SUMMARY
Rotavirus (RV) infection causes acute infantile diarrhoea in humans and animals and remains a major concern for vaccine development. The close proximity of humans to animals may foster cross-species infection resulting in the emergence of novel/unusual strains by genetic reassortment. In this study, we characterized 500 diarrhoeal samples for group A rotaviruses (RVA) from children (n = 290), piglets (n = 95) and calves (n = 115) in Northeast India during 2012–2013. The data showed that 142/500 (28·4%) faecal samples were positive for RVA with the highest level of infection detected in piglets (57/142, 40·1%) followed by children (51/142, 35·9%) and calves (34/142, 23·9%). Sequence-based G- and P-typing showed G1P[8] (25%) and G1P[7] (35%) were the prevailing genotypes in both humans and animals. Single cases of unusual genotypes, i.e. G9P[8], G5P[8] in humans and G1P[13], G1P[23] and G3P[7] in animals were also identified. Cluster analyses of the sequences showed regional strains were genetically closer to their homologous strains. However, human G5P[8] and porcine G1P[8] strains showed homology to heterologous hosts of their prototype strains. The subsequent global spread of unusual RV strains may result in their establishment over time, presenting challenges to future vaccine evaluation programmes. More studies on emerging genotypes are required to elucidate how RVA strains evolve post-vaccination. This study supports the need for continuous surveillance of RVA infections after detecting from diverse hosts in a common setting.
Key words: Genotyping, interspecies transmission, rotavirus, surveillance, unusual strain, vaccine
INTRODUCTION
Group A rotavirus (RVA) is a major pathogen that causes acute dehydrating infantile diarrhoea in humans and domestic animals [1]. The disease burden and associated morbidity/mortality is higher in developing countries where there are poor hygiene conditions. In India, of 334 000 diarrhoeal-attributed deaths occurring annually in children aged <5 years, 98 000 cases were due to RVA alone [2, 3]. Nevertheless, RVA infection also leads to immense economic losses in the livestock industries, with high mortality rates in neonatal piglets and calves [4, 5]. The virus belongs to the family Reoviridae and is enclosed in three concentric layers comprising 11 double-stranded (ds) RNA segments, encoding six structural proteins (VP1–4, VP6, VP7) and six non-structural proteins (NSP1–NSP5/6). Based on the VP6 capsid gene, rotaviruses (RVs) are classified into eight major species, i.e. A–H [6]. A sequence-based classification system proposed by the Rotavirus Classification Working Group (RCWG) further categorized RVA to G (glycoprotein) and P (protease-sensitive) genotypes for the VP7 and VP4 outer capsid proteins, respectively [7]. These segments are analysed as the VP4/VP7 segments possess immunogenic epitopes which are relevant for immune protection and vaccine development [1, 8]. For broad genomic analyses, 27 G and 37 P genotypes have been identified globally [8, 9] where different G–P combinations distributed among humans and animals are referred to either as usual or unusual genotypes [10, 11]. The usual genotypes in humans constitute G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8], which account for about 90% of RV infections globally. The unusual genotypes are fewer, corresponding to about 4·9% of RV infections worldwide [10, 12, 13]. Earlier studies in hospitalized children have reported RVA prevalence and strain diversity across India, where G1 strains were predominant along with significant occurrence of G9, G2 and G12 viruses [14–16]. A study on human RVA episodes in Northeast (NE) India estimated the incidence of uncommon genotypes G4P[4], G4P[6], G10P[6], G9P[19] with a first report of NE Indian G12 strains and a few non-typable RVAs [16]. Due to its segmented nature, the RVA genome allows genetic reassortment during mixed infections, more generally in human–animal close settings inducing progenies with unusual or atypical phenotypes [10]. Delineation of human RVAs in animal lineages suggests that interspecies transmission and reassortment events play a major role in neonatal RVA infections [16–19]. This provides evidence that animals are a source of virus and/or of genetic material transmission generating novel strains and raising concerns regarding vaccination programmes.
Two different RV vaccines, Rotarix (GlaxoSmithKline Biologicals, USA) and RotaTeq (Merck & Co. Inc., USA), are in use for immunizing children in different countries [20, 21]. In India, the vaccines are not included in the National Immunization Programme and only a few children receive the vaccine through the private health sector. The efficacy of vaccines can be affected by several factors, such as malnutrition, presence of maternal antibodies and the genotype of circulating strains [22, 23]. Thus, surveillance of locally prevailing RV genotypes helps in the development of more effective vaccine candidates to reduce the impact of this infectious disease. The main occupation in NE India, an agrarian region, involves rearing animals. The close contact of animals with their human handlers can lead to interspecies transmission of the viruses. Therefore, rural regions with poor sanitation may act as a potential hub for the emergence of new virus strains.
The majority of studies carried out earlier in NE India focused on children hospitalized with acute gastroenteritis; however, the incidence of RVA in animals remains largely unknown. Thus, in order to gain insight into the genetic diversity of the RVA strains detected in human, bovine and porcine neonates, we analysed the VP4 and VP7 sequences focusing on the emergence of novel/unusual genotypes.
METHODS
Virus detection
A total of 500 stool samples from piglets (n = 95), calves (n = 115) and children (n = 290) from NE India were collected during 2012–2013. The livelihood of the rural population mostly involves backyard animal rearing practices. Samples from children suspected of RV infection were collected following the World Health Organization's standard case definition of a child aged <5 years, admitted to a hospital for treatment, while a confirmed case was a suspected case whose stool sample demonstrated the presence of RV by enzyme immunoassay. The faecal samples from piglets aged <1 month and calves aged <3 months were collected several days after onset of clinical disease (watery diarrhoea). Samples were collected from the patient together with written consent from the patient's parents/guardians and the animal handlers. Preliminary screening of stool samples for the presence of RVA antigen was performed using the Premier Rotaclone (Meridian Biociences Inc., USA) enzyme immunosorbent assay (EIA) according to the manufacturer's instructions.
RNA extraction and reverse transcription–polymerase chain reaction (RT–PCR)
Subsets of EIA-positive samples were confirmed by RT–PCR targeting the full-length VP7 and partial VP4 gene to avoid a false-positive result. RVA dsRNA was extracted using QIAamp viral RNA mini kit (Qiagen, Germany) according to the manufacturer's protocol and one-step group A-specific RT–PCR was performed (1× RT buffer, 1·2 mm MgCl2, 3% DMSO, 200 µm dNTP mix, 20 pmol each primer, 5 U MuLV RT, 1 U Pfu Taq, 20 U RNAsin). Primers used to amplify full-length VP7 and a partial VP4 genes (VP8*) are given in Table 1 [24, 25]. A lyophilized group A porcine RV was reconstituted to recover RNA for a positive control and the first faecal samples of a caesarean-derived newborn piglet was used as a negative control.
Table 1.
Set of primers used for VP7 and VP4 gene amplification
| Primer | Primer sequence | Position (nt) |
|---|---|---|
| VP7 gene | Porcine/human | |
| Beg9 (+) | 5′-GGCTTTAAAAGAGAGAATTTCCGTCTGG-3′ | 1–28 |
| End9 (−) | 5′-GGTCACATCATACAATTCTAATCTAAG-3′ | 1062–1036 |
| Bovine | ||
| Bov9Com3 | 5′-TCACATCATACAACTCTAATCT-3′ | 1038–1060 |
| Bov9Com5 | 5′-TGTATGGTATTGAATATACCA-3′ | 50–71 |
| VP4 gene | Porcine/human/bovine | |
| Con 3 (+) | 5′-TGGCTTCGCTCATTTATAGACA-3′ | 11–32 |
| Con 2 (−) | 5′-ATTTCGGACCATTTATAACC-3′ | 868–887 |
Sequence analysis
The purified VP7 and VP4 amplicons were directly sequenced in an automated DNA sequencer (ABI 3500, Applied Biosystems, USA) at the sequencing service of the Department of Biotechnology, Assam University. Sequence alignments and editing were conducted using ClustalX [26] and Chromas 2·3 (http://technelysium.com.au/chromas.html), respectively.
The sequences were analysed using the National Centre for Biotechnology Information (NCBI, National Institutes of Health, Bethesda, MD) Basic Local Alignment Search Tool (BLAST) server on GenBank database release 143·0 [27, 28] and G–P genotypes were classified as proposed by the RCWG using RotaC v. 2.0 (http://rotac.regatools.be/) [29]. Molecular phylogenetic analysis was performed using the maximum-likelihood method based on the Tamura–Nei model [30] with MEGA6 [31]. The VP7 and VP4 sequences from group C rotavirus ‘Bristol’ strain retrieved from GenBank were used as an outgroup during phylogenetic tree constructions. The trees were drawn to scale with branch lengths measured as number of substitutions per site and were bootstrapped with 1000 replicates.
Nucleotide sequence accession numbers
The full-length VP7 and the partial VP4 (VP8*) gene sequences of the strains detected during this study were submitted to GenBank database under accession numbers: VP7: (C4b-KJ701389), (C4a-KJ701390), (B7-KJ701391), (C13-KJ701392), (C5-KJ701393), (B6-KJ701394), (B5-KJ701395), (F1-KJ701396), (F11-KJ701397), (C1-KJ701398), (F7-KJ701399), (F3-KJ701400), (B4-KJ701401), (B3-KJ701402), (B2-KJ701403), (F8-KJ701404), (F6-KJ701405), (F2-KJ701406), (F5-KJ701407), (F4-KJ701408); VP4: (P15-KJ701369), (P20-KJ701370), (P17-KJ701371), (P6-KJ701372), (P7-KJ701373), (P13-KJ701374), (P11-KJ701375), (P3-KJ701376), (P4-KJ701377), (P9-KJ701378), (P12-KJ701379), (P2-KJ701380), (P1-KJ701381), (P8-KJ701382), (P10-KJ701383), (P22-KJ701384), (P23-KJ701385), (P14-KJ701386), (P16-KJ701387), (P19-KJ701388). The GenBank accession numbers for the genome segments from each reference strain used in the analyses are presented in Supplementary Table S1.
RESULTS
RV occurrence and seasonal distribution
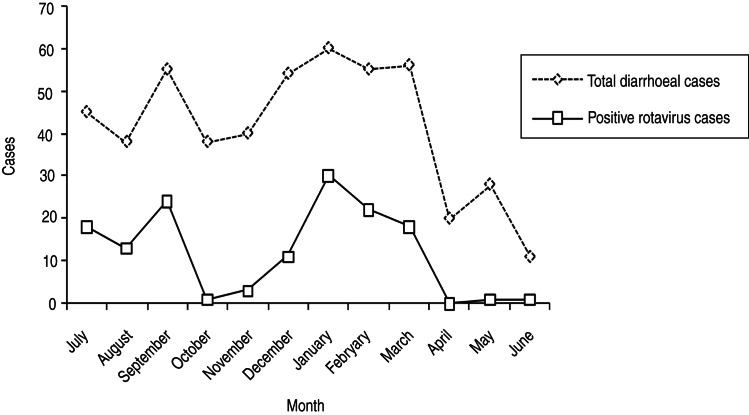
Screening of 500 diarrhoeal samples collected between 2012 and 2013 detected 142 (28·4%) cases positive for RVA major inner capsid protein (VP6 antigen) in the mAb-EIA test common to all known genotypes. The maximum incidence was recorded in piglets (57/142, 40·1%) followed by children (51/142, 35·9%) and calves (34/142, 23·9%). RVA infection was higher in children aged 16–48 months and those of piglets and calves aged 16–28 days. The frequency of RVA incidence was found to be rather similar in all hosts where infections appeared to occur throughout the year, with increased episodes during the rainy season (July–September) and a sudden decline during the autumn months (October–November). The infection started with a steady increase towards winter with peak number of cases during January–March. After that, a continual decrease was seen in the spring months reaching a low of 0–1 cases in May–June (Fig. 1).
Fig. 1.
Seasonality of rotavirus diarrhoeal cases in NE Indian children and animal neonates, 2012–2013. (Represented by monthly positive case variation that peaked during winter and the rainy season.)
Based on the archived stock availability and high OD value of EIA-positive samples, 70 specimens were subjected to RVA-specific RT–PCR for VP7 and VP4 gene primary amplification and successive sequence-based genotyping analysis. Of 40 RT–PCR-positive samples, VP7 and VP4 sequences of 20 representative strains (8 porcine, 6 human, 6 bovine) were analysed to determine their G and P genotypes (Table 2).
Table 2.
Molecular combinations of G and [P] types of reference rotavirus strains detected between 2012 and 2013 in NE Indian children and animal neonates with acute diarrhoea
| Strain | Host | VP7 sequences | VP4 sequences | Genotype combination | ||
|---|---|---|---|---|---|---|
| GenBank accession no. | G types | GenBank accession no. | P types | |||
| IA-06RVB | Bovine | KJ701402 | G1 | KJ701369 | P[7] | G1P[7] |
| IA-1009RVB | Bovine | KJ701401 | G1 | KJ701370 | P[7] | G1P[7] |
| IA-C5RVP | Porcine | KJ701389 | G1 | KJ701371 | P[7] | G1P[7] |
| IA-981RVB | Bovine | KJ701403 | G1 | KJ701372 | P[7] | G1P[7] |
| IA-C4RVP | Porcine | KJ701390 | G1 | KJ701373 | P[7] | G1P[7] |
| IA-970RVB | Bovine | KJ701391 | G1 | KJ701374 | P[7] | G1P[7] |
| IA-09RVP | Porcine | KJ701404 | G1 | KJ701375 | P[13] | G1P[13] |
| IA-14RVP | Porcine | KJ701399 | G1 | KJ701376 | P[13] | G1P[13] |
| IA-1035RVH | Human | KJ701396 | G1 | KJ701377 | P[8] | G1P[8] |
| IA-47RVP | Porcine | KJ701397 | G1 | KJ701378 | P[8] | G1P[8] |
| IA-967RVB | Bovine | KJ701394 | G1 | KJ701379 | P[23] | G1P[23] |
| IA-1013RVP | Porcine | KJ701400 | G1 | KJ701380 | P[23] | G1P[23] |
| IA-1034RVH | Human | KJ701406 | G5 | KJ701381 | P[8] | G5P[8] |
| IA-49RVH | Human | KJ701398 | G1 | KJ701382 | P[8] | G1P[8] |
| IA-1019RVH | Human | KJ701408 | G9 | KJ701383 | P[8] | G9P[8] |
| IA-911RVH | Human | KJ701392 | G1 | KJ701384 | P[8] | G1P[8] |
| IA-509RVH | Human | KJ701393 | G1 | KJ701385 | P[8] | G1P[8] |
| IA-15RVP | Porcine | KJ701405 | G1 | KJ701386 | P[13] | G1P[13] |
| IA-919RVP | Porcine | KJ701407 | G3 | KJ701387 | P[7] | G3P[7] |
| IA-77RVB | Bovine | KJ701395 | G1 | KJ701388 | P[7] | G1P[7] |
Genotype distribution
The current study reported four different RVA G genotypes G1, G3, G5 and G9 circulating in humans and animals in NE India (Table 2). The common G1 genotype (85%, 17/20) was detected from four humans (IA-49RVH, 509RVH, 911RVH, 1035RVH), six bovine (IA-06RVB, 1009RVB, 981RVB, 970RVB, 967RVB, 77RVB) and seven porcine (IA-C5RVP, C4RVP, 09RVP, 14RVP, 47RVP, 1013RVP, 77RVB) specimens. This was followed by detection of a single G9 (IA-1019RVH) and G5 (IA-1034RVH) strain, each from a human specimen along with a single G3 strain (IA-919RVP) from a porcine specimen.
Similarly, VP4 genotyping confirmed detection of common P[8] (35%, 7/20) strains from six human specimens (IA-1035RVH, 1034RVH, 49RVH, 1019RVH, 911RVH, 509RVH) and one porcine specimen (IA-47RVP). Genotype P[7] was detected from five bovine (IA-06RVB, 1009RVB, 981RVB, 970RVB, 77RVB) and three porcine (IA-C5RVP, C4RVP, 919RVP) (40%, 8/20) strains. Genotype P[13] (15%, 3/20) was characterized from three porcine RV strains (IA-14RVP, 1035RVH, 15RVP), and P[23] (10%, 2/20) was detected in two strains, each from a bovine (IA-967RVB) and a porcine (IA-1013RVP) specimen (Table 2). The genotyping data of regional RVA strains demonstrated combined G and P genotypes of G1P[8] (25%, 5/20) characterized from four human and one porcine strain; G1P[7] (35%, 8/20) from five bovine and three porcine strains; G1P[13] (15%, 3/20) from three porcine RVs; G1P[23] (10%, 2/20) from two strains (one bovine and one porcine); G3P[7] from a single porcine strain; G9P[8] and G5P[8], each from two different human RVs (5%, 1/20). Overall, we identified unusual G1P[8], G1P[7], G1P[13] and G1P[23] RV genotypes from porcine and bovine neonates, a rare G5P[8] genotype from an infected child and a few common RVA strains circulating in NE India (Table 2).
RVA VP7 and VP4 gene sequence analysis
Phylogenetic trees based on VP7 and VP4 sequences revealed clustering of regional RVA isolates with their homologous strains of different lineages isolated from various geographical regions (Figs 2 and 3). The G1 strains (irrespective of hosts) of lineage 1 shared 99% sequence identity with Indian human strains (RVA/Human-wt/IND/NIV-074150/2007/G1 and RVA/Human-wt/IND/NIV-0716541/2007/G1); however, the G1 isolates of lineage 2 shared 95% homology with a prototype Japanese human strain (RVA/Human-wt/JPN/AU19/1998/G1P[6]). The regional G3 porcine strain of lineage 4 showed 95% sequence homology and clustered with the prototype porcine strain RVA/Pig-wt/CHN/HeN3/2011/G3reported from China. The human G5 isolate of lineage 1 demonstrated 92% sequence similarity to a Chinese G5 porcine strain (RVA/Pig-wt/CHN/HLJsh-2/2011/G5) as well as with a Chinese human prototype strain (RVA/Human-wt/CHN/LL4260/2001/G5P[6]). The sequence analysis of the G9 isolate revealed high genetic similarity of 99% to its prototype USA strain RVA/Human-wt/USA/DC3/2009/G9P[8] of lineage 1.
Fig. 2.
(a–d) Phylogenetic trees of the VP7 gene. Trees were built using MEGA6 software with the maximum-likelihood method; bootstrap values (1000 replicates) of ⩾70% are shown at the branch nodes. The regional genotypes are highlighted in bold next to a solid circle (•): (a) G1, (b) G3, (c) G5, (d) G9.
Fig. 3.
(a–d) Phylogenetic trees of partial the VP4 gene. Trees were built using MEGA6 software with the maximum-likelihood method; bootstrap values (1000 replicates) of ⩾70% are shown at the branch nodes. The regional genotypes are highlighted in bold next to a solid circle (•): (a) P[8], (b) P[7], (c) P[13], (d) P[23].
Similarly, in the P[8] strains of diverse sub-genotypic lineages, regional human isolates were 99% homologous to human prototype strains RVA/Human-wt/PRY/3SR/2002/G9P[8] and RVA/Human-wt/PRY/11SR/2002/G1G9P[8] from Paraguay; RVA/Human-wt/BEL/BE00021/2007/G1P[8] from Belgium; RVA/Human-wt/CMR/MRC-DPRU1424/2009/G9P[8] from Cameroon; RVA/Human-wt/USA/2009727093/2009/G9P[8] and RVA/Human-wt/USA/DC8/2009/G9P[8] from the United States. Interestingly, besides the human isolates, the regional P[8] porcine strain RVA/Pig-wt/IND/IA-47RVP/2012/G1P[8] of lineage 4, clustered among human strains from countries like India, United States, Cameroon, Belgium, showed 99% sequence homology. The porcine and bovine P[7] RVAs of lineage 1 showed 92% genetic closeness to its prototype strains (RVA/Pig-wt/IND/RU172/2006/G12P[7] and RVA/Cow-lab/GBR/PP-1/G3P[7]) reported from India and Great Britain respectively. The regional porcine P[13] strains demonstrated 87% sequence similarity to their prototype Japanese strain (RVA/Pig-wt/JPN/pig9-112d/2003/G11P[13]) and prototype Indian strains (RVA/Pig-wt/IND/HP113/2007/G6P13 and RVA/Pig wt/IND/HP140/2007/G6P13). For the P type P[23], the isolated porcine strain had 87% sequence homology and clustered with the Brazilian prototype strain RVA/Pig-wt/BRA/PGRV16/2011/G5P[23] of lineage 1; however, the bovine P[23] strain showed 96% homology clustered among lineage 2 with a porcine strain (RVA/Pig-wt/CHN/NMTL/2008/G9P[23]) reported from China.
DISCUSSION
The present study was a sequence-based investigation of RVA G and P genotypes circulating in humans and animals in NE India. Globally and particularly in the Asian countries, RVA infections occur in the winter season. In NE India, RVA also peaked during winter as well as in the rainy season and children aged <2 years and animal neonates aged <2 months were most susceptible. Reports of RVA incidence in different geographical regions identified G1, G2, G3, G4, G9, P[8], P[4], and P[6] in human RV episodes [13, 19] and in combination, more than 90% of human RVA cases have been attributed to G1P[8], G2P[4], G3P[8], and G4P[8] genotypes. We also identified the common G1P[8] and G9P[8] genotypes in human RVA episodes along with an unusual G5P[8] genotype. Previous studies on eastern and NE Indian human RVAs detected G4, G9, G10, G12, P[4], P[6], P[7] and P[9] strains [16–18]; however, the majority of the strains were genetically distant with regional RVA isolates. Evidence of G1 RVAs in combination with a human P[8] VP4 specificity in animals was atypical with limited reports of its incidence in animal diarrhoeal cases worldwide [32–34]. The detection of certain unusual/rare RVA genotypes (G1P[8], G1P[7], G1P[13], G1P[23], G3P[7]) among the farm animals during the study was intriguing as there have been infrequent reports of their emergence from the region. The rationale behind such enormous diversity could be due to the expression of the natural variation of distinct RVA genotypes in RV-infected children or animal neonates over time particularly during mixed infections. The phenomenon was observed in the study sites, where animals were kept backyard in straw-lined sheds which allowed easy contact with waste matter from human handlers and vice versa. Besides, RVs are known for their environmental resilience and have also been detected in dust and dried faeces [35]. This suggests that close proximity contributed to the transmission of resident RVA strains to the incoming animals or their human handlers. Few reports from India have demonstrated evidence of zoonotic transmission of RVAs in diverse host species [16, 19, 36]. Our study also demonstrated many cases of zoonotic/interspecies transmission which might have been due to the close association of humans and animals. Thus, genetic reassortment of the RV genome during co-infection of a host is an important mechanism for generating viral diversity.
The cluster analysis of regional G and P genotypes revealed homology with isolates of different lineages from countries including China, Japan, USA, Cameroon, Canada, Brazil, Belgium and Paraguay. Phylogenetic analysis is establishing that these genotypes may emerge with immigration of people from such countries. The observed higher frequencies of RVA disease burden, with significant human health implications and considerable economic losses, can possibly be attributed to the absence of a national RV vaccination programme. Therefore, the emergence of divergent RV genotypes with different sub-genotypic lineages in NE Indian human and animal neonates underscores the need for a national RV vaccination programme. Although the existing RV vaccines do not cover the unusual/uncommon genotypes, the efficiency of these vaccines against diverse RVA genotypes has been established [37, 38]. Of note, an important limitation of this study is the sampling design, in which five/six provinces of NE India were explored. This clearly represented a small fraction of the population and, again, most likely underestimates the true RVA variability present in Indian humans and animals.
The availability of sequencing methods provided an alternative method to conventional PCR genotyping for the characterization of uncommon and non-typable strains. Overall, the study demonstrated extensive RV diversity and temporal/regional fluctuations in RVA genotype distribution in the three hosts, i.e. humans, cattle and pigs. The identification of unusual RVA genotypes from humans and animals sharing close settings provides evidence of interspecies transmission based on VP4 and VP7 sequencing. Advance studies with comprehensive analysis of whole genomic sequences will be required to continuously monitor distribution of novel/unusual RVA genotypes in the region. Continuous surveillance of RVA genotypes in humans and farm animals along with a regular RV vaccination programme could lead to successful control of RVA-attributed gastroenteritis.
ACKNOWLEDGEMENTS
The authors are grateful to the patients and animal handlers for their willingness to participate in providing samples and information about themselves and their animals. We gratefully acknowledge Professor Sailendra Kumar Das, Department of Microbiology, College of Veterinary Science, Khanapara for providing the lyophilized standard rotavirus reference strain. We also acknowledge Professor Sankar Kumar Ghosh, Department of Biotechnology, Assam University, Silchar for providing nucleotide sequencing facilities.
DECLARATION OF INTEREST
None.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268816000807.
click here to view supplementary material
REFERENCES
- 1.Parashar UD, et al. Global illness and deaths caused by rotavirus disease in children. Emerging Infectious Disease 2003; 9: 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tayeb HT, et al. Increased prevalence of rotavirus among children associated gastroenteritis in Riyadh Saudi Arabia. Virology Journal 2011; 8: 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris MK, et al. Rotavirus mortality in India: estimates based on a nationally representative survey of diarrhoeal deaths. Bulletin of the World Health Organization 2012; 90: 720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fagiolo A, et al. Buffalo pathologies. In: Borghese A, ed. Buffalo Production and Research. FAO, Rome: 2005, pp. 249–296. [Google Scholar]
- 5.Barman NN, et al. Prevalence of rotavirus, transmissible gastroenteritis virus and porcine epidemic diarrhoea virus antibodies in pigs of Assam, India. Indian Journal of Animal Science 2003; 73: 576–578. [Google Scholar]
- 6.Mihalov-Kovács E, et al. Candidate new rotavirus species in sheltered dogs, Hungary. Emerging Infectious Disease 2015; 21: 660–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matthijnssens J, et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Archives of Virology 2011, 156: 1397–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trojnar E, et al. Identification of an avian group a rotavirus containing a novel VP4 gene with a close relationship to those of mammalian rotaviruses. Journal of General Virology 2013; 94: 136–142. [DOI] [PubMed] [Google Scholar]
- 9.Matthijnssens J, et al. Uniformity of rotavirus strain nomenclature proposed by the rotavirus classification working group (RCWG). Archives of Virology 2011; 156: 1397–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iturriza-Gomara M, et al. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiology and Infection 2011; 139: 895–909. [DOI] [PubMed] [Google Scholar]
- 11.Martella V, et al. Zoonotic aspects of rotaviruses. Veterinary Microbiology 2010; 140: 246–255. [DOI] [PubMed] [Google Scholar]
- 12.Gentsch JR, et al. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. Journal of Infectious Disease 2005; 1: 146–1159. [DOI] [PubMed] [Google Scholar]
- 13.Santos N, Hoshino Y. Global distribution of rotavirus serotypes/ genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Reviews in Medical Virology 2005; 15: 29–56. [DOI] [PubMed] [Google Scholar]
- 14.Samajdar S, et al. Changing pattern of human group A rotaviruses: emergence of G12 as an important pathogen among children in eastern India. Journal of Clinical Virology 2006; 36: 183–188. [DOI] [PubMed] [Google Scholar]
- 15.Samajdar S, et al. Increase in prevalence of human group A rotavirus G9 strains as an important VP7 genotype among children in eastern India. Journal of Clinical Virology 2008; 43: 334–339. [DOI] [PubMed] [Google Scholar]
- 16.Mukherjee A, et al. Surveillance and molecular characterization of rotavirus strains circulating in Manipur. North-Eastern India: increasing prevalence of emerging G12 strains. Infection Genetics and Evolution 2010; 10: 311–320. [DOI] [PubMed] [Google Scholar]
- 17.Mullicka S, et al. , Hospital based surveillance and genetic characterization of rotavirus strains in children (<5 years) with acute gastroenteritis in Kolkata, India, revealed resurgence of G9 and G2 genotypes during 2011–2013. Vaccine 2014; 32S: A20–A28. [DOI] [PubMed] [Google Scholar]
- 18.Chakraborty P, et al. Restriction fragment length polymorphism analysis of rotavirus VP7-encoding gene from humans and animals of Northeast India: a relative study of Indian and global isolates. Epidemiology and Infection 2015; 143: 2503–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullick S, et al. Genomic analysis of human rotavirus strains G6P[14] and G11P[25] isolated from Kolkata in 2009 reveals interspecies transmission and complex reassortment events. Infection, Genetics and Evolution 2013; 14: 15–21. [DOI] [PubMed] [Google Scholar]
- 20.Linhares AC, et al. Efficacy and safety of an oral live attenuated human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in Latin American infants: a randomized, double-blind, placebo-controlled phase III study. Lancet 2008; 371: 1181–1189. [DOI] [PubMed] [Google Scholar]
- 21.Ciarlet M, Schödel F. Development of a rotavirus vaccine: clinical safety, immunogenicity, and efficacy of the pentavalent rotavirus vaccine, RotaTeq. Vaccine 2009; 27 (Suppl. 6): G72–G81. [DOI] [PubMed] [Google Scholar]
- 22.Matthijnssens J, et al. Rotavirus disease and vaccination: impact on genotype diversity. Future Microbiology 2009; 4: 1303–1316. [DOI] [PubMed] [Google Scholar]
- 23.Patel MM, Parashar UD. Assesing the effectiveness and public health impact of rotavirus vaccines after introduction in immunization program. Journal of Infectious Disease 2009; 200 (S1): S291–299. [DOI] [PubMed] [Google Scholar]
- 24.Gouvea V, et al. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. Journal of Clinical Microbiology 1990; 28: 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentsch JR, et al. Identification of group A rotavirus gene 4 types by polymerase chain reaction. Journal of Clinical Microbiology 1992; 30: 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JD, et al. Multiple sequence alignment using ClustalW and ClustalX. Current protocols in bioinformatics/editorial board. Baxevanis AD, et al. 2002; chapter 2: unit 2·3. [DOI] [PubMed]
- 27.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 1997; 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaffer AA, et al. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Research 2001; 29: 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maes P, et al. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiology 2009; 9: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 1993; 10: 512–526. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 2013; 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciarlet M, Liprandi F. Serological and genomic characterization of two porcine rotaviruses with serotype G1 specificity. Journal of Clinical Microbiology 1994; 32: 269–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blackhall JR, et al. A bovine rotavirus serotype 1: characterization of the virus and nucleotide sequence determination of the structural glycoprotein VP7 gene. Virology Journal 1992; 189: 833–837. [DOI] [PubMed] [Google Scholar]
- 34.Yuan L, et al. Rotavirus and reovirus. In: Straw B, Zimmerman JJ, D'Allaire S, Taylor DJ, eds. Diseases of Swine. Ames: Blackwell Publishing, 2006, pp. 435–454. [Google Scholar]
- 35.da Silva Soares L, et al. Diversity of rotavirus strains circulating in Northern Brazil after introduction of a rotavirus vaccine: high prevalence of G3P[6] genotype. Journal of Medical Virology 2013; 86: 1065–1072. [DOI] [PubMed] [Google Scholar]
- 36.Varshney B, et al. Prevalence of, and antigenic variation in, serotype G3 strains in diarrheic calves: implications for the origin of G10P11 or P11 type reassortant asymptomatic strains in newborn children in India. Archives of Virology 2002; 147: 143–165. [DOI] [PubMed] [Google Scholar]
- 37.Lamabadusuriya SP. Improving child survival through immunisation. Ceylon Medical Journal 1993; 38: 170–171. [PubMed] [Google Scholar]
- 38.Rose TL, et al. Rotavirus a genotype G1P [8]: a novel method to distinguish wild-type strains from the Rotarix vaccine strain. Memórias do Instituto Oswaldo Cruz 2010; 105: 1068–1072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268816000807.
click here to view supplementary material