Abstract
Pseudomonas pseudoalcaligenes POB310(pPOB) and Pseudomonas sp. strains B13-D5(pD30.9) and B13-ST1(pPOB) were introduced into soil microcosms containing 3-phenoxybenzoic acid (3-POB) in order to evaluate and compare bacterial survival, degradation of 3-POB, and transfer of plasmids to a recipient bacterium. Strain POB310 was isolated for its ability to use 3-POB as a growth substrate; degradation is initiated by POB-dioxygenase, an enzyme encoded on pPOB. Strain B13-D5 contains pD30.9, a cloning vector harboring the genes encoding POB-dioxygenase; strain B13-ST1 contains pPOB. Degradation of 3-POB in soil by strain POB310 was incomplete, and bacterial densities decreased even under the most favorable conditions (100 ppm of 3-POB, supplementation with P and N, and soil water-holding capacity of 90%). Strains B13-D5 and B13-ST1 degraded 3-POB (10 to 100 ppm) to concentrations of <50 ppb with concomitant increases in density from 106 to 108 CFU/g (dry weight) of soil. Thus, in contrast to strain POB310, the modified strains had the following two features that are important for in situ bioremediation: survival in soil and growth concurrent with removal of an environmental contaminant. Strains B13-D5 and B13-ST1 also completely degraded 3-POB when the inoculum was only 30 CFU/g (dry weight) of soil. This suggests that in situ bioremediation may be effected, in some cases, with low densities of introduced bacteria. In pure culture, transfer of pPOB from strains POB310 and B13-ST1 to Pseudomonas sp. strain B13 occurred at frequencies of 5 × 10−7 and 10−1 transconjugant per donor, respectively. Transfer of pPOB from strain B13-ST1 to strain B13 was observed in autoclaved soil but not in nonautoclaved soil; formation of transconjugant bacteria was more rapid in soil containing clay and organic matter than in sandy soil. Transfer of pPOB from strain POB310 to strain B13 in soil was never observed.
Bioremediation has become an accepted technology for restoration of contaminated environments. However, successful applications have primarily involved readily degradable organic compounds (4, 7, 45). Bioremediation is used infrequently with more recalcitrant pollutants, often because microorganisms indigenous to contaminated environments lack appropriate degradative capabilities (34, 52). In these cases, it may be possible to enhance bioremediation by adding microorganisms that have appropriate catabolic functions, a process referred to as bioaugmentation.
Attempts to demonstrate the potential for bioaugmentation in soils have resulted in successes and failures (52). For example, soil contaminated with pentachlorophenol was augmented with Flavobacterium (11), Arthrobacter (17), and Rhodococcus chlorophenolicus (5, 6) strains. In each case removal of pentachlorophenol was accelerated. Soil contaminated with 2,4,5-trichlorophenoxyacetic acid was augmented with Pseudomonas cepacia, and this resulted in a 95% reduction in the concentration of the herbicide (8). Application of Pseudomonas stutzeri and Pseudomonas aeruginosa to soil containing parathion resulted in complete degradation of the compound (3). In contrast, addition of microorganisms to soils contaminated with oil (33, 51) and coal tar (1) did not result in increases in the rates of contaminant removal. It is apparent that for bioaugmentation to be successful, the environmental conditions that control the survival and activity of introduced microorganisms need to be identified and properly managed.
In this study, three bacteria that degrade 3-phenoxybenzoic acid (3-POB) were added to soil microcosms containing 3-POB. 3-POB and its chlorinated analogs are metabolic products that are formed during degradation of pyrethroid insecticides (31, 46). They resemble diphenyl ether-based herbicides and can serve as model compounds for these biodegradatively recalcitrant chemicals (36). Pseudomonas pseudoalcaligenes POB310 was isolated for its ability to use 3-POB as a growth substrate. The initial catabolic step is an angular dioxygenation (18) catalyzed by POB-dioxygenase, which is encoded on a 40-kb, self-transmissible plasmid, pPOB (12). POB-dioxygenase also transforms certain mono- and dichlorinated analogs of 3-POB (23). However, transformation of the chlorinated analogs is unproductive and results in chlorophenols that are not further degraded. In response to this observation, the genes encoding POB-dioxygenase were transferred into a chlorophenol-degrading bacterium, Pseudomonas sp. strain B13 (15), which yielded two strains, Pseudomonas sp. strain B13-D5(pD30.9), which contains a constructed, non-self-transmissible plasmid (12), and Pseudomonas sp. strain B13-ST1(pPOB), which contains the original plasmid from strain POB310. The three bacteria were compared under different soil conditions with respect to survival, efficacy of removal of 3-POB, and transfer of plasmids containing POB-dioxygenase to a recipient bacterium.
MATERIALS AND METHODS
Chemicals.
The chemicals and antibiotics used were of the highest purity available from Aldrich Chemical Co. (Milwaukee, Wis.) and Sigma Chemical Co. (St. Louis, Mo.).
Bacteria and growth conditions.
A spontaneous, ampicillin-resistant strain of P. pseudoalcaligenes POB310(pPOB) (referred to below as strain POB310) was used in this study. Genes encoding POB-dioxygenase were cloned in Escherichia coli DH5α as a 4.5-kb fragment in pDSK519, which yielded pD30.9 (12). Pseudomonas sp. strain B13-D5(pD30.9) (referred to below as strain B13-D5) was formed by mobilization of pD30.9 into spontaneous nalidixic acid- and streptomycin-resistant Pseudomonas sp. strain B13 (referred to below as strain B13) by using E. coli HB101(pRK600) as the helper. Pseudomonas sp. strain B13-ST1(pPOB) (referred to below as strain B13-ST1) was formed by conjugative transfer of pPOB into spontaneous, rifampin-resistant strain B13. Bacteria were grown in M9 minimal medium (29) supplemented with trace elements (53) and 3-POB (5 mM). P. stutzeri(pRP4-4) was used to mobilize pD30.9 in some plasmid transfer experiments and was grown in medium containing benzoate (5 mM) plus tetracycline and ampicillin. Liquid cultures were shaken (200 rpm). Solid medium contained agar (15 g/liter) plus the following antibiotics when it was appropriate: tetracycline (30 μg/ml), ampicillin (50 μg/ml), nalidixic acid (50 μg/ml), streptomycin (50 μg/ml), and rifampin (100 μg/ml). All cultures were grown at 30°C in the dark.
The growth kinetics of strains POB310 and B13-D5 were determined in cultures (200 ml, 200 rpm, 21°C) containing 3-POB (100 ppm). Samples were removed periodically to assess the concentration of 3-POB and the biomass (dry weight), as calculated from the absorbance (optical density at 560 nm) of washed cells; 1 U of absorbance was equal to 299 ± 16 mg of strain POB310 per liter or 306 ± 9 mg of strain B13-D5 per liter. Values for maximum growth rates (μmax), affinity constants (Ks), and yield coefficients (Y) were obtained by fitting data to the Monod model by using nonlinear regression analysis and assuming that endogenous decay was negligible (kd = 0.01 h−1).
Soils.
Soils were obtained in the spring from the Cedar Creek Natural History Area (pale-brown, sandy Zimmermann soil; B21 horizon; depth, 18 to 38 cm) (20) and Fort Snelling State Park, both in Minnesota. The Cedar Creek soil contained (on a dry weight basis) 95% sand, 3% clay, and 2% silt; the organic matter content was 0.5%, and the total organic carbon content was 0.26%. The Fort Snelling soil contained 60% sand, 18% clay, and 22% silt; the organic matter content was 5.4%, and the total organic carbon content was 3.4%. The soils were sieved (2-mm mesh), stored at 4°C, and, when necessary, air dried and autoclaved twice (1 h, 145°C, 15 lb/in2). For some experiments the soil was stored for a maximum of 1 year. In some experiments, the Cedar Creek soil (containing 28.5 ppm of Bray phosphorus and 0.02% total nitrogen) was amended with the following nutrients: P-PO33− (300 mg/kg [dry weight] of soil [dws]) and N-NH4 (35 mg/kg [dws]).
Properties of 3-POB.
The solubility of 3-POB was determined by depositing a layer of the compound on the interior walls of 4-ml high-pressure liquid chromatography (HPLC) vials (nine replicates) via evaporation of methanolic solutions. The vials were filled with M9 buffer (pH 7.0), sealed with Teflon closures, and allowed to equilibrate for 3 days with agitation at 100 rpm. The aqueous concentrations of 3-POB were then determined by HPLC. To determine partitioning of 3-POB to soil, 2-g portions of Cedar Creek soil were placed into HPLC vials and dried at 105°C for 24 h, and 3-POB in methanol (20 μl) was added to obtain final concentrations ranging from 1 to 20 ppm (dws) (in triplicate). The methanol was evaporated, and 2 ml of water containing NaN3 (10 g/kg) was added to each vial. After equilibration for 3 days with agitation at 100 rpm, particles were allowed to settle out (24 h); 1-ml aliquots of the supernatants were transferred to microcentrifuge tubes containing 0.5 ml of acidified acetonitrile (2% H3PO4). Solids were removed by centrifugation (10,000 × g, 10 min), and the concentrations of 3-POB were determined by HPLC. Controls contained no soil. Octanol-water partition coefficients (KOW) for 3-POB were estimated by the fragment method (24) by starting with an average KOW of 1.39 × 104 for diphenyl ether (2, 9).
Soil microcosms.
Soil microcosms consisted of 100-ml serum bottles containing 20 g of soil to which 3-POB was added 24 h before bacteria were added. Bacteria were grown to the mid-log phase, collected by centrifugation (5,000 × g, 20 min), washed in M9 buffer, and mixed into the microcosm soil; the liquid volumes were adjusted so that the level was 75% of the water-holding capacity, a level that allowed microbial degradative activity to occur (see Fig. 7). Control microcosms contained autoclaved soil plus NaN3 (10 g/kg). The microcosms were covered with Parafilm and incubated in the dark at 21°C. One-half of the contents of each microcosm was used to enumerate bacteria, and the other half was used to measure the concentrations of 3-POB. The data presented below are averages of values from triplicate microcosms.
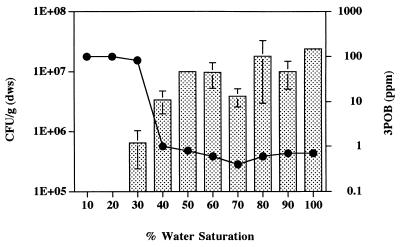
FIG. 7.
Cedar Creek soil samples with moisture contents ranging from 10 to 100% of the water-holding capacity were amended with 3-POB (100 ppm) and inocluated with strain B13-D5 (106 CFU/g [dws]). After 17 days, the densities of strain B13-D5 (bars) and the concentrations of 3-POB (●) were assessed. Error bars indicate standard errors.
Conjugative transfer of plasmids in soil.
Microcosms were established as described above with bacteria [plasmid donor, plasmid recipient, and P. stutzeri(pRP4-4)] that were mixed separately into the soil. Control microcosms contained either donor or recipient strains or no bacteria.
Enumeration of bacteria.
Bacteria were extracted from soil (10 g [dry weight] by agitation for 30 min in 10 ml of water containing 0.1 M (NH4)2HPO4, collected by centrifugation (5,000 × g, 20 min) of the extraction liquid, suspended in buffer, and serially diluted. Aliquots were spread onto solid medium containing substrates and antibiotics appropriate for selective growth of bacterial strains. The resulting plates were incubated at 30°C for 7 days, and then the numbers of bacterial CFU per gram (dws) were determined. Periodically, the identities of putative colonies of strains B13-D5 and B13-ST1 and transconjugant bacteria were confirmed by performing colony hybridization experiments (22) in which nucleotide probes specific for strain B13 (48) and the genes encoding POB-dioxygenase (32) were used.
Chemical analyses.
3-POB was extracted from soil and analyzed by HPLC by using methods described previously (21). The detection limit for 3-POB was 50 ppb (dws). The data presented below are averages of values from triplicate determinations.
RESULTS
Growth kinetics with 3-POB as the substrate.
The growth of strains POB310 and B13-D5 in liquid culture followed Monod kinetics (data not shown) with the following kinetic parameters (means ± standard errors): for strain POB310, μmax = 0.31 ± 0.02 h−1, Ks = 242 ± 12 mg/liter, and Y = 0.32 ± 0.02 g of biomass/g of 3-POB; for strain B13-D5, μmax = 0.45 ± 0.01 h−1, Ks = 150 ± 8.6 mg/liter, and Y = 0.44 ± 0.03 g of biomass/g of 3-POB.
Behavior of phenoxybenzoate compounds in soil.
A linear isotherm was obtained from the plot of aqueous concentrations (expressed in milligrams per liter) versus sorbed concentrations (expressed in milligrams per kilogram) of dissociated 3-POB at pH 7 (data not shown). The solubility of 3-POB was 4.60 ± 0.08 g/liter, KOW was 0.60, and Kd was 0.41 ± 0.04 liter/kg (mean ± 95% confidence interval).
Degradation of 3-POB in Cedar Creek soil.
When no bacteria were added, removal of 3-POB followed first-order kinetics (data not shown), and the rate was 3.86 × 10−3 ± 0.35 × 10−3 day−1 (mean ± standard error), which corresponded to a half-life of 180 days. Repeated attempts to isolate bacteria that used 3-POB as a sole growth substrate from Cedar Creek soil were unsuccessful.
Survival and activity of POB310 in Cedar Creek soil.
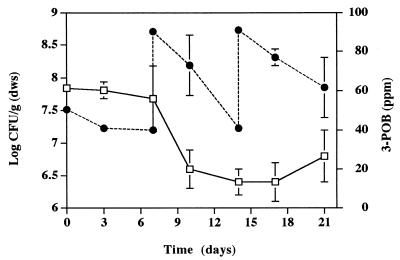
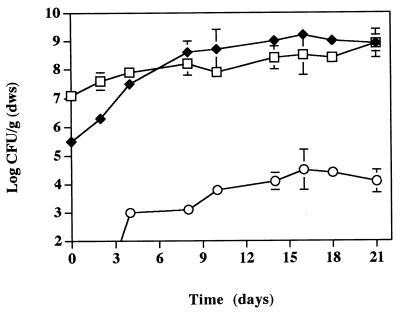
The survival and degradative activity of strain POB310 were assessed by using soil microcosms. The bacterial densities (108 CFU/g [dws]) decreased to nondetectable levels as a first-order process (data not shown). The die-off coefficient was 0.604 ± 0.006 day−1, and the corresponding half-life was 1.15 days. In contrast, strain POB310 remained detectable in soil that contained 50 ppm of 3-POB (Fig. 1). Removal of 3-POB from the soil was incomplete after 7 days; degradation of two subsequent additions of 3-POB (100 ppm) on days 7 and 15 was also incomplete; the minimum level of 3-POB observed was 40 ppm.
FIG. 1.
Density of strain POB310 (□) and degradation of 3-POB (●) in microcosms containing Cedar Creek soil. 3-POB was added to a final concentration of 100 ppm on days 7 and 14. Error bars indicate standard deviations.
Transconjugant bacteria, which were formed by transfer of pPOB to strain B13, were not observed in microcosms containing strains POB310 and B13 (108 CFU of each strain/g [dws]). Transconjugant bacteria were observed in filter matings at a frequency of 4.8 × 10−7 transconjugant/donor. Further experiments did not include strain POB310 due to its relatively poor activity with respect to degradation of 3-POB in soil.
Survival and activity of strains B13-D5 and B13-ST1 in Cedar Creek soil.
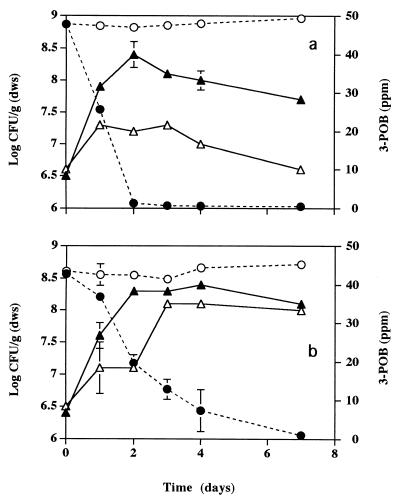
When strain B13-D5 was added to soil, the density of this organism increased by a factor approaching 1 log unit (Fig. 2a); in autoclaved soil, the increase in density was closer to 2 log units (Fig. 2b). In soil containing 50 ppm of 3-POB, a greater increase in bacterial density was observed as 3-POB was degraded to nondetectable levels (<50 ppb) (Fig. 2a). In autoclaved soil containing 50 ppm of 3-POB, degradation occurred at a slower pace (Fig. 2b). Both the higher densities of bacteria and the decreased rate of removal of 3-POB in autoclaved soil may have been due to bacterial utilization of organic carbon that was released during the autoclaving process (28). Strain B13-ST1 behaved like strain B13-D5 with respect to changes in density and degradation of 3-POB in both soil and autoclaved soil (data not shown).
FIG. 2.
Microcosms were established with Cedar Creek soil (a) and autoclaved Cedar Creek soil (b). The density of strain B13-D5 increased in soils to which 3-POB was not added (▵). Greater increases in density (▴) occurred in soil containing 3-POB which was degraded to nondetectable levels (●). Control microcosms contained 3-POB (○) and no strain B13-D5. Error bars indicate standard deviations.
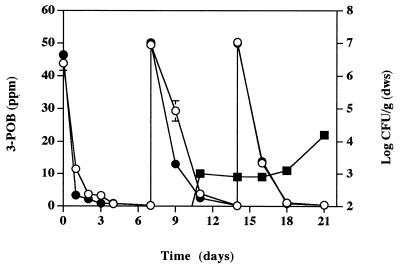
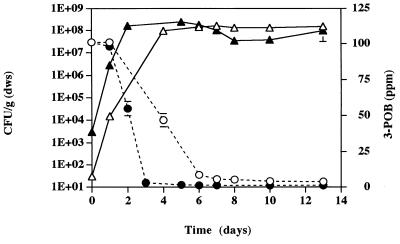
We prepared additional microcosms containing 50 ppm of 3-POB and strains B13-ST1 (5 × 106 CFU/g [dws]) and B13 (108 CFU/g [dws]). Degradation of 3-POB was complete (concentration, <50 ppb) within 1 week (Fig. 3). The density of strain B13-ST1 increased to 5 × 108 CFU/g (dws), and the density of strain B13 remained unchanged (data not shown). Transconjugant bacteria, which were formed by transfer of pPOB to strain B13, were not observed (<200 CFU/g [dws]) during this time. Subsequent additions of 3-POB were made on days 7 and 14; degradation of 3-POB was complete 7 days after each addition. Transconjugant bacteria were observed on day 11 in microcosms containing autoclaved soil (Fig. 3); the density of these bacteria was 104 CFU/g (dws) by day 21. Transconjugants were not observed in nonautoclaved soil. In filter matings, transconjugant bacteria occurred at a frequency of 0.9 transconjugant per donor.
FIG. 3.
Degradation of 3-POB by strain B13-ST1 in microcosms containing Cedar Creek soil (○) and autoclaved Cedar Creek soil plus strain B13 (●). 3-POB (50 ppm) was added on days 0, 7, and 14. Transconjugant bacteria (■) were observed only in microcosms containing autoclaved soil. Error bars indicate standard deviations.
We also prepared microcosms containing 50 ppm of 3-POB, strains B13-D5 (5 × 106 CFU/g [dws]) and B13 (108 CFU/g [dws]), and P. stutzeri(pRP4-4) (108 CFU/g [dws]) (data not shown). The density of B13-D5 increased to 5 × 108 CFU/g (dws) and degradation of 3-POB was complete (concentration, <50 ppb) during the first week of incubation. Two subsequent additions of 3-POB (50 ppm) on days 7 and 14 were also rapidly degraded. Transconjugant bacteria were not observed in either autoclaved or nonautoclaved soil. In filter matings in which strains B13-D5 and B13 and P. stutzeri(pRP4-4) were used, transconjugants occurred at a frequency of 1.3 × 10−6 transconjugant per donor.
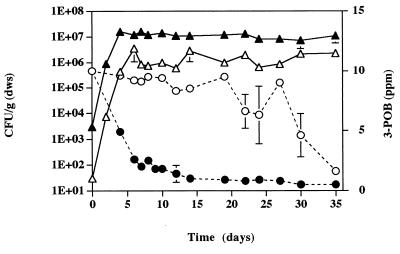
Additional microcosms containing strains B13-ST1 and B13 were established by using autoclaved Fort Snelling soil (concentration of 3-POB, 50 ppm). The densities of both bacterial strains increased in these experiments (Fig. 4). Degradation of 3-POB was complete in 1 day, as was degradation of further additions of 3-POB (50 ppm) on days 7 and 14 (data not shown). Transconjugant bacteria were observed on day 4; the density of these bacteria increased to 5 × 104 CFU/g (dws) by day 16.
FIG. 4.
Densities of strains B13-ST1 (□) and B13 (⧫) and transconjugant bacteria (○) in microcosms containing Fort Snelling soil. 3-POB (50 ppm) was added on days 0, 7, and 14 and was degraded to levels below the limit of detection (<50 ppb) within 24 h. Error bars indicate standard deviations.
Effect of inoculum size and substrate concentration on degradation of 3-POB.
Cedar Creek soil containing 100 ppm of 3-POB was inoculated with strain B13-D5 at calculated densities of 10 and 1,000 CFU/g (dws). After enumeration of the bacteria in the soil, these values were corrected to 30 and 3,000 CFU/g (dws). The density of strain B13-D5 increased rapidly; net doubling times were 4.7 and 4.9 h, respectively, and the densities peaked near 108 CFU/g (dws). The increases in density were accompanied by corresponding decreases in concentrations of 3-POB (Fig. 5). An initial lag in degradation of 3-POB corresponded to the time required to reach the density of cells (106 CFU/g [dws]) that was able to cause a detectable loss of 3-POB. In microcosms inoculated with 3,000 CFU/g (dws), this threshold value was attained in less time, and degradation of 3-POB was faster than degradation in the microcosms that received 30 CFU/g (dws).
FIG. 5.
Densities of strain B13-D5 in microcosms containing Cedar Creek soil that received inocula containing 30 (▵) and 3,000 (▴) CFU/g (dws). Degradation of 3-POB (100 ppm) was faster in microcosms that received the larger inoculum (●) than in microcosms that received the smaller inoculum (○). Error bars indicate standard errors.
The densities of strain B13-D5 also increased when the soils initially contained a lower concentration of 3-POB (10 ppm). However, the net doubling times were longer (8.5 h at a density of 30 CFU/g [dws] and 7.8 h at a density of 3,000 CFU/g [dws]), and the peak cell densities were lower (4 × 106 and 107 CFU/g [dws]) (Fig. 6). Degradation of 3-POB was more rapid in microcosms that received 3,000 CFU/g (dws), probably because at a concentration of 30 CFU/g (dws) strain B13-D5 barely attained the density required for degradation of 3-POB to be observed.
FIG. 6.
Densities of strain B13-D5 in microcosms containing Cedar Creek soil that received inocula containing 30 (▵) and 3,000 (▴) CFU/g (dws). Degradation of 3-POB (10 ppm) was faster in microcosms that received the larger inoculum (●) than in microcosms that received the smaller inoculum (○). Error bars indicate standard errors.
Effects of nutrients and water saturation.
Microcosms were established with 10 and 1,000 CFU of strain B13-D5 (suspended in water) per g (dws) plus 3-POB at concentrations of 10 and 100 ppm. In soil that was not amended with nutrients, strain B13-D5 neither survived nor degraded 3-POB (Table 1). Addition of phosphorus alone supported limited growth of the bacterium; however, only a relatively small fraction of 3-POB was degraded. In contrast, addition of both phosphate and nitrogen resulted in almost complete degradation of 3-POB. To test the effect of water saturation levels on survival and activity, strain B13-D5 was added (106 CFU/g [dws]) to soils with water contents ranging from 10 to 100% of the water-holding capacity (Fig. 7). Strain B13-D5 did not survive at water saturation levels that were ≤20% of the water-holding capacity; water saturation levels that were ≥40% of the water-holding capacity enhanced both survival of strain B13-D5 and degradation of 3-POB.
TABLE 1.
Effect of nutrient supplementation on survival of strain B13-D5 and removal of 3-POB in phosphate-buffered (pH 7.0) Cedar Creek soil
| Microcosma | Inoculum density (CFU/g [dws]) | Nutrient supplement(s)
|
|||||
|---|---|---|---|---|---|---|---|
| None
|
Phosphate
|
Phosphate + nitrogen
|
|||||
| Density (CFU/g) | 3-POB concn (ppm) | Density (CFU/g) | 3-POB concn (ppm) | Density (CFU/g) | 3-POB concn (ppm) | ||
| A | 10 | BDLb | 99 ± 0.9c | BDL | 96 ± 2.3c | 2.6 × 107 | 2.8 ± 0.8c |
| 1,000 | BDL | 99 ± 0.9 | 1.8 × 106 | 94 ± 4.6 | 1.1 × 107 | 0.8 ± 0.2 | |
| B | 10 | BDL | 9.9 ± 0.1 | BDL | 9.1 ± 0.1 | 1.5 × 106 | 1.6 ± 0.3 |
| 1,000 | BDL | 9.6 ± 0.2 | 8.0 × 104 | 7.4 ± 0.2 | 6.6 × 106 | 0.9 ± 0.6 | |
Microcosm A received 100 ppm of 3-POB and was incubated for 24 days. Microcosm B received 10 ppm of 3-POB and was incubated for 41 days. Data were obtained after incubation.
BDL, below the detection limit (<100 CFU/g [dws]).
Mean ± standard error.
DISCUSSION
Nonindigenous microorganisms may survive and become established in a habitat if they encounter an environmental pollutant that serves as a noncompetitive growth substrate (16, 27, 52). The diaryl ether compound 3-POB apparently has qualities necessary for providing such a niche, including (i) bioavailability due to a low tendency to sorb to soil (solubility, 4.60 ± 0.08 g/liter; KOW = 0.60; Kd = 0.41 ± 0.04 liter/kg) and (ii) persistence (half-life, 180 days) in soil to which degradative microorganisms are not added. These characteristics make 3-POB a noncompetitive growth substrate. Thus, 3-POB was used as a model compound for bioagumentation in this study.
The effectiveness of bioaugmentation can be measured by factors such as survival of the introduced organism, the stability of the genes encoding appropriate catalytic functions, and the degree of contaminant removal. If introduced microorganisms perform poorly in the target environment, survival and degradative performance may be improved by preadaptation to environmental conditions (30). An alternative is to transfer the genes encoding specific catalytic functions to a more suitable host. In such cases, the potential for the genes to transfer to indigenous organisms becomes a factor for consideration.
Strain POB310 was isolated for its ability to degrade 3-POB (18). However, initial experiments revealed that strain POB310 survived poorly in soils and that the genes encoding POB-dioxygenase were readily lost by segregation of pPOB (38). Thus, while 3-POB provided a niche for the bacterial strains tested, strain POB310 did not occupy this niche (Fig. 1) as successfully as strains B13-D5 (Fig. 2) and B13-ST1 (Fig. 3), both of which degraded 3-POB at faster rates and to greater extents, with resulting increases in cell density.
Strain B13, the parent of strains B13-D5 and B13-ST1, is fairly robust and survives in a variety of habitats (26, 35, 39); this strain was selected to host the genes encoding POB-dioxygenase in part because of this robust nature. Thus, it was not too surprising that strains B13-D5 and B13-ST1 outperformed strain POB310. Strain B13-D5 also exhibited a slightly greater affinity (lower Ks) for 3-POB and had a better growth yield, two factors that could have contributed to the improved performance.
The proficiency of strain B13-D5 was exemplified when an inoculum containing 30 CFU/g (dws) resulted in complete degradation of 3-POB and an increase of 6 to 7 orders of magnitude in bacterial density (Fig. 5 and 6). This suggests that in certain cases, bioaugmentation may be achieved more cost effectively by using relatively few bacteria in the inoculum. In a similar study, Pseudomonas cepacia BR16001 was introduced into soil containing 2,4-dichlorophenoxyacetic acid (220 ppm) at densities of 1 and 100 CFU/g of soil (10), and the amount of time required to completely degrade 2,4-dichlorophenoxyacetic acid was reduced from 47 days to 28 and 6 days, respectively. However, bacterial growth was not monitored.
The growth and degradative activity of strain B13-D5 in Cedar Creek soil were enhanced by adding nutrients (Table 1). Phosphate alone had a marginal effect; simultaneous addition of phosphate and nitrogen resulted in marked increases in degradation of 3-POB and concomitant increases in cell density. In similar studies, nutrient amendments were observed to have stimulatory effects on the degradative activities of microorganisms that were indigenous (37) and introduced into target environments (19, 42). Strain B13-D5 was also resilient to water stress and degraded 3-POB at water saturation levels that ranged from 40 to 100% of the soil water-holding capacity (Fig. 7). Normally, the survival and degradative activity of bacteria are compromised in soils with low water saturation levels (40, 44). The performance of strain B13-D5 (i.e., degradation of 3-POB and concomitant increases in cell density) suggests that prudent selection of bacteria for bioaugmentation may result in bioremediation in soils once thought to be less than hospitable to microbial survival.
When we compared transfer events for the plasmids used in this study, pD30.9 was not observed to transfer from strain B13-D5 to strain B13 in soil. However, in filter matings in which P. stutzeri(pRP4-4) was present, a frequency of transfer for pD30.9 of 10−6 transconjugant per donor was observed. In soil, this frequency would have yielded transconjugant densities that were below the limit of detection (<100 CFU/g [dws]), thus making it unlikely that transconjugants would have been observed.
In soil, transconjugant bacteria were formed by transfer of pPOB from strain B13-ST1 to strain B13 (Fig. 3 and 4) but not by transfer of pPOB from strain POB310 to strain B13. This difference may have been due to the relative ease with which intraspecies transfer of plasmids occurs compared to interspecies transfer (41, 43, 49, 50) and could account for the frequencies observed in filter matings in which pPOB was transferred to strain B13 at frequencies of 5 × 10−7 from strain POB310 and 0.9 from strain B13-ST1. Based on these frequencies, a POB310 density of 108 CFU/g (dws) would be necessary to observe transconjugant bacteria in soil; this density was not attained in these experiments (Fig. 1). In contrast, a density for strain B13-ST1 of only 5 × 102 CFU/g (dws) would be necessary to observe transconjugant bacteria in soil.
It is important to note that transconjugant bacteria were observed only in autoclaved soil that received multiple additions of 3-POB. This treatment could allow the levels of transconjugant bacteria to increase from initially low levels to detectable levels (Fig. 3 and 4) without competition and predation from indigenous microorganisms. It also could allow for growth of the plasmid donor, a factor that may be important for conjugative transfer of plasmids (43). However, it is unclear how important growth of the donor bacterium actually is, as other researchers have found that growth and formation of transconjugant bacteria are not related (13, 14, 25).
Strains B13-ST1 and B13 grew to higher densities and transconjugants appeared earlier in autoclaved Fort Snelling soil than in Cedar Creek soil (Fig. 3 and 4). The Fort Snelling soil contained more organic matter (5.4 versus 0.5%) and clay (18 versus 3%) than the Cedar Creek soil. Both of these parameters may increase the frequency of conjugative plasmid transfer by promoting cell-to-cell contact in microcolonies that develop on clay and organic aggregates (47). In sandy soils bacteria are less likely to be attracted to microhabitats, which leads to fewer instances of the cell-to-cell contact that is necessary for conjugation to occur.
ACKNOWLEDGMENTS
This research was supported by grant BCS-9318788 from the National Science Foundation and by grants-in-aid 15855 and 16269 from the University of Minnesota Graduate School.
We thank S. Thiem for the nucleic acid probe for strain B13.
REFERENCES
- 1.Aamand J G, Bruntse G, Jepsen M, Jorgensen C, Jensen B K. Degradation of PAHs in soil by indigenous and inoculated bacteria. In: Hinchee R E, Fredrickson J, Alleman B C, editors. Bioaugmentation for site remediation. Columbus, Ohio: Batelle Press; 1995. pp. 121–127. [Google Scholar]
- 2.Banerjee S, Yalkowsky S H, Valvani S C. Water solubility and octanol/water partition coefficient of organics. Limitations of the solubility-partition coefficient correlation. Environ Sci Technol. 1980;14:1227–1229. [Google Scholar]
- 3.Barles R W, Daughton C G, Hsieh D P H. Accelerated parathion degradation in soil inoculated with acclimated bacteria under field conditions. Arch Environ Contam Toxicol. 1979;14:1227–1229. doi: 10.1007/BF01054867. [DOI] [PubMed] [Google Scholar]
- 4.Bragg J R, Prince R C, Harner E J, Atlas R M. Effectiveness of bioremediation for the Exxon Valdez oil spill. Nature. 1994;368:413–418. [Google Scholar]
- 5.Briglia M, Middeldorp P J M, Salkinoja-Salonen M S. Mineralization performance of Rhodococcus chlorophenolicus strain PCP-1 in contaminated soil simulating site conditions. Soil Biol Biochem. 1994;26:377–385. [Google Scholar]
- 6.Briglia M, Nurmiaho-Lassila E-L, Vallini G, Salkinoja-Salonen M S. The survival of the pentachlorophenol-degrading Rhodococcus chlorophenolicus PCP-1 and Flavobacterium sp. in natural soil. Biodegradation. 1990;1:273–281. [Google Scholar]
- 7.Bulman T L, Newland M. In situ bioventing of a diesel fuel spill. Hydrol Sci. 1993;8:297–308. [Google Scholar]
- 8.Chatterjee D K, Kilbane J J, Chakrabarty A M. Biodegradation of 2,4,5-trichlorophenoxyacetic acid in soil by a pure culture of Pseudomonas cepacia. Appl Environ Microbiol. 1982;44:514–516. doi: 10.1128/aem.44.2.514-516.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiou C T, Freed V H, Schmedding D W, Kohnert R L. Partition coefficient and bioaccumulation of selected organic chemicals. Environ Sci Technol. 1977;11:475–478. [Google Scholar]
- 10.Comeau Y, Greer C W, Samson R. Role of inoculum preparation and density on the bioremediation of 2,4-D-contaminated soil by bioaugmentation. Appl Microbiol Biotechnol. 1993;38:681–687. [Google Scholar]
- 11.Crawford R L, Mohn W W. Microbiological removal of pentachlorophenol from soil using Flavobacterium. Enzyme Microb Technol. 1985;7:617–620. [Google Scholar]
- 12.Dehmel U, Engesser K-H, Timmis K N, Dwyer D F. Cloning, nucleotide sequence, and expression of the gene encoding a novel dioxygenase involved in metabolism of carboxydiphenyl ethers in Pseudomonas pseudoalcaligenes POB310. Arch Microbiol. 1995;163:35–41. doi: 10.1007/BF00262201. [DOI] [PubMed] [Google Scholar]
- 13.DeRore H, Demolder K, de Wilde K, Topp E, Houwen F, Verstraete W. Transfer of the catabolic plasmid RP4::Tn4371 to indigenous soil bacteria and its effect on respiration and biphenyl breakdown. FEMS Microbiol Ecol. 1994;15:71–78. [Google Scholar]
- 14.DiGiovanni J, Neilson W, Pepper I L, Sinclair N A. Gene transfer of Alcaligenes eutrophus JMP134 plasmid pJP4 to indigenous soil recipients. Appl Environ Microbiol. 1996;62:2521–2526. doi: 10.1128/aem.62.7.2521-2526.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorn E, Hellwig M, Reineke W, Knackmuss H-J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99:61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- 16.Dybas M J, Tatara G M, Knoll W H, Mayotte T J, Criddle C S. Niche adjustment for bioaugmentation with Pseudomonas sp. strain KC. In: Hinchee R E, Fredrickson J, Alleman B C, editors. Bioaugmentation for site remediation. Columbus, Ohio: Battelle Press; 1995. pp. 77–84. [Google Scholar]
- 17.Edgehill R U. Removal of pentachlorophenol from soil by Arthrobacter strain ATCC 33790. In: Hinchee R E, Fredrickson J, Alleman B C, editors. Bioaugmentation for site remediation. Columbus, Ohio: Batelle Press; 1995. pp. 85–90. [Google Scholar]
- 18.Engesser K-H, Fietz W, Fischer P, Schulte P, Kanckmuss H-J. Dioxygenolytic cleavage of aryl ether bonds: 1,2-dihydro-1,2-dihydroxy-4-carboxybenzophenone as evidence for initial 1,2-dioxygenation in 3- and 4-carboxy biphenyl ether degradation. FEMS Microbiol Lett. 1990;69:317–322. doi: 10.1016/0378-1097(90)90087-7. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein R M, Mallory L M, Alexander M. Reasons for possible failure of inoculation to enhance biodegradation. Appl Environ Microbiol. 1985;50:977–983. doi: 10.1128/aem.50.4.977-983.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grigal D F, Chamberlain L M, Finney H R, Wroblewski D V, Gross E R. Miscellaneous report 123. Minneapolis/St. Paul: Agricultural Experiment Station, University of Minnesota; 1974. [Google Scholar]
- 21.Halden R U, Halden B G, Dwyer D F. Removal of dibenzofuran, dibenzo-p-dioxin, and 2-chlorodibenzo-p-dioxin from soils inoculated with Sphingomonas sp. strain RW1. Appl Environ Microbiol. 1999;65:2246–2249. doi: 10.1128/aem.65.5.2246-2249.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halden R U, Mundfrom G W, Peters E G, Dwyer D F. Monitoring the fate and activity of diaryl ether-degrading bacteria in soil. In: Wukash R F, editor. 50th Purdue Industrial Waste Conference Proceedings. Chelsea, Mich: Ann Arbor Press, Inc.; 1995. pp. 57–64. [Google Scholar]
- 23.Halden, R. U., E. G. Peters, B. G. Halden, and D. F. Dwyer. Transformation of mono- and dichlorinated phenoxybenzoates by POB-dioxygenase in Pseudomonas pseudoalcaligenes POB310 and a modified, diaryl ether-mineralizing bacterium. Submitted for publication. [PubMed]
- 24.Hansch C, Leo A. Substituent constants for correlation analysis in chemistry and biology. J. New York, N.Y: Wiley; 1979. [DOI] [PubMed] [Google Scholar]
- 25.Henschke R B, Schmidt F R J. Plasmid mobilization from genetically engineered bacteria to members of the indigenous soil microflora in situ. Curr Microbiol. 1989;20:105–110. [Google Scholar]
- 26.Krumme M L, Smith R L, Egestorff J, Thiem S A, Tiedje J M, Timmis K N, Dwyer D F. Behavior of pollutant-degrading microorganisms in aquifers: predictions for genetically engineered organisms. Environ Sci Technol. 1994;28:1134–1138. doi: 10.1021/es00055a025. [DOI] [PubMed] [Google Scholar]
- 27.Liu S, Suflita J M. Ecology and evolution of microbial populations for bioremediation. Trends Biotechnol. 1993;11:344–352. doi: 10.1016/0167-7799(93)90157-5. [DOI] [PubMed] [Google Scholar]
- 28.Lynch J M. Limits to microbial growth in soil. J Gen Microbiol. 1982;128:405–410. [Google Scholar]
- 29.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 30.Megharaj M, Wittich R-M, Blasco R, Pieper D H, Timmis K N. Superior survival and degradation of dibenzo-p-dioxin and dibenzofuran in soil by soil-adapted Sphingomonas sp. strain RW1. Appl Microbiol Biotechnol. 1997;48:109–114. [Google Scholar]
- 31.Miyamoto J. The chemistry, metabolism and residue analysis of synthetic pyrethroids. Pure Appl Chem. 1981;53:1967–2002. [Google Scholar]
- 32.Mundfrom G W. M.S. thesis. Minneapolis: University of Minnesota; 1997. [Google Scholar]
- 33.Nerella S, Wright A L, Weaver R W. Microbial inoculants and fertilization for bioremediation of oil in wetlands. In: Hinchee R E, Fredrickson J, Alleman B C, editors. Bioaugmentation for site remediation. Columbus, Ohio: Batelle Press; 1995. pp. 31–38. [Google Scholar]
- 34.Norris L A. The movement, persistence, and fate of phenoxy herbicides and TCDD in the forest. Residue Rev. 1981;80:65–135. [Google Scholar]
- 35.Nüßlein K, Maris D, Timmis K N, Dwyer D F. Expression and transfer of engineered catabolic pathways harbored by Pseudomonas spp. introduced into activated sludge microcosms. Appl Environ Microbiol. 1992;58:3380–3386. doi: 10.1128/aem.58.10.3380-3386.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono H, Nakanisi J. Herbicide, CNP contamination in the Lake Kasumigaura basin. Water Res. 1987;21:669–675. [Google Scholar]
- 37.Palumbo A V, Scarborough S P, Pfiffner S M, Phelps T J. Influence of nitrogen and phosphorous on the in situ bioremediation of trichloroethylene. Appl Biochem Biotechnol. 1995;51:635–647. [Google Scholar]
- 38.Peters E G. M.S. thesis. Minneapolis: University of Minnesota; 1997. [Google Scholar]
- 39.Pipke R, Wagner-Döbler I, Timmis K N, Dwyer D F. Survival and function of a genetically engineered pseudomonad in aquatic sediment microcosms. Appl Environ Microbiol. 1992;58:1259–1265. doi: 10.1128/aem.58.4.1259-1265.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postma J, van Veen J A. Habitable pore space and survival of Rhizobium leguminosarium biovar trifolii introduced into soil. Microb Ecol. 1990;19:149–161. doi: 10.1007/BF02012096. [DOI] [PubMed] [Google Scholar]
- 41.Rafil F, Crawford D L. Transfer of conjugative plasmids and mobilization of a nonconjugative plasmid between Streptomyces strains on agar and in soil. Appl Environ Microbiol. 1988;54:1334–1340. doi: 10.1128/aem.54.6.1334-1340.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramadan M A, El-Tayleb O M, Alexander M. Inoculum size as a factor limiting success of inoculation for biodegradation. Appl Environ Microbiol. 1990;56:1392–1396. doi: 10.1128/aem.56.5.1392-1396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramos-Gonzalez M-I, Duque E, Ramos J L. Conjugational transfer of recombinant DNA in cultures and in soils: host range of Pseudomonas putida TOL plasmids. Appl Environ Microbiol. 1991;57:3020–3027. doi: 10.1128/aem.57.10.3020-3027.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rattray E A S, Prosser J I, Glover L A, Killham K. Matric potential in relation to survival and activity of a genetically modified microbial inoculum in soil. Soil Biol Biochem. 1992;24:421–425. [Google Scholar]
- 45.Riser Roberts E. Bioremediation of petroleum contaminated sites. Boca Raton, Fla: CRC Press; 1992. [Google Scholar]
- 46.Roberts T R. The metabolism of the synthetic pyrethroids in plants and soils. Prog Pestic Biochem. 1981;1:115–146. [Google Scholar]
- 47.Stotzky G, Devanas M A, Zeph L R. Methods for studying bacterial gene transfer in soil by conjugation and transduction. Adv Appl Microbiol. 1990;35:57–169. doi: 10.1016/s0065-2164(08)70243-0. [DOI] [PubMed] [Google Scholar]
- 48.Thiem S M, Krumme M L, Smith R L, Tiedje J M. Use of molecular techniques to evaluate the survival of a microorganism injected into an aquifer. Appl Environ Microbiol. 1994;60:1059–1067. doi: 10.1128/aem.60.4.1059-1067.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Elsas J D, Govaert J M, van Veen J A. Transfer of plasmid pFT30 between bacilli in soil as influenced by bacterial population dynamics. Soil Biol Biochem. 1987;19:639–647. [Google Scholar]
- 50.van Elsas J D, Trevors J T, Starodub M E, van Overbeek L S. Transfer of plasmid RP4 between pseudomonads after introduction into soil: influence of spatial and temporal aspects of inoculation. FEMS Microbiol Ecol. 1990;73:1–11. [Google Scholar]
- 51.Venosa A D, Suidan M T, Haines J R, Wrenn B A, Strohmeier K L, Eberhart B L, Kadkhodyan M, Holder E, King D, Anderson B. Field bioremediation study. In: Hinchee R E, Fredrickson J, Alleman B C, editors. Bioaugmentation for site remediation. Columbus, Ohio: Batelle Press; 1995. pp. 49–56. [Google Scholar]
- 52.Vogel T M. Bioaugmentation as a soil bioremediation approach. Curr Opin Biotechnol. 1996;7:311–316. doi: 10.1016/s0958-1669(96)80036-x. [DOI] [PubMed] [Google Scholar]
- 53.Wagner-Döbler I, Pipke R, Timmis K N, Dwyer D F. Evaluation of aquatic sediment microcosms and their use in assessing possible effects of introduced microorganisms on ecosystem parameters. Appl Environ Microbiol. 1992;58:1249–1258. doi: 10.1128/aem.58.4.1249-1258.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]