cell extracts.
E. coli cells were grown at 37°C for 18 h, harvested by centrifugation, and washed with 100 mM sodium phosphate buffer (pH 6.0). The cell paste was resuspended in the same buffer containing 0.1 mM phenylmethysulfonyl fluoride (PMSF) to 1/10 of the original culture volume, and the cells were broken by sonication. The cell debris and unbroken cells were removed by centrifugation at 22,000 × g for 20 min.
Enzyme assay and determination of d-glucose, d-gluconate, 2KDG, 5KDG, 2KLG, 25DKG, and IA.
The ketogluconate reductase activities were assayed as described previously (27) with 20 mM substrate and 0.1 mM NADH or NADPH. The reaction was monitored for the initial decrease in absorbance at 340 nm (ɛ = 6.22 mM−1 cm−1). One unit of activity corresponds to the production of 1 μmol of NADP+ or NAD+ per min. The protein concentration of each sample was determined with the bicinchoninic acid protein assay kit (Pierce). Glucose was determined with a glucose analyzer (model 2300 STAT; Yellow Springs Instrument Co.). d-Gluconate, 2KDG, 5KDG, 2KLG, 25DKG, and IA in the reactions were determined by high-pressure liquid chromatography with an HPX-87C column (Bio-Rad) at 30°C at a flow rate of 0.5 ml of 0.008 N H2SO4 per min as the eluent.
DNA preparation, manipulation, and sequence analysis.
Total DNA from E. coli was prepared by using Qiagen Genomic Tips. DNAs of the vector plasmids were prepared by a rapid alkaline lysis procedure (9), with a QIAprep Spin Miniprep kit (Qiagen). General DNA manipulations were carried out as described by Maniatis et al. (26). DNA sequencing of both strands was performed with an ABI 373 automated sequencer with dye-labelled terminators (Applied Biosystems Division of Perkin-Elmer). Oligonucleotides were synthesized by Bioneer (Chungweon, Korea). Sequence analysis, comparisons, and CLUSTAL alignments were performed, in part with LASERGENE (DNASTAR Inc., Madison, Wis.). Comparisons were also performed via BLAST and FASTA analysis of the SWISS-PROT and Protein Information Resource sequences. The PROSITE database was used for motif searches.
PCR cloning of the yqhE, yafB, and yjgU genes.
Design of the primers was based on the published yqhE, yafB, and yjgU nucleotide sequences (GenBank accession no. AE000383, AE000129, and AE000497, respectively) (10). Two PCR primers used for yqhE were 5′-TGAACGCGTCTAGAACATCACTG-3′ and 5′-CTTCCGGCTCTAGATGATGATGT-3′, those used for yafB were 5′-TCGCGGGTTCTAGACCCGTCCGT-3′ and 5′-GTGTTTGTCCGTCTAGATGATCGACA-3′, and those used for yjgU were 5′-AATACCTGTCTAGAGGTCACTCGT-3′ and 5′-ATACCATTTTGTCTAGAGTGCAGG-3′ (the XbaI restriction site is underlined, and the point-mutated position is shown in boldface). The primers contained several point mutations introducing an XbaI site at the 5′ and 3′ ends. All PCRs were carried out in a GeneAmp PCR 2400 system (Perkin-Elmer) with 30 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 65°C, and extension for 1 min at 72°C, followed by a 3-min extension period at 72°C. The resulting PCR products were cloned into plasmid pUC19 cleaved with XbaI to produce plasmids pYQHE, pYAFB, and pYJGU. The plasmids were sequenced to confirm that the sequences of the inserts were identical to those of the yqhE, yafB, and yjgU genes.
Purification of 25DKGR-A.
25DKGR-A was isolated by using the IMPACT (intein-mediated purification with an affinity chitin-binding tag) one-step protein purification system (New England Biolabs). The DNA fragment containing the coding region of the yqhE gene was synthesized by using two primers, 5′-AGGAACATATGGCTAATCCAACCGTTATTAAG-3′ and 5′-GAGAAGCTCTTCCGCAGCCGCCGAACTGGTCAGGATC-3′ (the NdeI and SapI restriction sites are underlined, and the point-mutated position is shown in boldface). In the system, cloning into the SapI site leaves no additional residues attached to the C terminus of the target protein. After being digested with NdeI and SapI, the fragment was cloned into the vector pTYB1 (New England Biolabs), which contains an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T7 promoter. The resulting plasmid, pTY-YQH, containing the yqhE gene fused to the intein gene at the 3′ end, was transformed into E. coli ER2566. For the purification of the recombinant protein, the resulting E. coli strain, ER2566(pTY-YQH), was grown in LB medium to an optical density at 600 nm of 0.5. Expression of the proteins was induced by the addition of 1 mM IPTG for 4 h. The cells were then resuspended in cell lysis buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 0.1 mM EDTA, and 0.1% Triton X-100) and disrupted by sonication. Cell debris was removed by centrifugation, and the supernatant was loaded onto the chitin columns. The columns were washed two times with cell lysis buffer. The native proteins were cleaved and eluted with cleavage buffer (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 0.1 mM EDTA, and 30 mM dithiothreitol).
Purification of 25DKGR-B.
Overproduction of 25DKGR-B was achieved by constructing plasmid pKK-YAF. To construct pKK-YAF, the yafB gene was amplified by PCR with two primers, 5′-TAAAAGAGGGAATTCATGGCTATCCC-3′ and 5′-GCTGTCAGAGAAGCTTAATCCCAT-3′ (the EcoRI and HindIII restriction sites are underlined, and the point-mutated position is shown in boldface), introducing an EcoRI and a HindIII site to yafB. The resulting DNA fragment was ligated to the EcoRI and HindIII cloning sites of plasmid pKK223-3 (Pharmacia), yielding plasmid pKK-YAF. For protein purification, the E. coli DH5α(pKK-YAF) cells were harvested after 4 h of induction with IPTG. The cells were harvested by centrifugation, washed two times with 20 mM Tris-HCl buffer (pH 8.3), resuspended in the same buffer with 0.1 mM phenylmethylsulfonyl fluoride and streptomycin (0.1 mg/ml), and disrupted with an ultrasonic oscillator. Cell debris was removed by centrifugation at 15,000 × g for 30 min. The resulting supernatant as a crude enzyme solution was used for the purification of the reductase. The dialyzed sample was put on a DEAE-Toyopearl column (2.5 by 4 cm) that had been equilibrated with 20 mM Tris-HCl buffer (pH 8.3). The column was washed with the same buffer, and proteins were eluted with a linear gradient of 0.1 to 0.5 M NaCl in the same buffer at a flow rate of 1.19 ml/min. Fractions that contained 25DKGR-B activity were pooled and dialyzed overnight against 20 mM Tris-HCl buffer (pH 7.0). The dialyzed sample was placed on a Blue-Sepharose CL-6B column (1.5 by 7 cm) equilibrated with 20 mM Tris-HCl buffer (pH 7.0). The column was washed with the same buffer, and proteins were eluted with a linear gradient of 0 to 0.3 M NaCl in the same buffer. The active fractions eluted from the affinity column were combined, concentrated with Centriprep 10 (Amicon) to 2.0 ml, and put on a Bio-Sil SEC-250 (Bio-Rad) gel filtration column (0.78 by 30 cm) equilibrated with 20 mM Tris-HCl buffer (pH 7.0) containing 0.15 M NaCl. The active fractions were pooled and stored at 4°C.
Purification of N-terminal His6-tagged 5KDGR.
Overproduction of His6-tagged 5KDGR was achieved by constructing plasmid pQE-YJG. To construct pQE-YJG, the yjgU gene was amplified by PCR with two primers, 5′-GAATAAGGATCCGAACGATCTATTTTCACTGGCAGGAA-3′ and 5′-GTAGGGGGGAAGCTTAAACAGCCAC-3′ (the BamHI and HindIII restriction sites are underlined, and the point-mutated position is shown in boldface), introducing a BamHI and a HindIII site to yjgU. The resulting DNA fragment was ligated to the BamHI and HindIII cloning sites of plasmid pQE31 (Qiagen), yielding plasmid pQE-YJG. In this plasmid, yjgU is fused N-terminally in frame to the six-His tag-coding region of plasmid pQE31, leading to expression under the control of the IPTG-inducible promoter. Overproduction of His6-tagged 5KDGR was achieved in E. coli DH5α(pQE-YJG) after IPTG (1 mM) induction in LB medium (containing 100 μg of ampicillin per ml) for at least 1 h. For protein purification, the cells were harvested after 4 h of induction with IPTG. The His6-tagged fusion protein was purified from recombinant E. coli cells with Ni-nitrilotriacetic acid (NTA) resin (Qiagen). Centrifugations were carried out at 4°C, and column chromatographies were carried out at room temperature. For His6-tagged 5KDGR purification, the cell pellet from a 500-ml culture of E. coli DH5α(pQE-YJG) was suspended in 20 ml of 50 mM sodium phosphate (pH 8.0)–0.3 M NaCl and sonicated on ice. The resulting cell lysate was centrifuged at 16,000 × g, and the supernatant was passed directly over a column containing 1.6 ml of Ni-NTA resin (Qiagen). After the column was washed with 30 ml of 50 mM sodium phosphate buffer (pH 8.0) containing 0.3 M NaCl–10% glycerol, the N-terminal His6-tagged 5KDGR was eluted with 4 ml of 50 mM Na-citrate buffer (pH 6.0). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was done by Laemmli’s method (23).
Chemicals.
Calcium 25DKG and sodium 2KLG were obtained from Dong-A Pharmaceutical Company (Seoul, Korea). Calcium 2KDG and potassium 5KDG were purchased from Sigma Chemical Company.
RESULTS AND DISCUSSION
Identification of yqhE and yafB genes encoding 25DKGR-A and 25DKGR-B.
In the previous work, we identified a gene (yiaE) encoding 2KR catalyzing the reduction of 25DKG to 5KDG and of 2KLG to IA, as well as of 2KDG to d-gluconate (37), which suggested strongly that the other ketogluconate reductases, 5KDGR and 25DKGR, and the related ketogluconate metabolism could also exist in E. coli. To find the possible ketogluconate reductases in E. coli, we searched homologous proteins in the protein databases by running FASTA searches with the amino acid sequences of 5KDGR (SP:P50199) of G. oxydans (21) and 25DKGR (PIR:I40838) of Corynebacterium sp. (6) as query sequences. As a result, our attention was drawn to two hypothetical oxidoreductases (SP:Q46857 and SP:P30863), encoded by yqhE and yafB, having 49.8 and 42% similarity, respectively, to 25DKGR and an oxidoreductase (SP:P39345), encoded by yjgU, showing 56% similarity to 5DKGR. Of the three genes, the yjgU gene was found to encode 5KDGR and was renamed idnO recently (8). To clarify the functions of the hypothetical oxidoreductases encoded by yqhE and yafB, we attempted to clone these putative genes. We amplified each of these genes by PCR under conditions that minimized errors and cloned them in the plasmid vector pUC19. The reductase activities in crude cell extracts of several different clones of each construct were determined by using 2KDG, 5KDG, 2KLG, or 25DKG as a substrate in the presence of NADPH or NADH. The presence of pYQHE and pYAFB containing yqhE and yafB genes, respectively, increased specific 25DKGR activity by about 10-fold, and the reaction products were identified as 2KLG. This result suggested that both yqhE and yafB genes encoded 25DKGRs, which were named 25DKGR-A and 25DKGR-B, respectively. As expected, the clone pYJGU containing the idnO gene also increased 5KDGR-specific activity by about 10-fold. The reductases in E. coli were purified further to confirm the substrate specificity.
Purification of 25DKGR-A, 25DKGR-B, and His6-tagged 5KDGR and their substrate specificity.
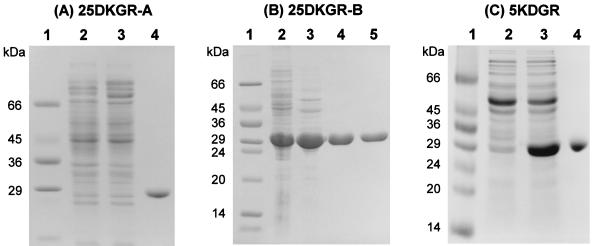
25DKGR-A was purified by using the IMPACT (intein-mediated purification with an affinity chitin-binding tag) one-step protein purification system. The N-terminal His6-tagged 5KDGR was isolated by using metal-chelate affinity chromatography on an Ni-NTA column instead of using the IMPACT system, since Leu at the C terminus of 5KDGR decreases in vitro cleavage with dithiothreitol at 4°C, resulting in a decrease in the yield of mature protein. Since the N-terminal or C-terminal His-tagged 25DKGR-B was catalytically inactive and the intein-mediated purification procedure was also inapplicable, 25DKGR-B was purified to homogeneity by column chromatographies on DEAE-Toyopearl, Blue-Sepharose CL-6B, and Bio-Sil SEC-250 from the crude cell extracts of the E. coli clone. Upon SDS-PAGE, the purified proteins yielded a single band when stained with Coomassie blue (Fig. 1). 2KDG, 25DKG, 2KLG, 5KDG, d-fructose, and l-sorbose were examined as substrates. Consistent with earlier results with crude cell extracts, purified 25DKGR-A and 25DKGR-B were highly specific for 25DKG in the presence of NADPH, and 2KDG, 2KLG, 5KDG, d-fructose, and l-sorbose were inactive when assayed at pH 6.0 in the presence of NADH or NADPH. In the enzymatic reduction of 25DKG, the reaction products with purified 25DKGR-A and 25DKGR-B were identified as exclusively 2KLG. His6-tagged 5KDGR was also highly specific for 5KDG, and the reaction product was identified as d-gluconate. The other substrates, 2KDG, 25DKG, 2KLG, d-fructose, and l-sorbose, showed no reduction activity with NADH or NADPH. The His6-tagged 5KDGR was active with either NADPH (threefold-lesser extent than NADH) or NADH, whereas 25DKGR-A and 25DKGR-B were active with only NADPH as a preferred electron donor. This result indicates that 25DKGRs and 5KDGR of E. coli have similar substrate specificities with 25DKGRs of Corynebacterium sp. (27, 34) and 5KDGR of G. oxydans (1, 3, 21), respectively. The native 25DKGR-A, 25DKGR-B, and His6-tagged 5KDGR had apparent molecular weights of about 30,000, 30,000, and 54,000, respectively, as determined by molecular exclusion chromatography with Bio-Sil SEC-250 (Bio-Rad). In SDS-PAGE, all three enzymes showed protein bands with a molecular weight of about 29,000 (Fig. 1). The molecular weights (31,003, 29,401, and 27,562) of 25DKGR-A, 25DKGR-B, and 5KDGR, respectively, calculated by deduced amino acid sequences were in good agreement with the molecular masses obtained by SDS-PAGE. These data suggest that 25DKGR-A, 25DKGR-B, and His6-tagged 5KDGR are monomeric, monomeric, and dimeric proteins, respectively. The optimum pHs for reduction were 7.5, 7.0, and 8.0, respectively.
FIG. 1.
SDS-PAGE monitoring of purification of 25DKGR-A (A), 25DKGR-B (B), and His6-tagged 5KDGR (C) from E. coli ER2566(pTY-YQH), DH5α(pKK-YAF), and DH5α(pQE-YJG), as described in Materials and Methods. (A) Lane 1, molecular mass markers (Sigma); lane 2, noninduced cells; lane 3, cells induced with IPTG; lane 4, eluate from chitin column. (B) Lane 1, molecular mass markers; lane 2, cells induced with IPTG; lanes 3 to 5, active fractions after DEAE-Toyopearl chromatography (lane 3), Blue-Sepharose CL-6B (lane 4), and Bio-Sil SEC-250 gel filtration (lane 5). (C) Lane 1, molecular mass markers; lane 2, noninduced cells; lane 3, cells induced with IPTG; lane 4, eluate from Ni-NTA column.
Comparison of ketogluconate reductases.
The ketogluconate reductases for which the amino acid sequences are known were compared with those of E. coli including 2KR (37). The alignments of amino acid sequences are compared in Table 1. The reductases having the same substrate specificity showed relatively high homology. The PROSITE database was used for motif searches. As a result, the signatures were identified as the aldo- and ketoreductase family for 25DKGR-A and 25DKG-B, the short-chain dehydrogenase-reductase family for 5KDGR, and the d-isomer-specific 2-hydroxyacid dehydrogenase family for 2KR (37). The alignment indicates conserved motifs that may be involved in substrate binding or catalysis, including signature motifs. The aldo- and ketoreductase family has three consensus patterns located in the N-terminal, central, and C-terminal sections, and the (α/β) eight-barrel fold provides a common scaffold for NAD(P)(H)-dependent catalytic activity, with substrate specificity determined by variation of loops on the C-terminal side of the barrel (19).
TABLE 1.
Statistical comparison of ketogluconate reductasesa
| Reductase (sp.) | % Identity
with reductase (sp.):
|
||||||
|---|---|---|---|---|---|---|---|
| 2KR (Erwinia herbicola) | 25DKGR-A (E. coli) | 25DKGR-A (Corynebacterium sp.) | 25DKGR-B (Corynebacterium sp.) | 25DKGR-B (E. coli) | 5KDGR (E. coli) | 5KDGR (G. oxydans) | |
| 2KR (E. coli) | 69.1 | 10.9 | 11.2 | 10.5 | 12.7 | 11.8 | 12.9 |
| 2KR (Erwinia herbicola) | 12.0 | 11.6 | 12.7 | 12.0 | 12.2 | 13.7 | |
| 25DKGR-A (E. coli) | 47.4 | 40.1 | 31.8 | 10.2 | 11.7 | ||
| 25DKGR-A (Corynebacterium sp.) | 37.7 | 37.1 | 12.1 | 12.9 | |||
| 25DKGR-B (Corynebacterium sp.) | 28.8 | 11.0 | 12.1 | ||||
| 25DKGR-B (E. coli) | 12.6 | 11.3 | |||||
| 5KDGR (E. coli) | 55.5 | ||||||
Accession numbers are as follows: 2KR of E. coli (SP:P37666), 25DKGR-A and -B of E. coli (SP:Q46857 and P30863, respectively), 25DKGR-A and -B of Corynebacterium sp. (PIR: I40838 and A45961, respectively), 5KDGR of E. coli (GB:AAC77223.1), and 5KDGR of G. oxydans (SP:P50199). Values presented in the table represent percent identities.
Proposed pathways for ketogluconate metabolism in E. coli.
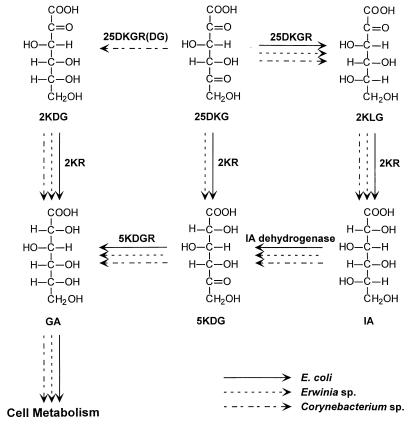
The pathways involved in the metabolism of ketogluconates by Erwinia sp. (35) and Corynebacterium sp. (32) have been investigated by use of a combination of enzyme assays and isolation of mutants. The presence of the ketogluconate reductase genes (yiaE, yqhE, yafB, and idnO) and the enzymatic activities described above suggest the pathway shown in Fig. 2 as the catabolic pathway for ketogluconates in E. coli. In Erwinia sp., 25DKG is converted to 5KDG, but in Corynebacterium sp., 25DKG is converted to 2KDG before being converted to d-gluconate. Interestingly, in E. coli, 25DKG can be catabolized by sequential reductions to d-gluconate via 5KDG. Then the resulting d-gluconate from ketogluconates is metabolized via the Entner-Doudoroff pathway, with the pentose phosphate pathway playing a secondary role (16). The other possible catabolic pathway, of 25DKG to d-gluconate via 2KLG, IA, and 5KDG, needs the idonate dehydrogenase (5KR[I]) catalyzing the oxidation of IA to 5KDG, which is known in Erwinia and Corynebacterium spp. Recently, 5KDGR of E. coli was identified by sequence analysis of the GntII system for gluconate metabolism (8). The GntII system encodes a pathway for catabolism of IA, in which IA is converted to 5KDG by IA dehydrogenase before being reduced to d-gluconate by 5KDGR.
FIG. 2.
Possible pathways for ketogluconate metabolism in E. coli. The pathways were compared with those of Erwinia sp. and Corynebacterium sp.
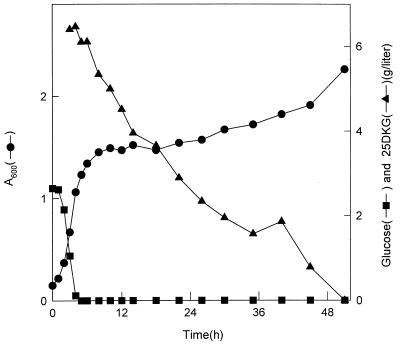
Even though the genes encoding 25DKGR-A and 25DKGR-B exist in E. coli, the W3110 strain showed poor growth on 2KDG (37), 5KDG, 25DKG, or 2KLG as the sole carbon source (data not shown), which may have been due to poor uptake of ketogluconates into the cell or redox imbalance. This assumption is consistent with experiments with acetic acid bacteria having NAD(P)H-dependent ketogluconate reductases, which showed slow growth on 2KDG or 5KDG (30). It has been suggested elsewhere that the ketogluconate reduction reactions in acetic acid bacteria may be important in maintaining redox balance rather than in providing a carbon source (35). E. coli grows well on IA, which is converted to d-gluconate by redox-coupled interconversion via intermediate 5KDG (8). The regulation of the sugar acid regulons allows E. coli to cometabolize the sugar acids, even in the presence of glucose (28). Therefore, the ketogluconate metabolism may provide E. coli with the metabolic advantage necessary for it to compete with other bacteria by maintaining redox balance. Because E. coli shows poor growth on 25DKG as the sole carbon source, cultivation in minimal medium containing both glucose (2 g/liter) and 25DKG (6 g/liter) was tried (Fig. 3). 25DKG was added into the medium after 4 h of cultivation with glucose. The level of 25DKG in the medium decreased after the exhaustion of glucose, and the cell optical density was reincreased after 18 h of cultivation. This observation indicates that the ketogluconate metabolism in E. coli may serve as a subsidiary metabolism in certain ecological environments.
FIG. 3.
Growth of E. coli in M9 minimal medium. E. coli W3110 was grown on M9 minimal medium containing glucose (2 g/liter). After 4 h of cultivation, 25DKG (6 g/liter) was added to the medium.
The GntII system encoding a pathway for catabolism of IA in E. coli consists of an operon, which encodes IA dehydrogenase, 5KDGR, an IA transporter, and an IA regulatory protein, respectively, in which the IA regulatory protein acts as a positive regulator, with IA and 5KDG serving as the inducers of the pathway (8). In the analysis of the genes in the vicinity of the genes encoding 2KR, 25DKGR-A, and 25DKGR-B, two genes, yqhC and yafC, encoding hypothetical transcriptional regulators adjacent to yqhE and yafB were found. Further, the yqhD gene encoding a hypothetical oxidoreductase was found to be located between the yqhE and yqhC genes. Two putative regulatory proteins may play a role in the regulation of yqhE and yafB gene expressions. Therefore, further studies should be focused on the identification of yafC, yqhC, and yqhD genes.
BLAST search of incomplete microbial genomes.
To investigate the possibility that other bacteria could also have genes encoding 2KR, 5KDGR, 25DKGR-A, and 25DKGR-B, amino acid sequences encoded by yiaE, idnO, yqhE, and yafB genes were subjected to a homology search with the tBLASTn program of the National Center for Biotechnology Information database of 26 incomplete microbial genome sequences of Actinobacillus actinomycetemcomitans, Bordetella pertussis, Borrelia burgdorferi, Campylobacter jejuni, Chlamydia trachomatis, Chlorobium tepidum, Clostridium acetobutylicum, Deinococcus radiodurans, Enterococcus faecalis, Helicobacter pylori, Methanobacterium thermoautotrophicum, Methanococcus jannaschii, Mycobacterium tuberculosis CSU 93, Mycobacterium tuberculosis H37Rv, Neisseria gonorrhoeae, Neisseria meningitidis MC58, Neisseria meningitidis serogroup Ai, Pseudomonas aeruginosa, Porphyromonas gingivalis W83, Pyrococcus furiosus, Staphylococcus aureus, Streptococcus pyogenes, Streptococcus pneumoniae, Thermotoga maritima, Vibrio cholerae, and Yersinia pestis. Even if these microbial genomes are not yet complete, several open reading frames (ORFs) encoding proteins showing high homologies with E. coli ketogluconate reductases were found in several strains. ORFs encoding proteins showing more than 30% identity with E. coli 2KR (8 ORFs), 5KDGR (14 ORFs), 25DKGR-A (14 ORFs), and 25DKGR-B (15 ORFs) were found. Especially in Y. pestis and P. aeruginosa, ORFs encoding putative proteins showing homologies with all four reductases were found.
The range of bacterial genome sequences available has grown rapidly and is likely to continue expanding. A search of published genome sequences, as well as partially completed genome sequences, indicates that putative ketogluconate reductases are present in several of these organisms. This is not surprising given that ketogluconates can serve as the sole source of carbon and energy for various bacteria. It has also been reported elsewhere that a large number of enterobacterial strains have the glucose oxidation pathway (11) and the ability to use 2KDG as a source of carbon and energy in a defined medium (12). It has also been postulated elsewhere that sugar acid metabolism is an important aspect of the ecology of E. coli (28). Although generally considered to be restricted to a few bacteria, the ketogluconate metabolism may be widespread in many species. Until now, complete genome sequences were available for only 16 microorganisms. More than 50 additional microbial genomes are scheduled to be completely sequenced by the year 2000 (14). The availability of the complete genome sequence should facilitate identification of how the metabolism is distributed in microorganisms. It may be possible that exploring the ketogluconate metabolism could provide a useful tool for classification and identification of microbial strains.
Application.
Current work concerns the study of the genes encoding enzymes involved in ketogluconate metabolism in E. coli by using a direct strategy for finding and characterizing unidentified genes from the E. coli genomic database. This finding should lead to further development of an E. coli host strain that lacks the catabolic pathway of ketogluconates produced when a membrane-bound gluconate dehydrogenase (38), 2KDG dehydrogenase, and 25DKGR are expressed for the conversion of glucose to 2KLG, a key intermediate in the biosynthesis of ascorbic acid (vitamin C). A process for the bioconversion of an ordinary microbial metabolite such as glucose into 2KLG with a single recombinant strain by combining the relevant traits of both Erwinia sp. and Corynebacterium sp. into a single organism has been tried (5, 6, 17). However, even when a 2KR gene of Erwinia sp. was disrupted, the production of an undesirable by-product, IA, could not be prevented, which was found to be due to the unexpected additional pathway of metabolism of ketogluconates by another 2KR (5). E. coli could be useful as a recombinant host strain for the production of 2KLG, because it does not grow well on ketogluconates. If needed, an E. coli mutant deficient in 2KR activity (37) could be used as a host strain.
25DKGR is an important enzyme in the bioconversion of 2KLG by the glucose pathway since the rate-limiting step may be the reduction of 25DKG to 2KLG by 25DKGR. Until now, the 25DKGR gene from Corynebacterium sp. has been used, and several studies have been carried out to increase the temperature stability of the enzyme (24, 29). Recently, the three-dimensional structure of 25DKGR was also determined to aid the elucidation of the structural determinants of specificity, catalysis, and stability for the enzyme (20). Though we do not know yet how stable and efficient two E. coli 25DKGRs are in the application and it is possible that their thermal stability might not prove to be satisfactory, two enzymes could be useful in enzyme improvement, considering that directed evolution with family shuffling of enzymes has been shown to be an effective method in recent studies (7, 15, 22, 25).
ACKNOWLEDGMENTS
We thank E. S. Choi for critical reading of the manuscript.
This investigation was supported by grant HS1810 from the Ministry of Science and Technology of Korea (MOST) and Korea Microbial Technology Inc. (KOMITECH).
REFERENCES
- 1.Adachi O, Shinagawa E, Matsushita K, Ameyama M. Crystallization and properties of 5-keto-d-gluconate reductase from Gluconobacter suboxydans. Agric Biol Chem. 1979;43:75–83. [Google Scholar]
- 2.Ameyama M, Adachi O. 2-Keto-d-gluconate reductase from acetic acid bacteria. Methods Enzymol. 1982;89:203–209. doi: 10.1016/s0076-6879(82)89033-2. [DOI] [PubMed] [Google Scholar]
- 3.Ameyama M, Adachi O. 5-Keto-d-gluconate reductase from Gluconobacter suboxydans. Methods Enzymol. 1982;89:198–202. [Google Scholar]
- 4.Ameyama M, Matsushita K, Shinagawa E, Adachi O. Sugar-oxidizing respiratory chain of Gluconobacter suboxydans. Evidence for a branched respiratory chain and characterization of respiratory chain-linked cytochromes. Agric Biol Chem. 1987;51:2943–2950. [Google Scholar]
- 5.Anderson, S., R. A. Lazarus, H. I. Miller, and R. K. Stafford. July 1991. U.S. patent 5,032,514.
- 6.Anderson S, Marks C B, Lazarus R, Miller J, Stafford K, Seymour J, Light D, Rastetter W, Estell D. Production of 2-keto-l-gulonate, an intermediate in l-ascorbate synthesis, by a genetically modified Erwinia herbicola. Science. 1985;230:144–149. doi: 10.1126/science.230.4722.144. [DOI] [PubMed] [Google Scholar]
- 7.Arnold F H, Moore J C. Optimizing industrial enzymes by directed evolution. Adv Biochem Eng Biotechnol. 1997;58:1–14. doi: 10.1007/BFb0103300. [DOI] [PubMed] [Google Scholar]
- 8.Bausch C, Peekhaus N, Utz C, Blais T, Murray E, Lowary T, Conway T. Sequence analysis of the GntII (subsidiary) system for gluconate metabolism reveals a novel pathway for l-idonic acid catabolism in Escherichia coli. J Bacteriol. 1998;180:3704–3710. doi: 10.1128/jb.180.14.3704-3710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coliK-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 11.Bouver O M, Lenormand P, Grimont P A. Taxonomic diversity of the d-glucose oxidation pathway in the Enterobacteriaceae. Intl J Syst Bacteriol. 1989;39:61–67. [Google Scholar]
- 12.Buissiere J, Brault G, Le Minor L. Utilization and fermentation of 2-ketogluconate by Enterobacteriaceae. Ann Microbiol (Paris) 1981;132:191–195. [PubMed] [Google Scholar]
- 13.Chong S, Mersha F B, Comb D G, Scott M E, Landry D, Vence L M, Perler F B, Benner J, Kucera R B, Hirvonen C A, Pelletier J J, Paulus H, Xu M Q. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 14.Clayton R A, White O, Fraser C M. Findings emerging from complete microbial genome sequences. Curr Opin Microbiol. 1998;1:562–571. doi: 10.1016/s1369-5274(98)80089-1. [DOI] [PubMed] [Google Scholar]
- 15.Crameri A, Raillard S A, Bermudez E, Stemmer W P. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391:288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 16.Fraenkel D G, Levisohn S R. Glucose and gluconate metabolism in an Escherichia colimutant lacking phosphoglucose isomerase. J Bacteriol. 1967;93:1571–1578. doi: 10.1128/jb.93.5.1571-1578.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grindley J F, Payton M A, van de Pol H, Hardy K G. Conversion of glucose to 2-keto-l-gulonate, an intermediate in l-ascorbate synthesis, by a recombinant strain of Erwinia citreus. Appl Environ Microbiol. 1988;54:1770–1775. doi: 10.1128/aem.54.7.1770-1775.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, K., H. van de Pol, J. Grindley, and M. A. Payton. July 1990. U.S. patent 4,945,052.
- 19.Jez J M, Bennett M J, Schlegel B P, Lewis M, Penning T M. Comparative anatomy of the aldo-keto reductase superfamily. Biochem J. 1997;326:625–636. doi: 10.1042/bj3260625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khurana S, Powers D B, Anderson S, Blaber M. Crystal structure of 2,5-diketo-d-gluconic acid reductase A complexed with NADPH at 2.1-Å resolution. Proc Natl Acad Sci USA. 1998;95:6768–6773. doi: 10.1073/pnas.95.12.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klasen R, Bringer Meyer S, Sahm H. Biochemical characterization and sequence analysis of the gluconate:NADP 5-oxidoreductase gene from Gluconobacter oxydans. J Bacteriol. 1995;177:2637–2643. doi: 10.1128/jb.177.10.2637-2643.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuchner O, Arnold F H. Directed evolution of enzyme catalysts. Trends Biotechnol. 1997;15:523–530. doi: 10.1016/S0167-7799(97)01138-4. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lazarus, R. A., M. Hurle, S. Anderson, and D. B. Powers. September 1992. U.S. patent 5,376,544.
- 25.MacBeath G, Kast P, Hilvert D. Redesigning enzyme topology by directed evolution. Science. 1998;279:1958–1961. doi: 10.1126/science.279.5358.1958. [DOI] [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 27.Miller J V, Estell D A, Lazarus R A. Purification and characterization of 2,5-diketo-d-gluconate reductase from Corynebacteriumsp. J Biol Chem. 1987;262:9016–9020. [PubMed] [Google Scholar]
- 28.Peekhaus N, Conway T. What’s for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J Bacteriol. 1998;180:3495–3502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powers, D. B., and S. Anderson. July 1997. Patent WO 97/25432.
- 30.Shinagawa E, Chiyonobu T, Matsushita K, Adachi O, Ameyama M. Distribution of gluconate dehydrogenase and ketogluconate reductases in aerobic bacteria. Agric Biol Chem. 1978;42:1055–1057. [Google Scholar]
- 31.Sonoyama T, Kageyama B, Yagi S. Distribution of microorganisms capable of reducing 2,5-diketo-d-gluconate to 2-keto-l-gulonate. Agric Biol Chem. 1987;51:2003–2004. [Google Scholar]
- 32.Sonoyama T, Kageyama B, Yagi S, Mitsushima K. Biochemical aspects of 2-keto-l-gulonate accumulation from 2,5-diketo-d-gluconate by Corynebacteriumsp. and its mutants. Agric Biol Chem. 1987;51:3039–3047. [Google Scholar]
- 33.Sonoyama T, Kageyama B, Yagi S, Mitsushima K. Facultatively anaerobic bacteria showing high productivities of 2,5-diketo-d-gluconate from d-glucose. Agric Biol Chem. 1988;52:667–674. [Google Scholar]
- 34.Sonoyama T, Kobayashi K. Purification and properties of two 2,5-diketo-d-gluconate-reductases from a mutant strain derived from Corynebacteriumsp. J Ferment Technol. 1987;65:311–317. [Google Scholar]
- 35.Truesdell S J, Sims J C, Boerman P A, Seymour J L, Lazarus R A. Pathways for metabolism of ketoaldonic acids in an Erwiniasp. J Bacteriol. 1991;173:6651–6656. doi: 10.1128/jb.173.21.6651-6656.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yum D-Y, Bae S-S, Pan J-G. Purification and characterization of the 2-ketoaldonate reductase from Brevibacterium ketosoreductumATCC 21914. Biosci Biotechnol Biochem. 1998;62:154–156. doi: 10.1271/bbb.62.154. [DOI] [PubMed] [Google Scholar]
- 37.Yum D-Y, Lee B-Y, Hahm D-H, Pan J-G. The yiaE gene, located at 80.1 minutes on the Escherichia colichromosome, encodes a 2-ketoaldonate reductase. J Bacteriol. 1998;180:5984–5988. doi: 10.1128/jb.180.22.5984-5988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yum D-Y, Lee Y-P, Pan J-G. Cloning and expression of a gene cluster encoding three subunits of membrane-bound gluconate dehydrogenase from Erwinia cypripedii ATCC 29267 in Escherichia coli. J Bacteriol. 1997;179:6566–6572. doi: 10.1128/jb.179.21.6566-6572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]