Abstract
A 300-bp repetitive element was found in the genome of the white button mushroom, Agaricus bisporus, and designated Abr1. It is present in ∼15 copies per haploid genome in the commercial strain Horst U1. Analysis of seven copies showed 89 to 97% sequence identity. The repeat has features typical of class II transposons (i.e., terminal inverted repeats, subterminal repeats, and a target site duplication of 7 bp). The latter shows a consensus sequence. When used as probe on Southern blots, Abr1 identifies relatively little variation within traditional and present-day commercial strains, indicating that most strains are identical or have a common origin. In contrast to these cultivars, high variation is found among field-collected strains. Furthermore, a remarkable difference in copy numbers of Abr1 was found between A. bisporus isolates with a secondarily homothallic life cycle and those with a heterothallic life cycle. Abr1 is a type II transposon not previously reported in basidiomycetes and appears to be useful for the identification of strains within the species A. bisporus.
Transposable elements, or transposons, are mobile genetic elements that can spread through genomes by integration in nonhomologous regions. They can be present in very high copy numbers in eukaryotic genomes, making up 10% of the Drosophila genome and more than 50% of some plant genomes (17). Transposons have also been found in filamentous fungi (33). Eukaryotic transposons are subdivided into two classes based on their method of transposition. Class I elements transpose via an RNA intermediate and usually code for a number of genes, one of which is always a reverse transcriptase (2). For the transposition of class II transposons, a single protein is needed (i.e., a transposase that is involved in processing the donor and target DNA via a “cut and paste” process) (34). Despite their nonreplicative method of transposition, class II transposons can increase in number by transposition from replicated to not yet replicated DNA or by gene conversion (34). Both types of transposons have been found in fungi (for recent reviews, see references 8 and 25).
Here, we describe the isolation, characterization, and distribution of a class II transposon in the genome of the white button mushroom, Agaricus bisporus (Lange) Imbach. This basidiomycetous fungus is the most cultivated mushroom in the world, accounting for approximately 75% of the global production of edible mushrooms (2 million metric tons) (14). Unfortunately its slow growth on artificial media, difficulties in obtaining and regenerating protoplasts, low frequency of spore germination (12), and the lack of an efficient transformation system (1) hardly make A. bisporus a basidiomycete of choice for laboratory studies. In addition to these drawbacks, all cultivars and most wild isolates have a typical secondarily homothallic life cycle (35), which makes breeding difficult. Most basidia produce two spores, and the four postmeiotic nuclei are divided in such a way that most spores receive two nonsister nuclei which, upon germination, yield fertile heterokaryotic mycelium (42). The low frequency of basidia producing three or four spores makes it difficult to select single-spore isolates that produce homokaryons that can be used for crossbreeding (26). Recently, however, a distinct variety has been found with a heterothallic life cycle (i.e., most basidia bear four spores) (3). Fortunately for A. bisporus researchers, during the last few years, genome mapping (27, 41, 48) and identifying genes and understanding their expression have progressed (10, 44, 46), and a usable transformation system appears to have been developed (9).
In an earlier study (41), one of the probes used for mapping showed a polymorphism that was caused by an insertion. In this study, we show that this insertion is a repetitive element with a length of ∼300 bp. Approximately 15 copies are present in both parental genomes of the commercially cultivated A. bisporus strain Horst U1. Our objectives were to determine (i) the nature of this repetitive element and (ii) its occurrence in cultivars and wild strains of A. bisporus.
MATERIALS AND METHODS
Strains and DNA manipulation.
The commercial strain A. bisporus Horst U1 and its parental strains, H39 and H97, were obtained from the culture collection of the Mushroom Experimental Station, Horst, The Netherlands. Wild strains of A. bisporus were obtained from the Agaricus Resource Program (ARP) (28). All strains were maintained at 4°C on slant tubes of wheat extract agar as previously described (18, 41). Monokaryotic mycelia from wild strains were obtained either by protoplasting, as described by Sonnenberg et al. (39), or as a single-spore isolate from lamellae distributed through the ARP collection. The homokaryotic nature was verified by establishing homozygosity for linked markers and mating with compatible homokaryons, followed by fruiting trials of the resulting heterokaryons (41). All strains used are listed in Table 1. Escherichia coli LE 392 (Promega, Madison, Wis.) was used for phage amplification and λ DNA isolation. E. coli DH5α (GIBCO BRL Life Technology, Gaithersburg Md.) was used for plasmid transformation and propagation.
TABLE 1.
Homokaryons used in this study
| No.a | Strain | Strain derived from |
|---|---|---|
| Secondarily homothallic life cycle | ||
| 1 | c24/4b | CB-3d |
| 2 | b69/6b | BP-6e |
| 3 | b67/11b | RWK1550f |
| 4 | b64/8b | CB-2d |
| 5 | c34/7b | FS-43g |
| 6 | b70/7b | FS-20g |
| 7 | b66/9b | MK-1h |
| 8 | c25/2b | FS-25i |
| 9 | c29/6b | FS-27i |
| 10 | b86/1b | CB-1d |
| 11 | c10/3b | FS-26i |
| 12 | c35/3b | FS-54g |
| Heterothallic life cycle | ||
| 14 | b55/9c | JB-2j |
| 15 | b51/3c | JB-3j |
| 16 | b123/1c | JB-17k |
| 17 | b132/1c | JB-26k |
| 18 | b116/8c | JB-10k |
| 19 | b124/1c | JB-18k |
| 20 | b129/1c | JB-23k |
| 21 | b131/2c | JB-25k |
| 22 | b138/1c | JB-32k |
| 23 | b140/8c | JB-34k |
| 24 | b142/2c | JB-36k |
| 25 | b144/2c | JB-38k |
| 26 | b162/2c | JB-174k |
| 27 | b145/8c | JB-39k |
| 28 | b161/1c | JB-173k |
Strain number, referred to in legend to Fig. 5.
Protoclone (homokaryon obtained from heterokaryon via protoplasting).
Single-spore isolate.
Collected in The Netherlands.
Collected in Santa Cruz, Calif.
Collected in Alberta, Canada.
Collected in San Mateo, Calif.
Collected in Ontario, Canada.
Collected in San Francisco, Calif.
Collected in Riverside, Calif.
Collected in the Sonoran Desert, Calif.
Standard DNA manipulations were carried out essentially as described previously (36). Restriction enzymes and other enzymes used for DNA manipulations were purchased from GIBCO BRL Life Technology and used according to the supplier’s instructions. Probes were labelled with digoxigenin by using the Dig DNA labelling kit (Boehringer Mannheim, Mannheim, Germany). Hybridization was carried out overnight at 65°C in a standard hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% lauroylsarcosine, 0.02% sodium dodecyl sulfate, 1% digoxigenin-blocking reagent). Detection of hybrids was carried out according to the conditions recommended in the Dig chemiluminescent detection kit (Boehringer Mannheim). DNA sequences were determined with a Thermo Sequenase fluorescent-labelled primer cycle sequencing kit with 7-deaza-dGTP (Amersham, Buckinghamshire, United Kingdom) and an ALF automated sequencer (Pharmacia Biotech, Uppsala, Sweden). Alternatively, sequences were determined commercially (BaseClear, Leiden, The Netherlands).
Cloning of Abr1.
Genomic clones containing copies of Abr1 were obtained by screening a λEMBL3 genomic library of A. bisporus H39 by standard methods, with Abr1.1 as a probe. Subcloning was performed in pUC19 (GIBCO BRL Life Technology). A copy of Abr1 in a cDNA clone was obtained by screening a previously constructed cDNA expression library (11) by standard methods, with Abr1.1 as a probe. PCR was performed in a total volume of 25 μl, containing 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 1.5 mM MgCl2, 0.4 mM deoxynucleoside triphosphates (dNTPs), 0.001% (wt/vol) gelatin, 0.3 U of Taq polymerase (SuperTaq; Sphaero Q, Leiden, The Netherlands), and 50 ng of genomic DNA. For amplification of the locus 1N150 (= Abr1.1), primers 1N150 forw (5′-CAA TCT CAA GCT TGC CTG G-3′) and 1N150 rev (5′-AGG TGA CAT GTC AGA AGC GC-3′) were used at a 0.6 μM concentration. For amplification of Abr1.2, primers Abr1.2 forw (5′-TTG TCC GAG ACT TAC TCA CG-3′) and Abr1.2 rev (5′-CCT CGC GCA AGC AGA TAC AA-3′) were used at a 0.4 μM concentration. Amplification was achieved in a program of 1 min at 94°C, 2 min at 58°C, and 2 min at 72°C for 31 cycles. In the last cycle, the extension at 72°C was performed for 5 min. PCR fragments were cloned by using the pGEM-T system (Promega).
Analysis of sequences.
Sequences were analyzed by using the program BLAST (22). Multiple sequence alignment was performed with the program CLUSTAL W (43).
Chromosome-size DNA preparations and pulsed-field electrophoretic separation of chromosomes.
The preparation of chromosome-size DNA and separation of intact chromosomes by using the CHEF-DRII contour-clamped homogeneous electric field (CHEF) system were done as described previously (41).
Segregation analysis.
The segregation of the Abr1 copies was analyzed as described in reference 41.
Nucleotide sequence accession number.
The nucleotide sequences (accession numbers given in parentheses) of Abr1.1 (Y18555), Abr1.2 (AJ238112), Abr1.3 (AJ238114), Abr1.4 (AJ238111), Abr1.6 (AJ238110), Abr1.7 (AJ238113), and Abr1.9 (AJ238115) have been deposited in the EMBL data bank.
RESULTS
Isolation of Abr1.
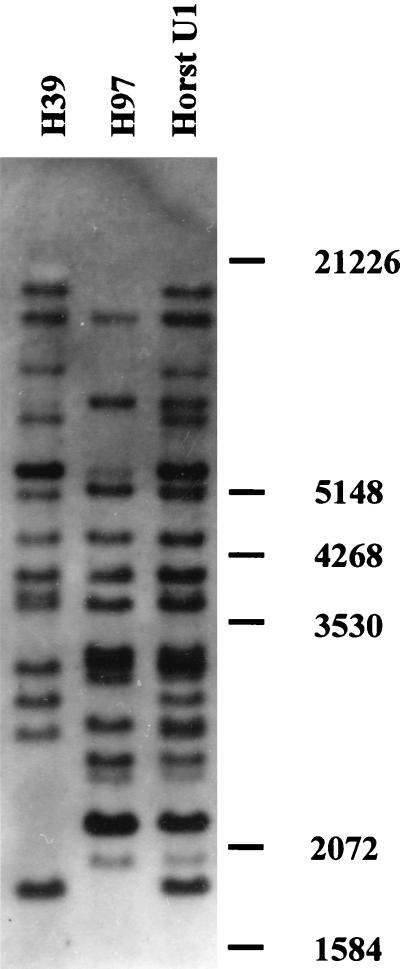
In previous studies, randomly cloned genomic EcoRI fragments were used as probes to construct a genomic map of A. bisporus (4, 27, 41). By Southern analysis, one 900-bp EcoRI fragment (probe p1n150) hybridized to a single band of EcoRI-digested genomic DNA in both parental lines of the commercial strain Horst U1. The length difference between the bands was ∼300 bp and may have been due to an insertion or a deletion. Sequence data from both ends of probe p1n150 were used to design primers that would amplify the major part of p1n150. The primers amplified an expected 900-bp fragment from the parental line H39 and a 1,200-bp fragment when genomic DNA of parental line H97 was used as a template. Both PCR products were cloned and used as probes in Southern analysis to confirm that the same region was amplified in both strains. The 900-bp fragment hybridized to a single band in both parental lines, with the band in H97 being 300 bp longer than the band in strain H39. When the 1,200-bp PCR product was used as a probe, however, numerous bands were seen in addition to the one expected. When sequenced, both products had similar sequences, except for a 313-bp insertion present in the 1,200-bp product. The insert was amplified by using primers for the sequences adjacent to it. When the cloned insert was used as a probe, 14 to 15 bands were seen in both parental strains on Southern blots of EcoRI-digested genomic DNA (Fig. 1), indicating that we had cloned a repetitive sequence present in similar copy numbers in both parental strains of Horst U1. The sequence was designated Abr1, for A. bisporus repeat 1.
FIG. 1.
Southern analysis of genomic DNA of strain Horst U1 and its parental lines, strains H39 and H97. EcoRI-digested DNA was separated on a 0.65% agarose gel, blotted onto a nylon membrane, and hybridized with Abr1. Molecular size markers (kilobases) are indicated on the right.
Additional copies and structural features of Abr1.
The first copy of Abr1 (Abr1.1) was used to screen a genomic library of strain H39. Subcloning and sequencing yielded five additional copies of Abr1. When Abr1.1 was used to screen a previously constructed cDNA derived from pinning fruit bodies (11), a partial cDNA of 670 bp was isolated that contained a complete copy of Abr1. The copy is located 233 bp upstream of the poly(A) tail. Primers flanking the Abr1 copy in the cDNA failed to amplify genomic DNA, indicating that one or both primers span an intron. Reverse transcription-PCRs (RT-PCRs) were done with the same primers with total RNA isolated from vegetative mycelium of the homokaryons H39 and H97 and the heterokaryon Horst U1 and from immature and mature fruit bodies of Horst U1. In each case, a single band of the expected length was obtained that hybridized to Abr1 and to a probe specific for the 233-bp sequence between Abr1 and the poly(A) tail (results not shown). This indicates that both alleles in strain Horst U1 contain the Abr1 insert and that both genes are transcribed. The inserted Abr1 copy contains several stop codons in each of the possible reading frames. If the element resides in the coding region of the gene, this might lead to a truncated protein.
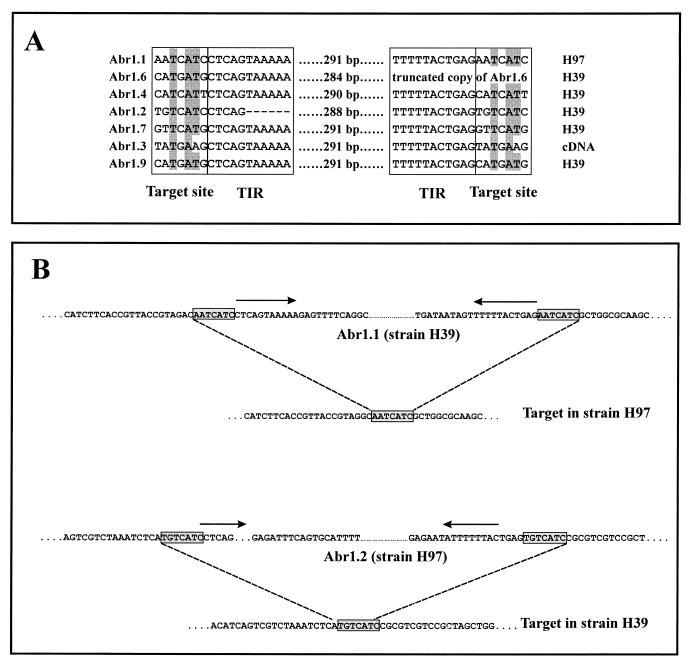
The sequences of the seven copies are highly conserved (89 to 97% identity when the deletions are not taken into account). No open reading frame of a significant length was found, and a database search revealed no similarity to known sequences. The aligned sequences (Fig. 2A) show that Abr1 has structural features of an inverted repeat transposable element (38, 49). Most copies have a terminal inverted repeat (TIR) of 10 bp flanked by a 7-bp direct repeat. Some copies have a deletion. Abr1.6 is missing the 3′ end including the right TIR, while Abr1.2 is lacking part of the left TIR. Within the sequence, several direct and inverted repeats were identified (data not shown). None of the sequenced copies of Abr1 contains an EcoRI site, which means that the number of bands seen on a Southern blot containing EcoRI-digested genomic DNA reflects the minimum number of Abr1 copies present in the genome.
FIG. 2.
Alignment of the borders of seven copies of Abr1. (A) The source of each copy is indicated at the end of each line. The target site duplications and the TIRs are indicated by boxes. The alignment of target sites of all of the Abr1 copies shows a clear conservation in sequence (shaded bases). Primers flanking Abr1.1 in strain H97 and Abr1.2 in strain H39 were used to amplify genomic DNA of both strains H39 and H97. Sequences of the amplified fragments (B) allowed the identification of the target site (boxed sequences) in the strain lacking the corresponding fragments of Abr1. The TIRs are indicated by arrows.
Chromosomal locations and target sites.
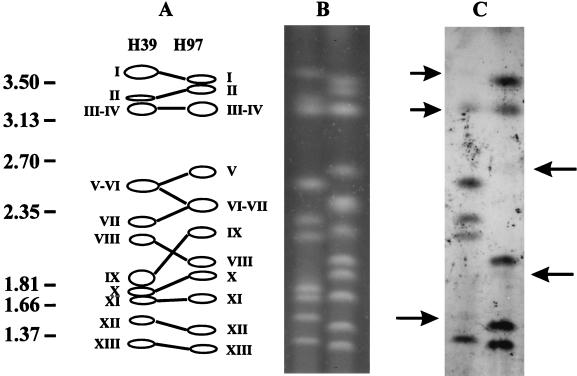
When Abr1 was used as a probe on CHEF-separated chromosomes, hybridization signals were obtained for several chromosomes of both parental strains H39 and H97 (Fig. 3). This hybridization pattern indicates that Abr1 is dispersed throughout the genome of A. bisporus. However, not all chromosomes hybridizing to Abr1 are identical for strains H39 and H97. The doublet containing chromosomes VI and VII in strain H97 does not hybridize to Abr1. Chromosome VII of strain H39, however, shows a clear hybridization signal. In addition, chromosome XII of strain H39 gives a weak hybridization signal, while chromosome XII of strain H97 shows a strong signal. Segregation analysis of a previously isolated set of offspring of strain Horst U1 (41) confirmed the results obtained with the CHEF analysis. When used as probe on a Southern blot containing EcoRI-digested genomic DNA, most bands of Abr1 mapped to different positions (not shown). Two of the four bands that have identical lengths in strain H39 and H97 were missing in some of the offspring of Horst U1. This result indicates that these bands of similar sizes represent different loci. For the two remaining bands with similar lengths, no segregation was found. These results suggest that the genomic locations of most of the copies of Abr1 differ for both strains. The segregation analysis also indicates that the differences in strength of the hybridization signals on different chromosomes are due to variation in Abr1 copy number on individual chromosomes. Transposable elements may duplicate their target sites after insertion, resulting in direct repeats adjacent to the inverted repeats of the element. One target site in strain H97 and the corresponding site in strain H39 with the integrated Abr1.1 copy have been sequenced (as the insertion in probe p1n150). The sequence data obtained after isolation of the Abr1.2 copy were used to amplify an additional genomic region in strain H97 homologous to the region that contains the Abr1.2 copy in strain H39. The fragment amplified with H97 DNA as template was, as expected, ∼300 bp smaller (data not shown). When the two sites containing Abr1 and their corresponding regions in the other parental strain missing the insert were compared, the site of integration could clearly be identified as the 7-bp sequence identical to the direct repeats flanking Abr1 (Fig. 2B). This strongly suggests that Abr1 originated from a transposition event. The seven target sites clearly show some sequence conservation (Fig. 2).
FIG. 3.
Analyses of chromosomal positions of copies of Abr1. Chromosomes of strains H39 and H97 were separated by CHEF (B) and blotted onto a nylon membrane and hybridized to Abr1 (C). Chromosomes that hybridized weakly are indicated by an arrow. In panel A, homologous chromosomes of H39 and H97 are indicated with the molecular size markers on the left (megabases).
Occurrence of Abr1 in commercial strains and wild strains collected in the field.
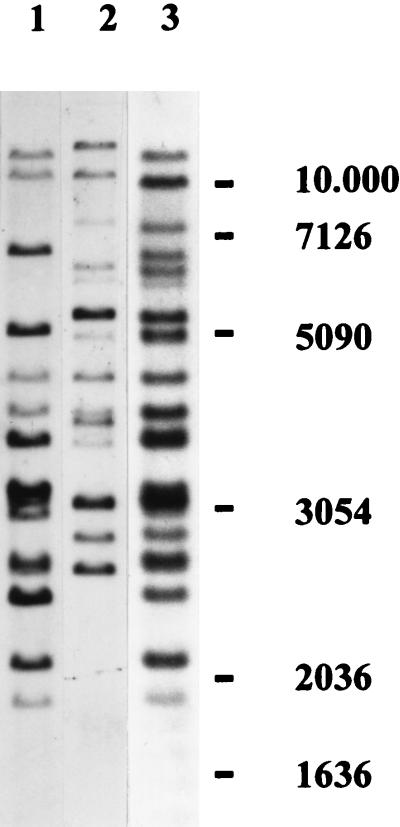
We examined eight traditional cultivars (in use mainly before 1980) and nine hybrid cultivars (that appeared after 1980) for their hybridization patterns on EcoRI-digested genomic DNA. The polymorphisms found in these lines were of three types (Fig. 4). The traditional cultivars could be separated into two distinct groups, type I and type II, with no variation in banding pattern within a group. This classification exactly coincides with the two types of strains that were used before the first hybrids were introduced (19), i.e., the “white” and the “off-white” strains (20). All present-day commercial hybrids were identical and showed a third type of hybridization pattern.
FIG. 4.
Southern analyses of EcoRI-digested genomic DNA of traditional and present-day commercial strains of A. bisporus hybridized to Abr1. The hybridization patterns were of three types, corresponding to different cultivar types. Lanes: 1, off-white traditional cultivars (Somycel 9.2, Somycel 76, Sinden A4, and Le Lion B62); 2, white traditional cultivars (Somycel 53, Sinden A1, Le Lion B14, and Les Miz 66); 3, present-day hybrids (Horst U1, Horst U3, Amycel 2800, Le Lion X8, Int. Spawn 643, Sylvan 100, Sylvan 608, Le Lion X20, Le Lion X22, and Sylvan 512). The molecular size markers are indicated on the right (kilobases). Homokaryotic strains H97 (single-spore isolate from the traditional off-white line Somycel 9.2) and H39 (single-spore isolate from the traditional white line Somycel 53), which were used to construct strain Horst U1, showed hybridization patterns as depicted in lanes 1 and 2, respectively.
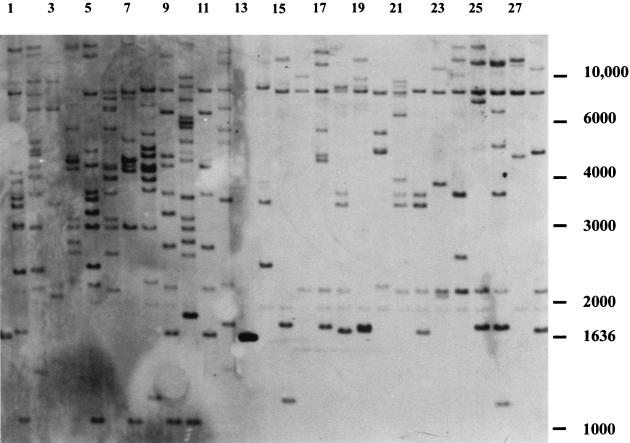
Twenty-seven homokaryons derived from field-collected lines present in the ARP collection (Table 1) were subjected to Southern analysis. All of these strains showed different banding patterns, indicating a high variability in genotypes. A clear difference in banding patterns was observed between strains with a secondary homothallic life cycle and those with a heterothallic mode of reproduction. To make the difference even clearer, both types of strains were grouped (Fig. 5). The number of bands in the two-spored varieties is approximately twice that in the four-spored varieties (12.6 and 6.6, respectively).
FIG. 5.
Southern analyses of EcoRI-digested genomic DNA of a number of wild varieties of A. bisporus hybridized to Abr1. Lanes: 1 to 12, A. bisporus var. bisporus (two-spored variety) corresponding to strain numbers 1 to 12, respectively, in Table 1; 14 to 28, A. bisporus var. burnettii (four-spored variety) corresponding to strain numbers 14 to 28 in Table 1. Lane 13 contains molecular size markers, one of which hybridizes to Abr1. The other bands of the molecular size markers are indicated to the right (kilobases).
DISCUSSION
Class II transposable elements transpose via a DNA intermediate. Most class II transposons found in fungi belong to the Fot1/Pogo, Tc1/Mariner, or hAT family (25). The relatively short TIRs, a target site duplication of 7 bp, and the presence of subterminal repeats indicate that Abr1 is likely a member of the hAT group (45), which has not been observed before in basidiomycetes. Furthermore, there is only one previous report of a class II transposon in basidiomycetes (21).
Its short length and lack of substantial coding regions suggest that Abr1 itself does not code for a transposase but depends for its mobility on a trans-acting transposase located at another chromosomal position. Well-known examples of such dependency are the Ds elements in the Ac/Ds system found in maize (15) and inactive P elements in Drosophila melanogaster (13). Typically, nonautonomous class II transposons are deletion derivatives of the autonomous element and thus form a heterogeneous collection with respect to their lengths. The seven isolated copies of Abr1 are almost identical in length, and the sequences are very similar, suggesting that the copies of Abr1 found in the genome of A. bisporus resulted from transposition of Abr1 as an entity. By using primers for PCR in the 5′ and 3′ regions of Abr1, no large fragments were obtained that could code for an active transposase. The absence of transposase activity is also supported by the fact that most Abr1 copies show a normal 1:1 segregation in offspring of strain Horst U1 (data not shown). The few copies that do not segregate 1:1 are located in genomic regions that have skewed segregation for other markers as well (data not shown).
The duplicated target site of Abr1 is conserved. This conservation might indicate that the transposase involved in the mobility of Abr1 interacts preferentially with particular sequences, as is found for bacteriophage Mu (32) and the bacterial transposon Tn10 (31). No conservation in the target site is known for previously isolated members of the hAT group of class II transposable elements (6, 34). The sequences surrounding the Abr1 copies show no obvious similarity (data not shown).
One Abr1 copy was found within a transcribed region of an unknown gene. Except from transposons isolated by gene tagging (7, 23), there is only one previous report of an insertion of a transposable element within a gene (21). In Phanerochaete chrysosporium, one allele of the lignin peroxidase gene, lipI1, contains a transposon-like element immediately adjacent to the fourth intron. This insertion leads to the inactivation of transcription of the gene. In A. bisporus, RT-PCR and hybridization experiments have shown that both constituent nuclei of strain Horst U1 contain Abr1 within a single-copy gene of unknown function. The RT-PCR experiments also show that the gene is constitutively expressed in both homokaryons, the heterokaryon, and fruit bodies. Since the inserted Abr1 copy contains several stop codons, a truncated protein might be produced. Since both alleles contain the insertion, either this truncation does not lead to an inactive product, or the gene has no essential function.
Within eight traditional cultivars, two different Abr1 genotypes were seen. These genotypes coincide with the phenotypes of traditional cultivar types used before 1980, the white and the off-white strains (20). A comparison of mitochondrial genotypes of the traditional cultivars has led in a previous study (40) to the same conclusion. This means that many strains marketed under different names are genetically very similar. The uniform banding pattern seen in hybrids when Abr1 is used as a probe indicates a similar situation for the present-day cultivars. The origin of most commercial hybrids is a well-kept secret, but the construction of Horst U1 is well documented (19, 24). Horst U1 was obtained by mating the infertile single-spore-derived culture H39 obtained from the white strain Somycel 53 with H97 obtained from the off-white strain Somycel 9.2. After the release of Horst U1, other hybrids appeared within a few months. The time of release of these “new” hybrids suggests that they are either copies or selections of fertile single-spore isolates derived from the first hybrids. In the latter case, an unchanged genotype is not surprising, since the typical life cycle of A. bisporus tends to maintain parental heterozygosity in the offspring (42).
Hybridization of Abr1 to separated chromosomes and segregation analysis have both shown that the genomic locations of most copies in the parental lines of strain Horst U1 are different. Since the parental lines are derived from Somycel 53 and Somycel 9.2, this conclusion also extends to the two types of traditional cultivars. The most plausible explanation for the different genomic positions is that the transposons spread in each strain independently. When the banding patterns of the traditional cultivars and the present-day hybrids are compared, no bands of new sizes are found, indicating that there have been no recent transpositions of Abr1. This result suggests that the two types of cultivars are not closely related. When combined with data on restriction fragment polymorphisms (41), the previous suggestions that all traditional white-colored cultivars of A. bisporus in the world (which include the white and off-white cultivars) are derived from the famous clump of “snow white” mushrooms that occurred on a bed of cream mushrooms in 1926 (30, 37) are most unlikely.
The uniformity of the Abr1 patterns found in the commercial strains contrasts with the high variability found in wild populations collected in the ARP (28). The large differences seen in banding patterns when Abr1 was used as a probe suggest that there is a high variation in genomic position and in copy number among the different isolates. The difference in the copy number of Abr1 in the bisporic and tetrasporic varieties is striking. The latter type is found in the Sonoran Desert in California, and a study of mitochondrial DNA variation has shown that this tetrasporic variety is genetically very distinct from commercial cultivars and wild bisporic isolates (47). Our findings support the hypothesis that the separation of these two morphological types may be ancient. If the banding patterns of Abr1 that we have identified in traditional cultivars and present-day hybrids are representative for all cultivars used in the last decades, Abr1 might be a powerful tool to determine the extent to which cultivar types have invaded natural populations that seem to be at risk of extinction (29).
REFERENCES
- 1.Binz T, D’Mello N, Horgen P A. A comparison of DNA methylation levels in selected isolates of higher fungi. Mycologia. 1998;90:785–790. [Google Scholar]
- 2.Boeke J D, Corces V G. Transcription and reverse transcription in retrotransposons. Annu Rev Microbiol. 1989;43:403–433. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- 3.Callac P, Billette C, Imbernon M, Kerrigan R W. Morphological, genetic, and interfertility analyses reveal a novel, tetrasporic variety of Agaricus bisporus from the Sonoran desert of California. Mycologia. 1993;85:835–851. [Google Scholar]
- 4.Callac P, Desmerger C, Kerrigan R W, Imbernon M. Conservation of genetic linkage with map expansion in distantly related crosses of Agaricus bisporus. FEMS Microbiol Lett. 1997;146:235–240. doi: 10.1111/j.1574-6968.1997.tb10199.x. [DOI] [PubMed] [Google Scholar]
- 5.Colot V, Haedens V, Rossignol J-L. Extensive, nonrandom diversity of excision footprints generated by Ds-like transposon Ascot-1 suggests new parallels with V(D)J recombination. Mol Cell Biol. 1998;18:4337–4346. doi: 10.1128/mcb.18.7.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig N L. Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 7.Daboussi M J, Langin T, Brygoo Y. Fot1, a new family of fungal transposable elements. Mol Gen Genet. 1992;232:12–16. doi: 10.1007/BF00299131. [DOI] [PubMed] [Google Scholar]
- 8.Daboussi M J, Capy P. Fungal transposable elements and genome evolution. Genetica. 1997;100:253–260. [PubMed] [Google Scholar]
- 9.De Groot M J A, Bundock P, Hooykaas P J J, Beijersbergen A G M. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16:839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- 10.De Groot P W J. Biochemical and molecular aspects of growth and fruiting of the edible mushroom Agaricus bisporus. Mycol Res. 1999;102:1297–1308. [Google Scholar]
- 11.De Groot P W J, Schaap P J, Sonnenberg A S M, Visser J, Van Griensven L J L D. The Agaricus bisporus hyp A gene encodes a hydrophobin and specifically accumulates in peel tissue of mushroom caps during fruit body development. J Mol Biol. 1996;257:1008–1018. doi: 10.1006/jmbi.1996.0219. [DOI] [PubMed] [Google Scholar]
- 12.Elliott T J. The general biology of mushrooms. In: Flegg P B, Spencer D M, Wood D A, editors. The biology and technology of the cultivated mushroom. New York, N.Y: John Wiley and Sons; 1985. pp. 9–22. [Google Scholar]
- 13.Engels W R. P elements in Drosophila melanogaster. Nature. 1989;331:368–370. [Google Scholar]
- 14.FAOSTAT. 1998. Agricultural database. [Online.] http://apps.fao.org/default.htm. [Update November 1998.] [June 1999, last date accessed.]
- 15.Federoff N V, Wessler S, Shure M. Isolation of the transposable maize controlling elements Ac and Ds. Cell. 1983;35:235–242. doi: 10.1016/0092-8674(83)90226-x. [DOI] [PubMed] [Google Scholar]
- 16.Finnegan D. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- 17.Freeling M. Plant transposable elements and insertion sequences. Annu Rev Plant Physiol. 1984;35:277–298. [Google Scholar]
- 18.Fritsche G. Cultivated fungi in Agaricales. In: Chang S T, Hayes W A, editors. The biology and cultivation of edible mushrooms. New York, N.Y: Academic Press; 1978. pp. 239–250. [Google Scholar]
- 19.Fritsche G. Breeding Agaricus bisporus at the Mushroom Experimental Station, Horst. Mushroom J. 1983;122:49–53. [Google Scholar]
- 20.Fritsche G, Sonnenberg A S M. Mushroom strains. In: Van Griensven L J L D, editor. The cultivation of mushrooms. Darlington, United Kingdom: Rustington; 1988. pp. 101–123. [Google Scholar]
- 21.Gaskell J, Van den Wymelenberg A, Cullen D. Structure, inheritance, and transcriptional effects of Pce1, an insertional element within Phanerochaete chrysosporium lignin peroxidase gene lipl. Proc Natl Acad Sci USA. 1995;92:7465–7469. doi: 10.1073/pnas.92.16.7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 23.Glayzer D C, Roberts I N, Archer D B, Oliver R P. The isolation of Ant1, a transposable element from Aspergillus niger. Mol Gen Genet. 1995;249:432–438. doi: 10.1007/BF00287105. [DOI] [PubMed] [Google Scholar]
- 24.Imbernon M, Callac P, Gasqui P, Kerrigan R W, Velco A J. BSN, the primary determinant of basidial spore number and reproductive mode in Agaricus bisporus, maps to chromosome I. Mycologia. 1996;88:749–761. [Google Scholar]
- 25.Kempken F, Kück U. Transposons in filamentous fungi—facts and perspectives. BioEssays. 1998;20:652–659. doi: 10.1002/(SICI)1521-1878(199808)20:8<652::AID-BIES8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 26.Kerrigan R W, Baller L M, Horgen P A, Anderson J B. Strategies for the efficient recovery of Agaricus bisporus homokaryons. Mycologia. 1992;84:575–579. [Google Scholar]
- 27.Kerrigan R W, Royer J C, Baller L M, Kohli Y, Horgen P A, Anderson J B. Meiotic behavior and linkage relationships in the secondarily homothallic fungus Agaricus bisporus. Genetics. 1993;133:225–236. doi: 10.1093/genetics/133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerrigan R W. Characteristics of a large collection of wild edible mushroom germ plasm: the Agaricus resource program. In: Samson R A, Stalper J A, van der Mei D, Stouthamer A H, editors. Culture collections to improve the quality of life. Baarn, The Netherlands: Centraal Bureau voor Schimmelcultures; 1996. pp. 302–308. [Google Scholar]
- 29.Kerrigan R W. The indigenous coastal Californian population of the mushroom Agaricus bisporus, a cultivated species, may be at risk of extinction. Mol Evol. 1998;7:35–45. [Google Scholar]
- 30.Lambert E B. Improving spawn culture of cultivated mushrooms. Mushroom Sci. 1959;4:33–51. [Google Scholar]
- 31.Lee S, Butler D, Kleckner N. Efficient Tn10 transposition into a DNA insertion hot spot in vivo requires the 5-methyl groups of symmetrically disposed thymines within the hot-spot consensus sequence. Proc Natl Acad Sci USA. 1987;84:7876–7880. doi: 10.1073/pnas.84.22.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizuuchi M, Mizuuchi K. Target site selection in transposition of phage mu. Cold Spring Harbor Symp Quant Biol. 1993;58:515–523. doi: 10.1101/sqb.1993.058.01.058. [DOI] [PubMed] [Google Scholar]
- 33.Oliver R P. Transposons in filamentous fungi. In: Stahl U, Tudzynski P, editors. Molecular biology of filamentous fungi. Proceedings of the EMBO-Workshop, Berlin 1991. 1992. pp. 3–11. [Google Scholar]
- 34.Plasterk H A. Mechanisms of DNA transposition. In: Sherratt D J, editor. Mobile genetic elements. Oxford, United Kingdom: IRL Press; 1995. [Google Scholar]
- 35.Raper C A, Raper J R, Miller R E. Genetic analysis of the life cycle of Agaricus bisporus. Mycologia. 1972;64:1088–1117. [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.San Antonio J P. Origin and improvement of spawn of the cultivated mushroom Agaricus brunnescens Peck. Hortic Rev. 1984;6:85–118. [Google Scholar]
- 38.Smit A F A, Riggs A D. Tiggers and other DNA transposon fossils in the human genome. Proc Natl Acad Sci USA. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonnenberg A S M, Wessels J G H, Van Griensven L J L D. An efficient protoplasting/regeneration system for Agaricus bisporus and Agaricus bitorquis. Curr Microbiol. 1988;17:285–291. [Google Scholar]
- 40.Sonnenberg A S M, Van Loon P C C, Van Griensven L J L D. Mitochondrial genotypes and their inheritance in the cultivated mushroom Agaricus bisporus. In: Van Griensven L J L D, editor. Genetics and breeding of Agaricus. Wageningen, The Netherlands: Pudoc; 1991. pp. 42–51. [Google Scholar]
- 41.Sonnenberg A S M, de Groot P W J, Schaap P J, Baars J J P, Visser J, Van Griensven L J L D. Isolation of expressed sequence tags of Agaricus bisporus and their assignment to chromosomes. Appl Environ Microbiol. 1996;62:4542–4547. doi: 10.1128/aem.62.12.4542-4547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Summerbell R C, Castle A J, Horgen P A, Anderson J B. Inheritance of restriction fragment length polymorphisms. Genetics. 1989;123:293–300. doi: 10.1093/genetics/123.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thurston C F. The structure and function of fungal laccases. Microbiology. 1994;140:19–26. [Google Scholar]
- 45.Warren W D, Atkinson P W, O’Brochta D A. The Hermes transposable element from the house fly, Musca domestica, is a short inverted repeat-type element of the hobo, Ac, and Tam3 (hAT) element family. Genet Res Cambr. 1994;64:87–97. doi: 10.1017/s0016672300032699. [DOI] [PubMed] [Google Scholar]
- 46.Wood D A, Perry C, Thurston C F, Matcham S E, Dudley K, Claydon N, Allan M. Molecular analysis of lignocellulolytic enzymes of the edible mushroom Agaricus bisporus. In: Kirk T K, Chang H-M, editors. Biotechnology in pulp and paper manufacture. Proceedings of the 4th International Conference on Biotechnology in the Pulp and Paper Industry. Boston, Mass: Butterworth-Heinemann; 1990. pp. 659–666. [Google Scholar]
- 47.Xu J, Kerrigan R W, Sonnenberg A S M, Callac P, Horgen P A, Anderson J B. Mitochondrial DNA variation in natural populations of the mushroom Agaricus bisporus. Mol Evol. 1998;7:19–33. [Google Scholar]
- 48.Xu J P, Kerrigan R W, Horgen P A, Anderson J B. Localization of the mating type gene in Agaricus bisporus. Appl Environ Microbiol. 1993;59:3044–3049. doi: 10.1128/aem.59.9.3044-3049.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeadon P J, Catcheside D E A. Guest, a 98 bp inverted repeat transposable element in Neurospora crassa. Mol Gen Genet. 1995;247:105–109. doi: 10.1007/BF00425826. [DOI] [PubMed] [Google Scholar]