Abstract
OBJECTIVE:
As data science and artificial intelligence continue to rapidly gain traction, the publication of freely available ICU datasets has become invaluable to propel data-driven clinical research. In this guide for clinicians and researchers, we aim to: 1) systematically search and identify all publicly available adult clinical ICU datasets, 2) compare their characteristics, data quality, and richness and critically appraise their strengths and weaknesses, and 3) provide researchers with suggestions, which datasets are appropriate for answering their clinical question.
DATA SOURCES:
A systematic search was performed in Pubmed, ArXiv, MedRxiv, and BioRxiv.
STUDY SELECTION:
We selected all studies that reported on publicly available adult patient-level intensive care datasets.
DATA EXTRACTION:
A total of four publicly available, adult, critical care, patient-level databases were included (Amsterdam University Medical Center data base [AmsterdamUMCdb], eICU Collaborative Research Database eICU CRD], High time-resolution intensive care unit dataset [HiRID], and Medical Information Mart for Intensive Care-IV). Databases were compared using a priori defined categories, including demographics, patient characteristics, and data richness. The study protocol and search strategy were prospectively registered.
DATA SYNTHESIS:
Four ICU databases fulfilled all criteria for inclusion and were queried using SQL (PostgreSQL version 12; PostgreSQL Global Development Group) and analyzed using R (R Foundation for Statistical Computing, Vienna, Austria). The number of unique patient admissions varied between 23,106 (AmsterdamUMCdb) and 200,859 (eICU-CRD). Frequency of laboratory values and vital signs was highest in HiRID, for example, 5.2 (±3.4) lactate values per day and 29.7 (±10.2) systolic blood pressure values per hour. Treatment intensity varied with vasopressor and ventilatory support in 69.0% and 83.0% of patients in AmsterdamUMCdb versus 12.0% and 21.0% in eICU-CRD, respectively. ICU mortality ranged from 5.5% in eICU-CRD to 9.9% in AmsterdamUMCdb.
CONCLUSIONS:
We identified four publicly available adult clinical ICU datasets. Sample size, severity of illness, treatment intensity, and frequency of reported parameters differ markedly between the databases. This should guide clinicians and researchers which databases to best answer their clinical questions.
Keywords: critical care, data science, data set, guide, ICU, systematic review
Intensive care medicine has long been at the forefront of clinical data science. This is facilitated by unique factors that include the automated, digital capture of vital signs or laboratory measurements and comprehensive capture of a patient’s physiologic state due to frequent recording of observations and interventions (1). Taken together, this results in high-resolution, high-quality, large-scale data enabling research questions that were previously challenging in terms of experimental design to be addressed. Indeed, intensive care medicine has often been called a natural habitat for developing artificial intelligence and, particularly, machine learning (2).
Historically, the early use of medical devices for monitoring started in the late 1960s and 1970s (3, 4). Over time, data availability has advanced due to an ever-increasing number of physiologic monitoring and intervention devices. As data availability continued to increase in subsequent decades, medical data science was significantly propelled by the worldwide web—which simplified data sharing—as well as the publication of electronic patient data through platforms such as PhysioNet (5). In the new millennium, the publication of the Medical Information Mart for Intensive Care (MIMIC) (6) has been a landmark in intensive care medicine and data science. MIMIC continuous to be expanded and improved over time and currently also captures waveform data, clinical notes, radiology images, and emergency department data. Together, all MIMIC versions have resulted in over a thousand scientific publications and conference proceedings so far. More recently, additional ICU databases have been published, thus providing alternative datasets for clinicians and data scientists to study (7–9). With the emergence of more clinical datasets, selecting the right data for the right clinical question is key (10). From a data perspective, some considerations include the setting (e.g., single-center vs multicenter data, academic vs communal hospital, and geographic location), data capture (i.e., fully automated or manual input), data completeness, as well as data richness. From a clinical perspective, researchers should consider the observational nature of the data and be mindful of statistical and epidemiological pitfalls, including selection bias (which patients get admitted to the ICU), temporal biases, or patient data not missing at random (11). In case of single-center data, researchers should recognize the limitations to generalizability, as patient population, local protocols, and treatment outcomes may diverge from (inter-) national standards and averages.
In this guide for clinicians and researchers, we aim to: 1) systematically search and identify all publicly available adult clinical ICU datasets, 2) compare their characteristics, data quality, and richness and critically appraise their strengths and weaknesses, and 3) provide clinicians and researchers with suggestions which datasets are appropriate for answering their clinical questions.
MATERIALS AND METHODS
Search Strategy and Inclusion Criteria
A systematic search was conducted to identify all relevant publications using: 1) publicly available, 2) adult, 3) patient-level, and 4) intensive care datasets. Where possible, the Meta-analysis Of Observational Studies in Epidemiology (MOOSE) criteria were followed as closely as possible, with some deviation as no classical meta-analysis was performed (12). The study protocol was drafted beforehand and registered on PROSPERO (https://www.crd.york.ac.uk/prospero/display_record.php?RecordID=223377) (13). The search terms were validated by a medical librarian (K.A.Z.); PubMed, ArXiv, MedRxiv, and BioRXiv were searched for relevant literature from inception to November 1, 2021. The full search strategy has been published on PROSPERO (13). Any types of publications were included. We excluded studies published in languages other than English, German, Dutch, and French. Last, a Google scholar search was performed to check for any databases that might not have been identified otherwise.
Data Extraction and Analysis
All hits were screened independently by two of the authors (C.M.S., T.A.D.). If deemed relevant after initial screening, full texts were reviewed by the authors. If disagreement on inclusion arose, reasons were discussed, and all issues were resolved between the two screening authors. No tie-breaking vote by a third author (P.E.) as defined in the protocol was required. All databases deemed relevant were included in the analysis. Access to databases was requested by the authors, and all necessary legal and ethical approvals were obtained.
A list of over 40 database elements was a priori defined and subsequently compiled for all databases. Detailed description of characteristics is available in Supplementary Table 1 (http://links.lww.com/CCM/H84). Database characteristics were, if available, extracted from the website of the datasets or accompanying publications, or otherwise extracted from the data set. Information on ICU patients, records, and laboratory measurements was compiled by the author team and results validated with the database administrators (L.A.C., M.F., P.J.T.). Data were extracted through Google BigQuery (14) and PgAdmin Version 4.24 (15) using both previously published or newly written queries. All code can be accessed through the following GitHub repository: https://github.com/tariqdam/review-ICU-datasets. Auxiliary data analysis steps were performed using R (16). Data analysis focused on descriptive statistics, that is, mean, median (for skewed data), sd, and frequency.
RESULTS
A total of 345 publications were identified from PubMed and preprint servers. After title and abstract screening, a total of 269 hits were full-text screened for eligibility. Main reason for exclusion was irrelevance, experimental data, nonpublic databases, COVID-19 data only, and neonatal/pediatric data only. After removal of duplicate publications and earlier versions of the same database (e.g., MIMIC-III), four unique databases were identified (Fig. 1). A Google scholar search did not result in any additional hits. Two additional datasets were found, yet these were not eligible (one billing codes only and one neonatal patients only). Studies included in the final analysis were those reporting on MIMIC-IV (Version 0.4, 2020 [17]), eICU Collaborative Research Database (eICU-CRD) (Version 2.0, 2019 [8]), Amsterdam University Medical Center data base (AmsterdamUMCdb) (Version 1.02, 2020 [9]), and High time-resolution intensive care unit dataset (HiRID) (Version 1.1, 2021 [7]).
Figure 1.
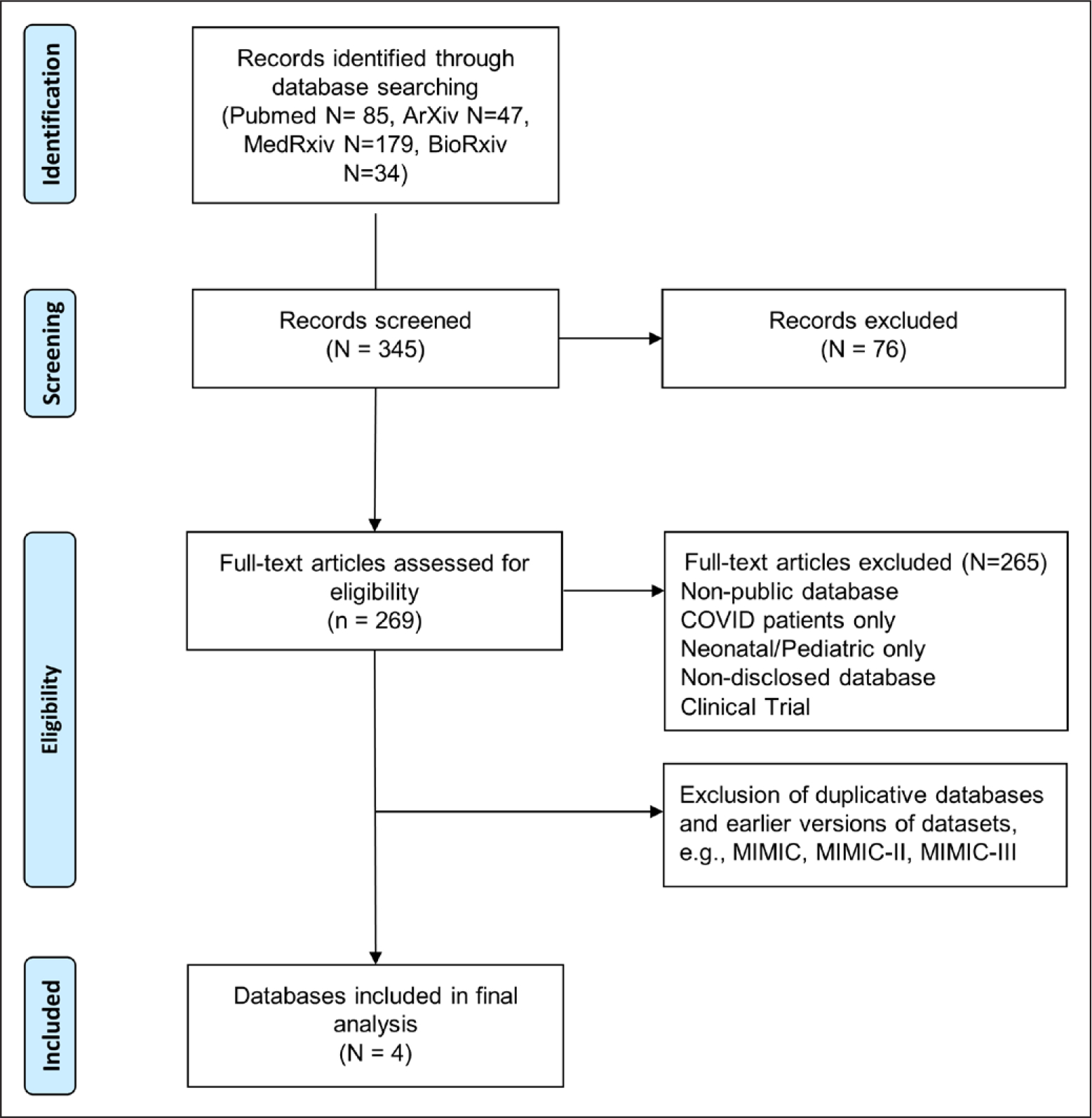
Step-by-step results of the systematic search and article selection process. After excluding nonrelevant databases and duplicates, a total of 4 databases was retrieved. Reporting based on Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (18). MIMIC = Medical Information Mart for Intensive Care.
Database characteristics, ICU patient characteristics, outcomes, and frequency of laboratory measurements and vital records were extracted as a priori defined. Results are summarized in Table 1 and visualized in Figure 2. Among the four datasets included, the number of admitted patients varied considerably, with AmsterdamUMCdb being the smallest (~23k patients) and eICU-CRD being the largest (~139k patients). Severity of illness as well as outcomes were also significantly different, as visible from divergent rates of mechanical ventilation (21.0–83.0%) and major differences in ICU mortality rates (5.5–9.9%). Although all databases contained vital sign recordings, laboratory measurements, and administered drugs, frequency of reporting differed. HiRID tended to have the most frequent vital sign recordings, with AmsterdamUMCdb coming in second, whereas MIMIC-IV has the lowest (31.7 ± 10.2 vs 17.0 ± 29.8 vs 1.1 ± 0.4 mean heart rate measurements per hour, respectively). Frequency of laboratory measurements was highest in HiRID for all studied measurements. For instance, serum sodium is reported with a mean frequency of 5.1 ± 3.4 times per day, whereas this was sparser in eICU-CRD (1.6 ± 1.5 times per day). Depending on the laboratory measurement, either MIMIC-IV or AmsterdamUMCdb had the second highest frequency per day. We summarized key epidemiological considerations, strengths, and weaknesses of all included databases in Table 2.
TABLE 1.
Comparison of Key ICU Database Characteristics
| Characteristics | Amsterdam UMCdb | eICU-CRD | HiRID | MIMIC-IV |
|---|---|---|---|---|
| Number of centers | 1 | 208 | 1 | 1 |
| Center location | Amsterdam, the Netherlands | United States | Bern, Switzerland | Boston, United States |
| Time period | 2003–2016 | 2014–2015 | 2005–2016 | 2008–2019 |
| ICU unique patient counta | 20,109 | 139,367 | 33,905b | 50,048 |
| ICU admissions/unique patient | 100% | 120.7% | 100.00% | 139.1% |
| ICU patient age, median (IQR) | 60–69 (50–59, 70–79)c | 65 (53–76) | 65 (55–75) | 64 (51–70) |
| Gender: male | 63.6% | 54.0% | 64.2% | 56.1% |
| Ethnicity: White | Not reported | 77.2% | Not reported | 65.6% |
| Ethnicity: African-American | Not reported | 10.6% | Not reported | 9.0% |
| Ethnicity: Other/unknown | Not reported | 12.1% | Not reported | 24.4% |
| Mortality: ICU | 9.9% | 5.5% | 6.1% | 8.6% |
| Mortality: ICU elective patients | 6.8% | 1.9% | Not reported | 4.3% |
| Mortality: ICU urgent patients | 18.3% | 6.2% | Not reported | 8.9% |
| Mortality: hospital | 13.3% | 9.0% | Not reported | 14.8% |
| Mortality: 28 d | 13.6%d | Not reported | Not reported | 11.6%d |
| Length of stay ICU, median (sd) | 1.0 (0.8–3.1) | 1.6 (0.8–3.0) | 1.0 (0.8–2.2) | 2.0 (1.1–3.9) |
| Patients with > 1 comorbidity | Not reported | 87.8% | Not reported | 83.8% |
| Severity of illness scores | APACHE II, SOFA | APACHE IV, APACHE IVa | APACHE II | APACHE III, Oxford Acute Severity of Illness Score, SOFA |
| APACHE admission, median (IQR) | APACHE II: 17 (13–22) | APACHE IVa: 51 (37–68) | APACHE II: 16 (12–22) | APACHE III: 41 (30–57) |
| SOFA first 24 hr, median (IQR) | 7 (4–9) | Not reported | Not reported | 2 (1–5) |
| Availability of radiology images | Not reported | Not reported | Not reported | Yese |
| Availability of clinical notes | Not reported | Deconstructed notes available | Not reported | Not publicly available |
AmsterdamUMCdb = Amsterdam University Medical Center data base, APACHE = Acute Physiology and Chronic Health Evaluation, eICU-CRD = eICU Collaborative Research Database, HiRID = High time-resolution intensive care unit dataset, MIMIC = Medical Information Mart for Intensive Care, SOFA = Sequential Organ Failure Assessment.
Proportions above 100% indicate multiple ICU admissions per patient.
Admissions received unique patient ids.
Median due to age categorization.
Inhospital mortality only.
Through Medical Information Mart for Intensive Care-CXR Database Version 2.0.
Overview of the database design and key outcomes. Frequencies are reported as mean (sd).
Figure 2.
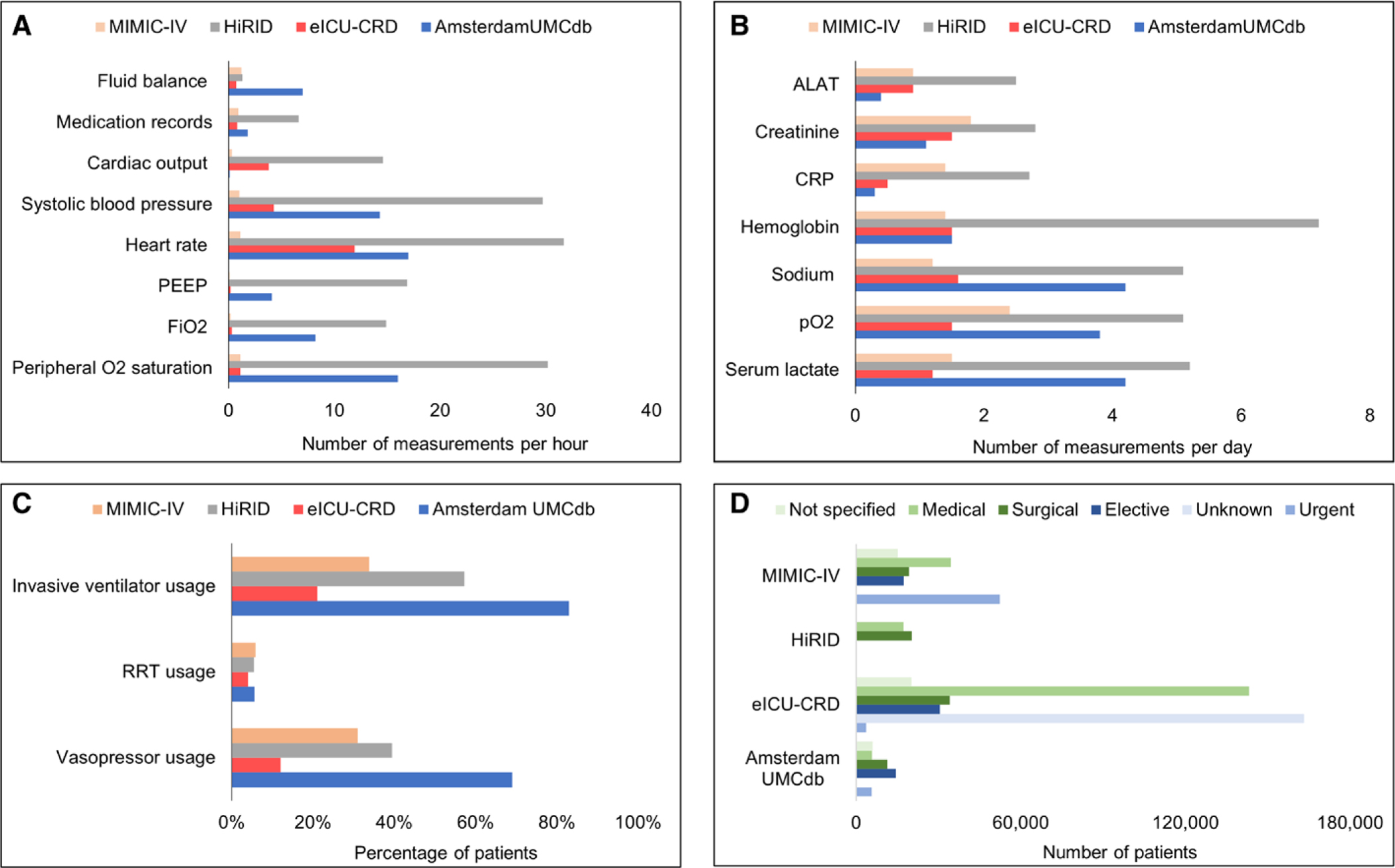
Visual comparison of ICU patient characteristics for each data set. A, Mean frequency of vital sign measurements per patient per hour. B, Mean frequency of laboratory measurements per patient per day. C, Percentage of patients receiving invasive treatments. D, Number of patients by reason for admission (green) and urgency of admission (blue). Standard deviations are available from Supplementary Table 2 (http://links.lww.com/CCM/H85). ALAT = alanine aminotransferase, AmsterdamUMCdb = Amsterdam University Medical Center data base, APACHE = Acute Physiology and Chronic Health Evaluation, CRP = C-reactive protein, eICU-CRD = eICU Collaborative Research Database, HiRID = High time-resolution intensive care unit dataset, MIMIC = Medical Information Mart for Intensive Care, PEEP = positive end-expiratory pressure, RRT = renal replacement therapy.
TABLE 2.
Subjective Summary of Key Epidemiological Considerations, Strengths, and Weaknesses of the ICU Databases
| Database, Location | Key Epidemiological Considerations | Key Strengths | Key Weaknesses |
|---|---|---|---|
|
| |||
| Medical Information Mart for Intensive Care-IV, Boston, MA | Highest proportion of urgent admissions (75%) | Most established data set Large GitHub repository with many well-established views |
Single, academic center |
| Mostly medical ICU patients (50%) | Includes electronic health record data, radiology images, and non-ICU labs | ||
|
| |||
| eICU-CRD, United States | Telemonitoring–only data set including community hospitals | Largest data set (~139k) | Data quality varies by center due to reporting differences |
| Least invasively treated patient cohort, i.e., vasopressors (12%) and mechanical ventilation (21%) | Multicenter database (208 centers) | Least rich data set | |
| Least sick patient cohort with lowest ICU mortality rate (5.5%) | |||
| Mostly medical ICU patients (73%) | |||
|
| |||
| Amsterdam UMCdb, Amsterdam, The Netherlands | Most invasively treated patient cohort, i.e., vasopressors (69%) and mechanical ventilation (83%) | Includes non-ICU labs | Single academic center |
| Sickest patient cohort with highest ICU mortality rate (9.9%) | No medical history available | ||
| Mostly surgical ICU patients (49%) | Smallest data set (~20k) | ||
|
| |||
| HiRID, Bern, Switzerland | Similar split surgical/medical ICU patients (54%/46%) | Least explored data set Highest frequency of laboratory measurements and vital signs |
Single academic center No medical history available |
Being the first published and widely used available data set, MIMIC-IV comes with an extensive online GitHub repository (https://github.com/) that includes numerous summarized views and code for many concepts. The latest version also captures laboratory measurements beyond the ICU stay, radiology images, and waveforms, thus making it the most comprehensive data set. Its key weakness is that it is derived from a single center.
On the other hand, eICU-CRD is a multicenter data set that includes more than 200 hospitals across the United States. Therefore, admission and treatment policies vary considerably across the ICUs, which can be an advantage for studies trying to identify optimal treatment policies. This variation also results in heterogeneous data reporting and less data richness.
AmsterdamUMCdb meanwhile has overall high data richness yet is also single center and has the smallest unique patient cohort of the included datasets (n = 20,109). Lack of medical history is a key weakness. Patients tend to be sickest and the most invasively treated in terms of mechanical ventilation and vasopressor use.
HiRID is the most recent published data set and thus the least explored. It is similar to the other European, single-center database, AmsterdamUMCdb, and also does not capture medical history. Each admission is assigned a separate patient ID, making it impossible to link patients across time, for example, to identify readmissions. Key advantages are high resolution in the time domain with monitoring data reported every 2 minutes and frequent laboratory measurements.
Other considerations for the choice of an appropriate data set include the availability of clinical notes (all datasets), loss of information due to deidentification (e.g., year of admission in MIMIC-IV), or availability of emergency department data (only MIMIC-IV). Overall, we judge all datasets to be readily accessible and easy to handle, with all being downloadable, available in Google BigQuery (14), and even accessible in R (16) directly though the ricu R package (19). No major differences in the credentialing process exist, with, for example, the Collaborative Institutional Training Initiative (CITI) human subjects research course (20) being accepted, although AmsterdamUMCdb requires a practicing intensivist as a reference. Unfortunately, no data set includes out-of-hospital mortality data. The individual datasets have been described in detail previously (7–9, 17, 21).
DISCUSSION
In this systematic review and comparison of publicly available adult ICU databases, we identified and analyzed four datasets. Although there is significant overlap on captured data elements, overall data richness, and quality, major differences between the datasets were identified. Patient characteristics seem to reflect differences in ICU organization, admission criteria, and treatment characteristics resulting in differences between countries and centers. Of note, all datasets are derived from highly developed, Western countries with Caucasian race majorities; thus, generalization to other countries worldwide might be limited. It must be emphasized that models and findings based on analysis of one or more of these datasets will not necessarily translate to other ICUs, or even within the same ICU where the data came from some other time period. Differences in practice patterns across ICUs and changes in treatment protocols within the same ICU over time limit generalizability across space and time. Ideally, as with clinical trials and observational studies, analyses of electronic health record datasets are locally and regularly performed to ascertain validity of previously published findings and models (22). Furthermore, there is a difference in the frequency of data capture of some features, such as laboratory measurements or vital signs (see Table 1). Taken together, these differences result in datasets with distinct strengths and weaknesses, as listed in Table 2.
Prior research already described how each of the databases was built, outlined their structures, and provided a limited, summary of patient characteristics (8, 23–25). To the best of our knowledge, no prior comprehensive comparison of ICU datasets has been performed. Of note, the definitions used in this article can differ from prior publications (8, 9, 21), as different researchers may have used slightly different definitions or querying approaches. Subsequently, results and values presented here should not be considered to be ground truth. Rather, our approach may be viewed as a reasonable strategy to compare relevant metrics across datasets.
When considering one of the datasets to address clinical research questions, the research question should be central in determining which data set to use. Considerations include the substantial differences in cohort size, treatment intensity, and mortality rates. In particular, we hope that researchers will become aware of shortcomings in the data early, so that missing data, for example, prior medical history, post-ICU mortality data, or unreported measurements, will not derail research efforts. Based on the divergent patient populations and treatment policies described above, we would argue that researchers should use at least two datasets to determine generalizability of the models. In general, we suggest eICU-CRD to be used where generalizability and variability are major concerns or when trying to research rare phenomena. HiRID and AmsterdamUMCdb are good choices for studies where granular and frequent measurements are required. MIMIC-IV is a particular good choice for starters or those looking for well-established concepts to be leveraged.
Researchers frequently try to find novel associations or even try to establish causal relationships from observational data. When using unfamiliar datasets, they should be aware and understand intrinsic limitations, biases, and practice differences embedded in the data. Although a detailed description of these is beyond the scope of this study, it should be highlighted that local ICU triage decision and treatment protocols will bias results and should be considered. If in doubt, authors should reach out to the database administrators for clarification.
CONCLUSION
We describe key features of publicly available adult ICU datasets and provide a summary of patient characteristics, data richness, and structural strengths and limitations. We hope that this article will guide researchers to decide on which database(s) to use to answer their clinical question. We believe that both intensive care medicine and data science will benefit from more high-resolution ICU datasets that also include pre-and postdischarge data, and add quality-of-life measurements and pre- and post-ICU hospital data.
Supplementary Material
Acknowledgments
Dr. Dam received funding from AmsterdamUMC and the Netherlands Organization for Health Research and Development (project number 10430012010003). Dr. Celi received support for article research from the National Institutes of Health. Dr. Faltys received funding from the Swiss National Fund. The remaining authors have disclosed that they do not have any potential conflicts of interest.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccmjournal).
REFERENCES
- 1.Dauvin A, Donado C, Bachtiger P, et al. : Machine learning can accurately predict pre-admission baseline hemoglobin and creatinine in intensive care patients. NPJ Digit Med 2019; 2:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fleuren LM, Dam TA, Tonutti M, et al. : The Dutch Data Warehouse, a multicenter and full-admission electronic health records database for critically ill COVID-19 patients. Crit Care 2021; 25:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caceres CA: Telemetry in medicine and biology. Adv Biomed Eng Med Phys 1968; 1:279–316 [PubMed] [Google Scholar]
- 4.Farrier RM: Electronic monitoring of the critically ill. Mil Med 1964; 129:343–348 [PubMed] [Google Scholar]
- 5.Moody GB, Mark RG, Goldberger AL: PhysioNet: A research resource for studies of complex physiologic and biomedical signals. Comput Cardiol 2000; 27:179–182 [PubMed] [Google Scholar]
- 6.Saeed M, Villarroel M, Reisner AT, et al. : Multiparameter intelligent monitoring in intensive care II: A public-access intensive care unit database. Crit Care Med 2011; 39:952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faltys M, Zimmermann M, Lyu X, et al. : HiRID, a high time-resolution ICU dataset. Available at: https://physionet.org/content/hirid/1.1.1. Accessed August 23, 2021
- 8.Pollard TJ, Johnson AEW, Raffa JD, et al. : The eICU collaborative research database, a freely available multi-center database for critical care research. Sci Data 2018; 5:180178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thoral PJ, Peppink JM, Driessen RH, et al. ; Amsterdam University Medical Centers Database (AmsterdamUMCdb) Collaborators and the SCCM/ESICM Joint Data Science Task Force: Sharing ICU patient data responsibly under the Society of Critical Care Medicine/European Society of Intensive Care Medicine Joint Data Science Collaboration: The Amsterdam University Medical Centers Database (AmsterdamUMCdb) example. Crit Care Med 2021; 49:e563–e577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosgriff CV, Celi LA, Stone DJ: Critical care, critical data. Biomed Eng Comput Biol 2019; 10:1179597219856564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan W, Beaulieu-Jones BK, Yu KH, et al. : Temporal bias in case-control design: Preventing reliable predictions of the future. Nat Commun 2021; 12:1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stroup DF, Berlin JA, Morton SC, et al. : Meta-analysis of observational studies in epidemiology: A proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000; 283:2008–2012 [DOI] [PubMed] [Google Scholar]
- 13.Sauer C, Elbers P, Dam T: Comparison of publicly available ICU data sets - a guide for clinicians and data scientists. PROSPERO 2021; CRD42021223377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.BigQuery – Google Cloud Platform. Available at: https://console.cloud.google.com/bigquery. Accessed August 23, 2021
- 15.pgadmin.org: pgAdmin - PostgreSQL Tools. Available at: https://www.pgadmin.org/. Accessed August 23, 2021
- 16.R Core Team: R: A language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2020. Available at: https://www.R-project.org. Accessed August 23, 2021 [Google Scholar]
- 17.Johnson A, Bulgarelli L, Pollard T, et al. : MIMIC-IV (version 1.0). PhysioNet. 2021. Available at: https://physionet.org/content/mimiciv/0.4/. Accessed May 5, 2021
- 18.Moher D, Liberati A, Tetzlaff J, et al. ; PRISMA Group: Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med 2009; 6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett N, Plečko D, Ukor I-F, et al. : ricu: R’s interface to intensive care data. arXiv:210800796 [q-bio, stat]. 2021. Available at: http://arxiv.org/abs/2108.00796. Accessed October 28, 2021
- 20.CITI PROGRAM: CITI - Collaborative Institutional Training Initiative. 2021. Available at: https://www.citiprogram.org/index.cfm?pageID=154&icat=0&ac=0. Accessed October 28, 2021
- 21.O’Halloran HM, Kwong K, Veldhoen RA, et al. : Characterizing the patients, hospitals, and data quality of the eICU collaborative research database. Crit Care Med 2020; 48: 1737–1743 [DOI] [PubMed] [Google Scholar]
- 22.Futoma J, Simons M, Panch T, et al. : The myth of generalisability in clinical research and machine learning in health care. Lancet Digit Health 2020; 2:e489–e492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyland SL, Faltys M, Hüser M, et al. : Early prediction of circulatory failure in the intensive care unit using machine learning. Nat Med 2020; 26:364–373 [DOI] [PubMed] [Google Scholar]
- 24.Johnson AE, Pollard TJ, Shen L, et al. : MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3: 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vistisen ST, Pollard TJ, Enevoldsen J, et al. : VitalDB: Fostering collaboration in anaesthesia research. Br J Anaesth 2021; 127:184–187 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


