SUMMARY
Preliminary evidence suggests that direct poultry contact may play a lesser role in transmission of avian influenza A(H7N9) than A(H5N1) to humans. To better understand differences in risk factors, we quantified the degree of poultry contact reported by H5N1 and H7N9 World Health Organization-confirmed cases. We used publicly available data to classify cases by their degree of poultry contact, including direct and indirect. To account for potential data limitations, we used two methods: (1) case population method in which all cases were classified using a range of sources; and (2) case subset method in which only cases with detailed contact information from published research literature were classified. In the case population, detailed exposure information was unavailable for a large proportion of cases (H5N1, 54%; H7N9, 86%). In the case subset, direct contact proportions were higher in H5N1 cases (70·3%) than H7N9 cases (40·0%) (χ2 = 18·5, P < 0·001), and indirect contact proportions were higher in H7N9 cases (44·6%) than H5N1 cases (19·4%) (χ2 = 15·5, P < 0·001). Together with emerging evidence, our descriptive analysis suggests direct poultry contact is a clearer risk factor for H5N1 than for H7N9, and that other risk factors should also be considered for H7N9.
Key words: Avian influenza, epidemiology, exposure, H5N1, H7N9
INTRODUCTION
Two novel influenza A viruses of direct avian origin have emerged in the past 20 years as unexpected causes of severe human disease. Influenza A(H5N1) emerged in humans in 1997 in Hong Kong SAR and has spread over several continents in bird and human populations, with over 840 World Health Organization (WHO)-confirmed cases to date [1]. Influenza A(H7N9) emerged in February 2013 in China and has now caused over 680 WHO-confirmed cases, with human and avian detections restricted mainly to China [1].
A significant body of literature examining H5N1 transmission shows that human infection is strongly associated with direct, close poultry contact. In their systematic review of 24 case-control and seroepidemiology studies, Van Kerkhove et al. [2] found that increased risk was usually associated most strongly with forms of direct contact and less strongly and more sporadically with forms of indirect and environmental contact. For instance, a matched case-control study conducted in Vietnam found substantially larger odds ratios for risk factors such as cooking unhealthy poultry and having unhealthy poultry within the household, compared to having unhealthy poultry in the wider neighbourhood [3].
Few equivalent epidemiological studies have been published examining risk factors for H7N9 transmission; however, preliminary evidence suggests that direct poultry contact may play a lesser role in H7N9 transmission, with indirect contact more associated with this virus. An initial case-control study found that, similar to H5N1, increased risk of H7N9 infection was associated with forms of direct poultry contact and, to a lesser extent, contact with poultry environments [4]. However, a later case-control study found that increased risk was associated with indirect poultry contact at home and visiting live-bird markets and not with forms of riskier direct contact [5]. Consistently, Wang et al. [6] found that H7N9 seroconversion rates in poultry workers were not significantly associated with forms of direct contact with poultry. A recent matched case-control study found that infection was associated with both direct and indirect contact, the latter being visits to live-bird markets without direct poultry contact [7]. Descriptive studies show that the majority of H7N9 cases report some form of poultry contact, with a wide range of behaviours involving direct and/or indirect contact [8, 9].
The findings of two studies comparing poultry exposures between H5N1 and H7N9 are also indicative of differing risk factors, although contact types (i.e. direct and indirect contact) are not reported, which is the focus of this study. Cowling et al. [10] examined all WHO-confirmed cases of H5N1 and H7N9 reported in China up to 24 May 2013 and observed that exposure to sick, dead or backyard poultry (which could be assumed to involve direct contact) was more frequent in H5N1 cases, while visiting live-bird markets (which could be assumed to involve indirect contact) was more frequent in H7N9 cases. Wang et al. [11] report a similar finding. The different temporal and spatial distribution of the viruses, with H7N9 showing a much higher incidence rate but far less spatial dispersion, could also be indicative of important differences in risk factors and mechanisms of spread [12].
The aim of our study was to better understand differences in transmission risk factors between H5N1 and H7N9 by systematically classifying the degree of poultry contact (including direct and indirect) reported by WHO-confirmed cases of H5N1 and H7N9, using publicly available information on case exposure history. We used two different contact quantification methods to account for potential limitations with the publicly available data and to determine useful methodologies for future applications.
METHODS
We used an algorithm (see Fig. 1) adapted from Rabinowitz et al. [13] to classify the degree of poultry contact reported by WHO-confirmed H5N1 and H7N9 cases, using publicly available information on their exposure history in the 14 days prior to illness onset. A case was considered as WHO-confirmed if H5N1 or H7N9 virus infection was confirmed by the WHO or by a WHO-certified laboratory and reported as such by the WHO [1].
Fig. 1.
Algorithm for classification of WHO-confirmed cases based on reported poultry contact.
Classification levels reflected the degree of transmission risk associated with different poultry contact behaviours (see Fig. 1 for specific behaviours). Direct contact was divided into medium- and high-risk contact, based on the possibility that the viral concentrations to which humans are exposed affect the risk of infection [2]. Indirect contact included proximity to poultry without any behaviours specified in the direct contact category. If multiple risk behaviours were recorded for a given case, classification was based on the highest risk behaviour.
To account for potential data limitations, we used two quantification methods. The case population method included all cases reported by WHO up to 30 September 2014 and used multiple data sources: WHO [1], ProMED-mail [14], and peer-reviewed papers. The adapted case subset method [13] was restricted to a sample of cases who had detailed exposure data from peer-reviewed papers.
To identify published papers, we searched EMBASE and Medline using the key terms ‘avian influenza’ and ‘risk factor’ or ‘disease transmission’ or ‘environment’. This yielded 1077 EMBASE entries and 251 Medline entries. These papers were assessed and selected if they were published by 31 May 2015, written in English and provided data on the exposure history of cases. The reference lists of these publications were also searched for other relevant papers. This yielded 28 H5N1 and 22 H7N9 papers (50 in total).
Case population method
We developed a database (in Microsoft Excel) containing case-by-case information on all human cases of H7N9 and H5N1 reported by the WHO up to 30 September 2014. To check the comprehensiveness of the database, cumulative case totals by month, year and country were calculated and compared to corresponding totals published by the WHO [15]. This indicated that the database was missing 21 H5N1 cases that were reported in Indonesia in 2009. These cases were added to the database using the Food and Agriculture Organization's EMPRES-i database [16]. Compared to WHO case totals, an additional five H5N1 cases remained missing from Vietnam and Indonesia. These could not be matched to EMPRES-i records and were therefore left as missing. No H7N9 cases were missing.
We selected a subset of 25 H5N1 and 18 H7N9 publications (43 in total) from the 50 identified in the literature search (86%) which contained identifying information on cases. ProMED-mail archives were searched using ‘avian influenza’ and ‘human’ as key words in the subject field. Due to a high volume of posts regarding H5N1 and limited research resources, posts made after 2005 were excluded for H5N1. The identified posts provided additional exposure information for 7/72 H5N1 cases (9·7%).
If there was conflicting information between sources, WHO reports were ranked highest, followed by published research and ProMED-mail. However, conflicting information was rare. Two authors (A.B. and C.M.B.) independently categorized, according to the classification described in Figure 1, the first year of cases for H5N1 (n = 52) and H7N9 (n = 373). The results were compared and there were no disagreements so one author (A.B.) categorized the remaining cases.
Case subset method
We selected a subset of publications from those identified in the literature search which reported on epidemiological investigations of cases and included detailed exposure information that allowed cases to be confidently categorized using the classification algorithm. For H5N1, 21 papers met these criteria; for H7N9, 22 papers (43 papers in total; 86% of 50 publications identified in the literature search).
Using the epidemiological information provided in these papers, two authors (A.B. and C.M.B.) independently categorized cases in 10 papers (five for each virus). The results were compared to reach a consensus. One author (A.B.) categorized the remaining papers. When cases were reported in more than one paper, all data were compiled and considered during classification so that cases were not duplicated.
Data analyses
The number and proportion of cases in each contact category were calculated. The proportion of cases in the medium- and high-risk categories were similar between H5N1 and H7N9. To simplify comparisons, cases in the medium- and high-risk categories were categorized as direct contact and analyses thereafter focused on direct contact rather than on the medium- and high-risk categories separately. Statistical analyses were not used to compare proportions in the case population because we assumed that WHO records provided complete information about virus occurrence (a census rather than a sample). For the case subset, we conducted χ2 tests to compare contact proportions between viruses and exact binomial tests of goodness of fit to compare direct and indirect contact proportions within each virus.
During classification of the case population, there were concerns regarding a substantial number of cases for which exposure information indicated only the presence of live poultry contact and not the degree of contact. Rather than classifying these cases as having indirect contact, as specified by our algorithm, we categorized these cases as ‘unspecified contact’ and conducted a sensitivity analysis to examine contact proportions if these unspecified contact cases were (i) all attributed to indirect contact category; (ii) all attributed to direct contact category; or (iii) attributed to indirect and direct contact categories in the same proportions observed in the case population.
RESULTS
Table 1 shows the frequency and percentage of cases in each poultry contact category in the case population and case subset. Supplementary Tables S1 and S2 show the number of cases allocated to each poultry contact type in the case subset by country/province and data source.
Table 1.
Frequency and percentage (in parentheses) of WHO-confirmed cases of H5N1 and H7N9 by level of poultry contact in the case population, the sensitivity analysis and the case subset, to 30 September 2014
| Contact type | H5N1 | H7N9 |
|---|---|---|
| Case population | ||
| Unknown | 217 (32·0) | 191 (42·3) |
| Zero | 2 (0·3) | 3 (0·7) |
| Unspecified contact | 146 (21·5) | 195 (43·1) |
| Low (indirect) | 119 (17·6) | 22 (4·9) |
| Medium | 142 (20·9) | 32 (7·1) |
| High | 52 (7·7) | 9 (2·0) |
| Medium + high (direct) | 194 (28·6) | 41 (9·1) |
| Total | 678 | 452 |
| Sensitivity analysis | ||
| Indirect + unspecified contact | 265 (39·1) | 218 (48·2) |
| Direct + unspecified contact | 340 (50·1) | 236 (52·2) |
| Indirect + proportional distribution of unspecified contact* | 170·3 (25·1) | 76·7 (17·0) |
| Direct + proportional distribution of unspecified contact* | 288·7 (42·6) | 181·3 (40·1) |
| Case subset | ||
| Unknown | 15 (8·6) | 7 (10·8) |
| Zero | 3 (1·7) | 3 (4·6) |
| Low (indirect) | 34 (19·4) | 29 (44·6) |
| Medium | 87 (49·7) | 18 (27·7) |
| High | 36 (20·6) | 8 (12·3) |
| Medium + high (direct) | 123 (70·3) | 26 (40·0) |
| Total | 175 | 65 |
* Unspecified contact cases were distributed to indirect and direct contact categories in same proportions as original indirect and direct contact categories in case population.
Case population
There were 678 H5N1 cases and 452 H7N9 cases in the case population. As shown in Figure 2a, proportions of unknown contact and unspecified contact were high for H5N1 (unknown 32·0%, unspecified 21·5%) and particularly for H7N9 (unknown contact 42·3%, unspecified 43·1%). Zero contact was infrequent for both viruses (0·3% for H5N1 and 0·7% for H7N9). This meant that proportions of direct and indirect contact were low for H5N1 (direct 28·6%, indirect 17·6%) and particularly for H7N9 (direct 9·1%, indirect 4·9%). Direct contact proportions exceed indirect contact proportions for both viruses.
Fig. 2.
Proportion of H5N1 (n = 678) and H7N9 (n = 452) cases in each poultry contact category in the case population. (a) Unspecified contact cases categorized separately; (b) unspecified contact cases added to indirect contact; (c) unspecified contact cases added to direct contact; (d) unspecified contact cases proportionally distributed to indirect and direct contact in same proportions as original indirect and direct contact categories in case population.
In the sensitivity analysis, attributing all unspecified contact cases to indirect contact resulted in higher proportions of indirect contact in H7N9 cases (48·2%) than H5N1 cases (39·1%) (Fig. 2b), while attributing unspecific contact cases to direct contact resulted in higher proportions of direct contact than indirect contact in H7N9 cases (direct 52·2%) and H5N1 cases (direct 50·1%) (Fig. 2c). This is due to the larger proportion of H7N9 cases with unspecified contact. Proportionally attributing unspecified contact cases to indirect and direct contact resulted in higher proportions of direct contact than indirect contact in both H7N9 cases (direct 40·1%, indirect 17·0%) and H5N1 cases (direct 42·6%, indirect 25·1%).
Case subset
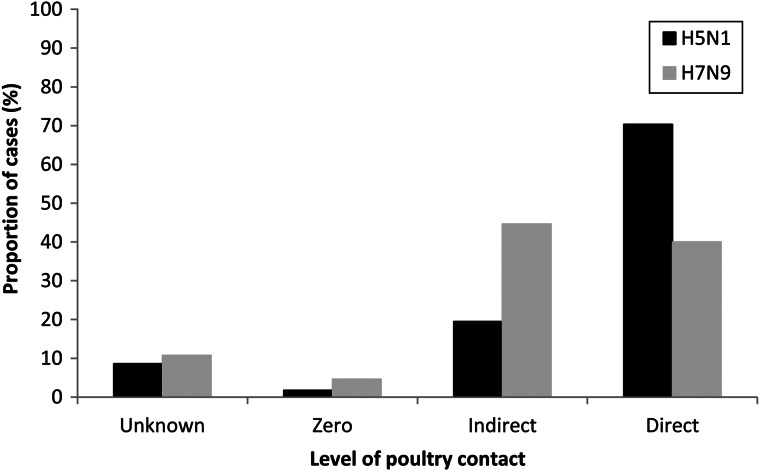
The case subset comprised 175 H5N1 cases and 65 H7N9 cases: 25·8% and 14·4% of the H5N1 and H7N9 case populations, respectively. As shown in Figure 3, cases with unknown contact were less common by this method and did not differ by virus (8·6% for H5N1 and 10·8% for H7N9) (χ2 = 0·3, P = 0·60). Few cases reported zero contact and proportions did not differ between H5N1 (1·7%) and H7N9 (4·6%) (χ2 = 1·6, P = 0·20). Indirect contact proportions were higher in H7N9 cases (44·6%) than H5N1 cases (19·4%) (χ2 = 15·5, P < 0·001), and direct contact proportions were higher in H5N1 cases (70·3%) than H7N9 cases (40·0%) (χ2 = 18·5, P < 0·001). For H7N9, the proportions of cases reporting direct contact or indirect contact did not differ significantly (P = 0·79). For H5N1, the proportion of cases reporting direct contact was greater than the proportion of cases reporting indirect contact (P < 0·001).
Fig. 3.
Proportion of H5N1 (n = 175) and H7N9 (n = 65) cases in each poultry contact category in the case subset.
DISCUSSION
We quantified the degree of poultry contact reported by WHO-confirmed cases of H7N9 and H5N1. In the subset of cases with detailed exposure data, we found considerable heterogeneity in contact proportions: the proportion of direct contact was substantially higher for H5N1 than for H7N9 and conversely the proportion of indirect contact was higher for H7N9 than H5N1. This finding is consistent with previous research suggesting that direct poultry contact may play less of a role in H7N9 transmission relative to H5N1 transmission [5–7, 10, 11].
H7N9 infection via indirect poultry contact is possible although the mechanisms are not well understood. Limited airborne transmission of H7N9 viruses has been shown in ferret experiments [17–19] and wind-mediated spread of virus-contaminated particulate matter has been evidenced for other low-pathogenic avian influenza viruses [20]. H7N9 may also survive for longer periods in the environment, increasing the risk of transmission via touching of contaminated surfaces, as the virus has been found to be somewhat tolerant to moderately acidic and alkali conditions which is unusual for Orthomyxoviridae viruses [21]. However, if indirect transmission is a risk factor, the physiology of the dose-response relationship would suggest that direct contact should pose a higher risk. There is also no systematic reason why people with H7N9 would be less likely to have direct contact with poultry than people with H5N1.
Live-bird markets, to which the majority of H7N9 cases (65%) report exposure [8], are plausible locations of transmission via indirect contact. These markets often have no physical segregation of poultry holding, slaughter and sale areas and likely have suboptimal disinfection practices [22]. Customers and workers may become exposed to contaminated poultry faeces, feathers, viscera and other secretions on surfaces, as well as contaminated dust or aerosols, even if they do not visit the poultry section of markets [22]. Given how common live-bird market exposure is in China, comparison of exposure between cases and healthy controls is important in determining risk. In relevant case-control studies, visiting live-bird markets, including visiting without direct poultry contact, is associated with infection [5, 7]. However, rates of detection of H7N9 in poultry and environmental samples at live-bird markets have been highly variable and dependent on sampling techniques [22, 23], leaving uncertainty as to the extent of viral contamination. Interestingly, we have also previously described lower prevalence of H7N9 compared to H5N1 in birds, despite a much higher incidence of H7N9 cases in humans [24].
It is also important to consider whether other animal vectors may play a role in transmitting H7N9 to humans. Indirect transmission routes, particularly airborne routes, are uncertain and if they do not carry significant risk, a large proportion of H7N9 cases in this study lacked a known exposure. Genetic and experimental inoculation studies have suggested that non-poultry terrestrial birds, including finches and sparrows, are vulnerable to H7N9 infection and may act as vectors for virus transmission to poultry and humans [25, 26].
To address continued uncertainty surrounding H7N9 transmission, epidemiological investigations of new H7N9 and H5N1 cases should remain a priority and should routinely investigate indirect and environmental routes of spread, including analysis of dusts, aerosols and surfaces. Investigations should also examine all forms of animal exposure and consider serological and disease surveillance of animals in direct or indirect contact with cases. The use of standardized epidemiological tools would increase comparability between the viruses. Better understanding of rates of exposure to poultry in the general population, especially in countries with high levels of poultry exposure, would also be useful. Existing surveys of poultry exposure in China are not easily comparable to case exposure proportions as they report select poultry practices rather than exposure types, use time periods not equivalent to the viral incubation period (e.g. the past year) and were usually conducted prior to the H7N9 epidemic [27–29].
For H7N9 prevention and control, our findings, together with other preliminary evidence [5–9], suggest that the present focus on avoiding direct contact with poultry (e.g. Chinese CDC guidelines [30]) should be reviewed and also other risk factors and corresponding mitigation strategies considered. There should also be greater emphasis placed on investigating indirect and environmental routes of spread, including analysis of dusts, aerosols and surfaces.
The utility of detailed and reliable publicly available data is being increasingly recognized in avian influenza research. Such data allows higher quality research to be conducted more rapidly, which is particularly important for emerging diseases where uncertainty hinders effective control. However, this study found important limitations associated with using publicly available data to investigate exposure in avian influenza cases. In the case population method, the information available from WHO [1], ProMED-mail [14] and the peer-reviewed literature was incomplete in that many cases lacked any information on poultry contact or had vague information that lacked details required for classification of the degree of contact (‘unspecified contact’ cases). The latter was particularly common in the WHO archives. These limitations affected H7N9 cases more than H5N1 cases, despite the exclusion of post-2004 ProMED-mail entries for H5N1. As illustrated in the sensitivity analysis, the assumptions made when categorizing the contact level of the ‘unspecified contact’ cases substantially impacted the contact proportions. Therefore, at present, this ‘case population’ method of measuring exposure is not useful. A recent study [31] similarly concluded that there is too much uncertainty associated with the publicly available data to draw conclusions regarding H7N9 transmission risk factors.
The limitations in the case population exposure data could be addressed through better reporting by public health authorities and researchers. With respect to the latter, detailed de-identified case histories could be published in online supplementary material. This data would allow more rapid, comprehensive and sophisticated analysis of exposure proportions. Restricting analyses to the subset of cases with detailed exposure data from the peer-reviewed literature (the ‘case subset’ method) was also an effective way of resolving these limitations and this method yielded useful epidemiological insights.
Our study is subject to some other limitations. The case subset was not randomly sampled from the case population and may not be representative. Recall bias may have favoured recall of high-level direct exposure for all cases, and indirect exposure for cases without obvious close contact with poultry. It would be useful to compare recall of direct and indirect poultry contact in the general population. Incomplete exposure data, along with probable differences in the measurement and reporting of exposure over time and place, may have caused misclassifications of poultry exposure and reduced the comparability of the viruses. However, this was likely a minor issue for the subset of cases drawn from the peer-reviewed literature for which detailed epidemiological information was available. The comparability of H5N1 and H7N9 may have also been reduced by differences in the poultry practices of different countries. Better understanding of poultry exposure in the general population would be useful to address this limitation.
In this study, data derived from the published research literature on the degree of poultry contact reported by WHO-confirmed H5N1 and H7N9 cases yielded useful epidemiological insights into potential differences in transmission risks. Other exposure data sources, including WHO [1] and ProMED-mail [14], were subject to substantial limitations that require attention before these sources can be used for reliably measuring poultry contact proportions. Our descriptive analysis found that the proportion of cases who reported direct contact was substantially higher for H5N1 than for H7N9. Together with emerging evidence, this suggests that direct poultry contact may be a clearer risk factor for H5N1 than for H7N9. We recommend further research be undertaken that involves detailed measurement of poultry exposure for avian influenza cases and that transmission mechanisms other than direct poultry contact also be considered for H7N9.
DECLARATION OF INTEREST
None.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268816001035.
click here to view supplementary material
REFERENCES
- 1.World Health Organization. Global outbreak and alert response network archives (http://www.who.int/csr/don/archive/year/2014/en/).
- 2.Van Kerkhove MD, et al. Highly pathogenic avian influenza (H5N1): pathways of exposure at the animal-human interface, a systematic review. PLoS ONE 2011; 6: e14582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinh PN, et al. Risk factors for human infection with avian influenza A H5N1, Vietnam, 2004. Emerging Infectious Diseases 2006; 12: 1841–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ai J, et al. Case-control study of risk factors for human infection with influenza A(H7N9) virus in Jiangsu Province, China, 2013. Eurosurveillance 2013; 18: e20510. [DOI] [PubMed] [Google Scholar]
- 5.Li J, et al. Case-control study of risk factors for human infection with avian influenza A(H7N9) virus in Shanghai, China, 2013. Epidemiology and Infection 2015; 143: 1826–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, et al. Seroprevalence to avian influenza A(H7N9) virus among poultry workers and the general population in southern China: a longitudinal study. Clinical Infectious Diseases 2014; 59: e76–e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu B, et al. Risk factors for influenza A(H7N9) disease – China, 2013. Clinical Infectious Diseases 2014; 59: 787–794. [DOI] [PubMed] [Google Scholar]
- 8.Li Q, et al. Epidemiology of human infections with avian influenza A(H7N9) virus in China. New England Journal of Medicine 2014; 370: 520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong Z, et al. Epidemiology of the avian influenza A(H7N9) outbreak in Zheijiang Province, China. BMC Infectious Diseases 2014; 14: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowling BJ, et al. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet 2013; 382: 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, et al. Geographic co-distribution of influenza virus subtypes H7N9 and H5N1 in humans, China. Emerging Infectious Diseases 2013; 19: 1898–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bui C, et al. A systematic review of the comparative epidemiology of avian and human influenza A H5N1 and H7N9 – lessons and unanswered questions. Transboundary and Emerging Diseases. Published online: 29 January 2015. doi: 10.1111/tbed.12327. [DOI] [PubMed]
- 13.Rabinowitz P, Perdue M, Mumford E. Contact variables for exposure to avian influenza H5N1 virus at the human–animal Interface. Zoonoses and Public Health 2010; 57: 227–238. [DOI] [PubMed] [Google Scholar]
- 14.ProMED-mail. Program for monitoring emerging diseases of the International Society for Infectious Diseases (http://www.promedmail.org/).
- 15.World Health Organization. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2014. 2 Oct. 2014 (http://www.who.int/influenza/human_animal_interface/EN_GIP_20141002CumulativeNumberH5N1cases.pdf).
- 16.Food and Agriculture Organization of the United Nations. EMPRES-i Global Animal Disease Information System (http://empres-i.fao.org/eipws3g/).
- 17.Belser JA, et al. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 2013; 501: 556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richard M, et al. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature 2013; 501: 560–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, et al. H7N9 Influenza viruses are transmissible in ferrets by respiratory droplet. Science 2013; 341: 410–414. [DOI] [PubMed] [Google Scholar]
- 20.Jonges M, et al. Wind-mediated spread of low-pathogenic avian influenza virus into the environment during outbreaks at commercial poultry farms. PLoS ONE 2015; 10: e0125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou S, et al. Inactivation of the novel avian influenza A (H7N9) virus under physical conditions or chemical agents treatment. Journal of Virology 2013; 10: e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang M, et al. Environmental sampling for avian influenza A(H7N9) in live-poultry markets in Guangdong, China. PLoS ONE 2015; 10: e0126335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perkins LE, Swayne DE. Pathobiology of A/Chicken/Hong Kong/220/97 (H5N1) avian influenza virus in seven gallinaceous species. Veterinary Pathology 2001; 38: 149–164. [DOI] [PubMed] [Google Scholar]
- 24.Bui C, et al. A meta-analysis of the prevalence of influenza A H5N1 and H7N9 infection in birds. Transboundary and Emerging Diseases. Published online: 10 January 2016. doi: 10.1111/tbed.12466. [DOI] [PubMed] [Google Scholar]
- 25.Jones JC, et al. Influenza A(H7N9) virus transmission between finches and poultry. Emerging Infectious Diseases 2015; 21: 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Zhang Z, Weng Z. Rapid reassortment of internal genes in avian influenza A(H7N9) virus. Clinical Infectious Diseases 2013; 57: 1059–1061. [DOI] [PubMed] [Google Scholar]
- 27.Liao Q, et al. Live poultry exposure, Guangzhou, China, 2006. Epidemics 2009; 1: 207–212. [DOI] [PubMed] [Google Scholar]
- 28.Peng Z, et al. Rural villagers and urban residents exposure to poultry in China. PLoS ONE 2014; 9: e95430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, et al. Human exposure to live poultry and psychological and behavioral responses to influenza A(H7N9), China. Emerging Infectious Diseases 2014; 20: 1296–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chinese Center for Disease Control and Prevention. Frequently asked questions on human infection with avian influenza A(H7N9) virus, China. 10 April 2013 (http://www.chinacdc.cn/en/research_5311/FAQ/201304/t20130418_80053.html).
- 31.Mao C, et al. An internet-based epidemiological investigation of the outbreak of H7N9 avian influenza A in China since early 2013. Journal of Medical Internet Research 2014; 16: e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268816001035.
click here to view supplementary material