Abstract
Plants depend on light for energy production. However, the UV component in sunlight also inflicts DNA damage, mostly in the form of cyclobutane pyrimidine dimers (CPD) and (6-4) pyrimidine–pyrimidone photoproducts, which are mutagenic and lethal to the plant cells. These lesions are repaired by blue-light-dependent photolyases and the nucleotide excision repair enzymatic systems. Here, we characterize nucleotide excision repair in Arabidopsis thaliana genome-wide and at single nucleotide resolution with special focus on transcription-coupled repair and the role of the CSA1 and CSA2 genes/proteins in dictating the efficiency and the strand preference of repair of transcribed genes. We demonstrate that CSA1 is the dominant protein in coupling repair to transcription with minor contribution from CSA2.
INTRODUCTION
Plants are widely exposed to solar ultraviolet (UV) radiation due to their sessile lifestyle and dependence on photosynthesis for energy. The UV exposure of cells leads to the formation of two different types of DNA damages: cyclobutane pyrimidine dimer (CPD) and pyrimidine-pyrimidone (6-4) photoproduct (6-4PP). These UV photoproducts block transcription and replication in plant cells thereby affecting their growth and development. Plants can eliminate UV photoproducts by nucleotide excision repair or blue-light-dependent photoreactivation. In excision repair, a DNA fragment containing the bulky DNA adduct is removed by concerted dual incision and the resulting gap is filled by polymerase and ligase activities (1). Two different excision repair pathways exist: while transcription-coupled repair (TCR) only eliminates DNA adducts stalling RNA polymerase II, global repair can remove DNA adducts present in any region of the genome (2). Only the damage recognition steps differ between TCR and global repair. The genome-wide excision repair map of Arabidopsis thaliana has shown that TCR in plants is much faster than global repair throughout the genome (3).
In mammalian TCR, the damage-stalled RNA polymerase II is recognized by Cockayne Syndrome A (CSA) and Cockayne Syndrome B (CSB) proteins leading to the recruitment of other excision repair factors to the damage site. CSA/ERCC8 (Cockayne Syndrome A) protein is a WD-repeat protein (4) which functions in TCR as a substrate-recognition component of DDB1-CUL4-Xbox E3 ubiquitin ligase complex (5). CSB (Cockayne Syndrome B) is a large protein with helicase motifs (6) but with a translocase (7) and not helicase activity. It directly interacts with RNA polymerase stalled at DNA damage and with the aid of CSA promotes transcription-coupled repair (TCR) which is concurrent with CSA-mediated RNA Pol II ubiquitylation and degradation (8). Two CSA homologs are found in Arabidopsis thaliana, named CSA1 (At1g27840) and CSA2 (At1g19750) (9). Both proteins localize to the nucleus and the Arabidopsis plants lacking CSA1 and CSA2 proteins have been shown to be more sensitive to UV-B (8). In addition, these proteins interact with CUL4-DDB1A complex (8) that plays a role in excision repair of UV damage in Arabidopsis (9).
Here, we generated genome-wide excision repair map of CPD damage in csa1, csa2, and csa1csa2 double-mutant Arabidopsis plants to determine the roles of the CSA1 and CSA2 proteins in TCR. Our results showed that the TCR activity in csa1 and csa1csa2 are almost completely lost, demonstrating the essential and dominant role of CSA1 in TCR. We detected a significant, but slight reduction in TCR levels of csa2 plants compared with WT plants. This implies that CSA2 has a minor effect on TCR activity. Our study contributes to understanding the molecular mechanism of TCR in plants that maintains biological processes under DNA-damaging environmental stress.
MATERIALS AND METHODS
Plant materials and growth conditions.
All experiments were performed on Arabidopsis thaliana ecotype Columbia (Col-0). The T-DNA insertion lines SALK_151258 (csa1) and SALK_144623 (csa2) (10) were obtained from Arabidopsis Biological Resource Center (ABRC). Homozygous insertion lines were determined by PCR-based genotyping using the primers of LP_csa1 (5′-GCTGCTGGAACTGAAGATGTC-3′), RP_csa1 (5′-CCAGCAGATGCTGCCTATAAC-3′), LP_csa2 (5′-TGGTTTGGTTTTGTTCTTTCG-3′), RP_csa2 (5′-CATGACAATTTGGTAGCCTGG-3′) and BP (5′-ATTTTGCCGATTTCGGAAC-3′) (Fig. 1).
Figure 1.

Multiple sequence alignment for Arabidopsis CSA1 and CSA2 and related proteins. 1000 most similar eukaryotic protein sequences to each of the Arabidopsis CSA1 and CSA2 proteins are aligned using MAFFT and conservation scores are calculated using Bio3D. Randomly selected 17 proteins are shown together with Arabidopsis CSA1 and CSA2, and human CSA, due to visual purposes. Similarly, the gap positions in Arabidopsis CSA1 and CSA2 sequences are not included in the figure. Dark colors indicate highly conserved residues while light colors indicate lower conservation among all protein sequences in the multiple sequence alignment. Red boxes show the differences between Arabidopsis CSA1 and CSA2 protein sequences.
To generate csa1csa2 double-mutant lines, CSA2 gene is targeted by CRISPR/Cas9 in csa1 background. For this purpose, the guide RNA (ATCTAGCGAGAAACCTTCAG) targeting CSA2 gene was designed using the CRISPR-Plant platform (11) and cloned into AtGGentry plasmid using Golden-Gate cloning. AtGGentry including the CSA2 gRNA construct was used as an entry clone to transfer the CSA2 gRNA into pCUT4-GW plasmid by Gateway Cloning (12). The resulting plasmid was transformed into Agrobacterium tumefaciens GV3101 by electroporation and this strain was used to transform csa1 plants by floral dip method (13). The CRISPR/Cas9-edited seeds were sterilized and grown on MS agar medium containing 10 μg ml−1 hygromycin. The seedlings that grew on selection media were genotyped by amplifying the DNA region including CRISPR target site by PCR and Sanger sequencing to obtain csa1csa2 double-mutant (Fig. 1C).
XR-seq library preparation.
The seeds were surface-sterilized and stratified for 2 days at 4°C and then grown on MS agar medium under long-day condition (16 h light/8 h dark) with a cool white fluorescent light at 24°C for 10 days. Ten-day-old seedlings were treated with 1 J (m2s)−1 UVC (254 nm) for 2 min (120 J m−2 UVC) and incubated under yellow light for 30 min. The XR-seq protocol was applied to prepare the libraries for each Arabidopsis T-DNA insertion line (3,14). The libraries were sequenced in the Illumina HiSeq 4000 and single-end 50-nt reads were generated.
XR-seq data processing.
Adapter sequences (TGGAATTCTCGGGTGCCAAGGAACTCCAGTNNNNNNACGATCTCGTAT GCCGTCTTCTGCTTG) were trimmed from 3′ ends of the raw XR-seq reads using Cutadapt (15) (v2.5). Trimmed reads were aligned on Arabidopsis TAIR10 reference genome using Bowtie2 (16) (v 2.3.5.1). Aligned reads were then filtered and unique reads were kept for further steps. Simulated XR-seq data were obtained using Boquila (https://github.com/CompGenomeLab/boquila) (v0.3.1).
Repair profiles on genic regions.
Each of Araport11 (17) protein-coding genes (~27 000) were divided into 100 windows and XR-seq read counts were obtained on each window and its opposite strand using BEDTools (18) intersect (v2.27.1). Similarly, 1 kb upstream and downstream of TSS and TES of each gene were divided into 50 windows and XR-seq read counts were obtained. RPKM normalization was applied for the read counts in each window and these normalized read counts were plotted.
TS/NTS ratios were calculated by counting and normalizing XR-seq reads intersecting with transcribed and nontranscribed strand of each protein-coding gene. Boxplots were generated using TS/NTS ratios of each of the protein-coding genes.
Repair profile screenshots.
BED files of the processed XR-seq data were converted to BedGraph format using BEDTools (18) genomecov utility (-bg -scale options) and RPM normalization was applied. BedGraph files were then converted to BigWig format using UCSC tools (19). Integrative Genomics Viewer (20) was used to visualize BigWig files and capture screenshot images.
Multiple sequence alignment and phylogenetic tree building.
BLAST (21) algorithm (v2.9.0) was used to compare all Uniprot eukaryotic protein sequences with CSA1 and CSA2, separately (-evalue 1E-06 - max_target_seqs 1000). The protein hits for CSA1 and CSA2 were merged and redundant sequences were eliminated. Multiple sequence alignment was performed using MAFFT (22) (v7.407) with --auto option. Conservation scores for the residues in the multiple sequence alignment was calculated using Bio3D (23). Phylogenetic tree was formed with the most similar 200 eukaryotic protein sequences to CSA1 and CSA2, using IQ-TREE2 (v2.0.6). -B 1000 option was used to obtain bootstrap values (24).
RESULTS
We examined sequence similarities between CSA1 and CSA2, as well as their similarities with other eukaryotic proteins. For that purpose, we extracted most similar eukaryotic protein sequences to each one of the CSA1 and CSA2 proteins. We performed multiple sequence alignment and built a phylogenetic tree. CSA1 and CSA2 clustered in an Arabidopsis clade in the phylogenetic tree, suggesting that CSA gene duplication has recently occurred in Arabidopsis (Fig. 2). Multiple sequence alignment revealed differences at 33 positions between CSA1 and CSA2, and these positions did not have high conservation scores among other proteins in the alignment (Fig. 1). The residues in the positions with high conservation scores were mostly shared within CSA1 and CSA2 protein sequences, suggesting that these two proteins are not significantly differentiated from each other. The presence of selection pressure against conserved CSA-family positions in CSA2 suggest that these two paralogous proteins likely preserve the same or similar functions, possibly in different tissues.
Figure 2.
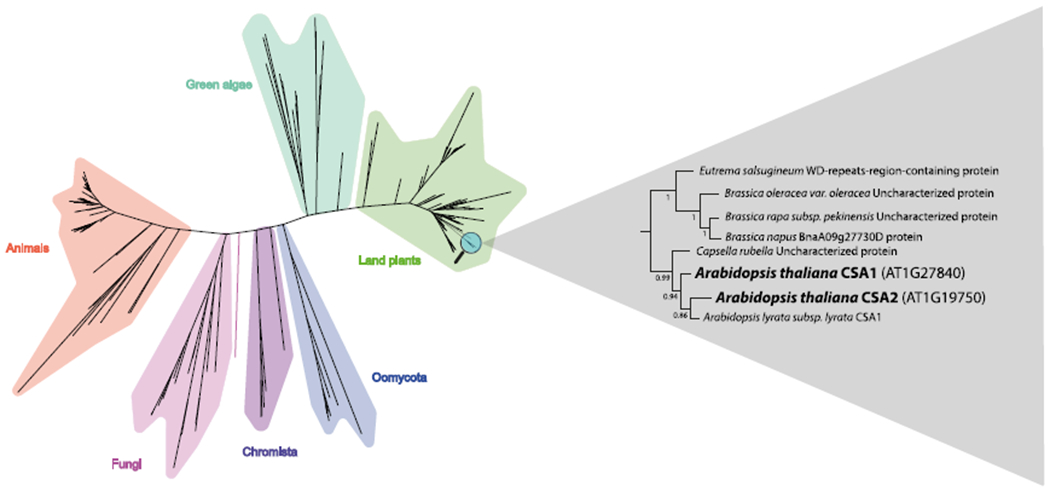
Conservation of CSA proteins in eukaryotes. Phylogenetic tree is formed using IQ-TREE2 (-B 1000). 200 eukaryotic protein sequences that are most similar to each of the Arabidopsis CSA1 and CSA2 proteins are included in the tree. Arabidopsis CSA1 and CSA2 proteins and their closest orthologs are shown in a separate tree with the bootstrap values.
To understand the role of Arabidopsis CSA homologs in TCR, we obtained and genotyped SALK_151258 (csa1) and SALK_144623 (csa2) homozygous mutant lines that include T-DNA insertion in the 5th exon of CSA1 gene and 2nd exon of CSA2 gene, respectively (Figure S1). We irradiated 10-days old seedlings of WT, csa1, and csa2 plants with UVC (254 nm) and kept them for 30 min under yellow light to prevent blue-light-mediated photoreactivation. Then, we isolated oligonucleotides with CPDs excised during excision repair and generated XR-seq libraries to create genome-wide excision repair maps of CPDs (14,25). We performed two experiments and prepared two libraries for each strain. We compared the aligned sequencing data sets across samples and observed that csa1 and csa1csa2 plants were similar to each other, while csa2 and wild-type samples were spread in two separate groups (Figure S2). We detected enrichment of “TT” at the position of 4–6 nucleotides from 3′ end of the oligonucleotides, as expected (26) (Fig. 3A). In each sample, the size distribution of excision products was 23–27 nucleotides in length consistent with our previous report (3), and a population of DNA fragments with lengths of 14–20, which presumably resulted from the degradation of excision products was also detected (Fig. 3B). Both the dinucleotide content and size distribution of excision products show that the dual incision mechanism in csa1 and csa2 are identical to WT.
Figure 3.
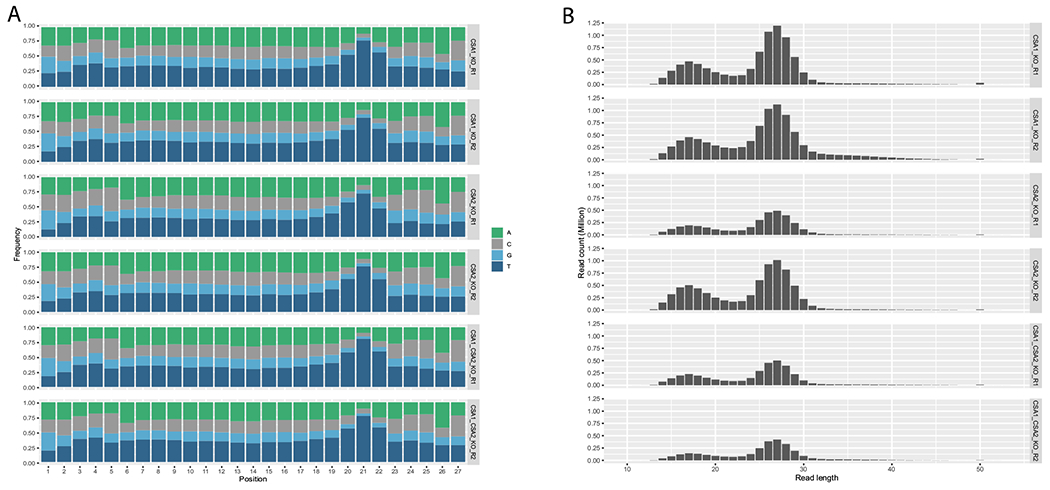
Nucleotide content and length distribution properties of XR-seq data. (A) Nucleotide frequencies of 27 nucleotide-long XR-seq reads coming from csa1, csa2, and csa1csa2 plants. (B) Length distribution of XR-seq reads for csa1, csa2, and csa1csa2 samples.
The involvement of human CSA protein in TCR and the role of Arabidopsis CSA homologs in UV tolerance (8) suggest that CSA1 and CSA2 might play roles in TCR. To test this possibility, we identified genome-wide TCR levels in csa1 and csa2 plants by analyzing the repair maps of CPDs in comparison with WT (Fig. 4). In our analysis, we detected a drastic loss of TCR signal in csa1 plants; however, TCR levels slightly, but significantly, decreased in csa2 plants compared with WT (Fig. 4A, TS/NTS: 92.544 vs. 2.335, and 56.739, respectively). In addition to excision repair levels, XR-seq indicates transcription rates of genes since TCR is strongly correlated with transcription rate (3). Based on XR-seq data, it can be extrapolated that the transcription level of CSA2 in 10-day old seedlings is much lower than CSA1 (Figure S3). This suggests that the minor effect of CSA2 might result from the low expression level of CSA2 gene in the whole plant, assuming that the steady-state of CSA1 and CSA2 mature mRNAs is similar and that the corresponding CSA1 and CSA2 protein are equally stable. In addition, simulations provided artificial XR-seq data for csa1 and WT samples, taking into account the nucleotide distributions in the real XR-seq reads. By comparing the repair profiles of the real and the simulated XR-seq data on the genes, we obtained an evidence that our results from the XR-seq data were not because of the dipyrimidine sequence content in genic regions (Fig. 4B, Figures S4 and S5). Our results indicate that while Arabidopsis TCR requires CSA1 protein, CSA2 protein has minor influence on TCR (Fig. 4, Figures S4 and S5).
Figure 4.
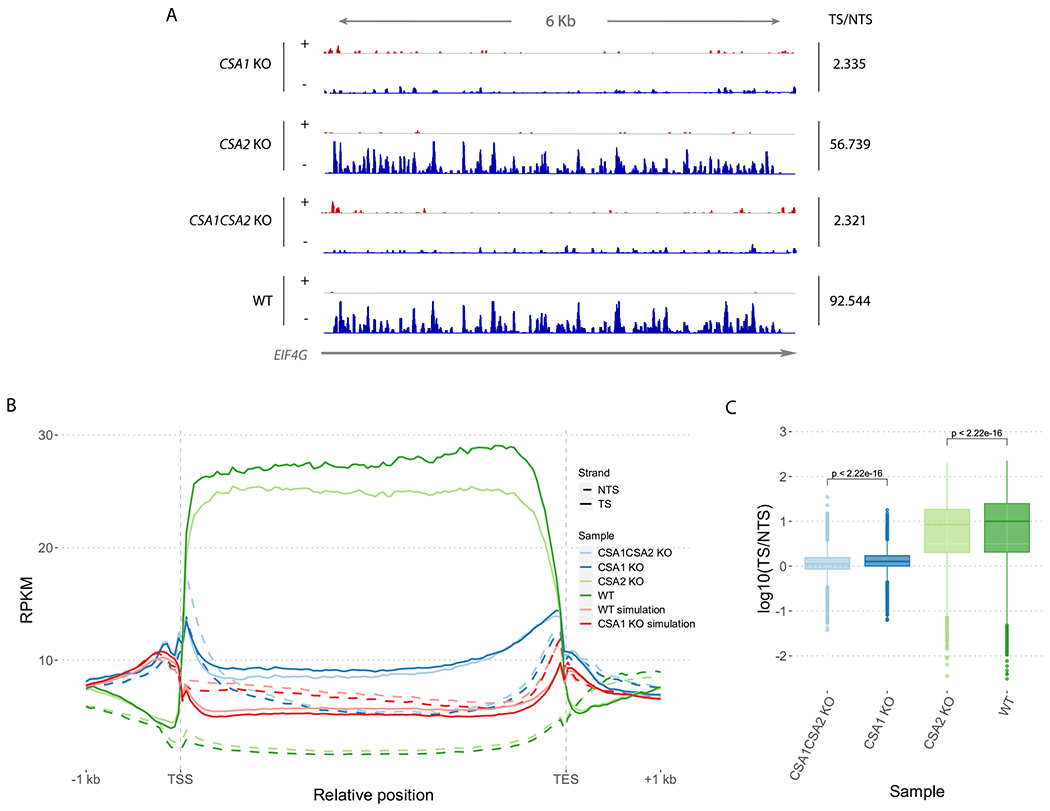
Repair profiles for XR-seq datasets. (A) Screenshot of repair profiles on EIF4G house-keeping gene. Integrative Genomics Viewer is used to visualize strand-specific XR-seq data of knock-out and wild-type samples. Plus and minus DNA strands are represented by + and − symbols. Gray horizontal line represents the EIF4G gene and the arrow indicates its direction on the genome. TS/NTS ratios on EIF4G gene for each sample are given on the right. (B) Normalized XR-seq read counts on Araport11 protein-coding genes. 1 kb upstream of TSS and downstream of TES are also included in the genic regions. XR-seq data for wild-type and knock-out samples as well as simulated XR-seq data for simulated csa1 and WT samples are included. Two replicates from each sample are merged. (C) TS/NTS ratios for XR-seq samples on Araport11 protein-coding genes. Merged replicates of knockout and WT samples are included. Significance of the difference between samples are determined using Wilcoxon test. RPKM: Reads Per Kilobase of transcript per Million mapped reads, TSS, Transcription start site; TES, Transcription end site.
Even though TCR levels declined dramatically, we still could detect a low level of TCR in csa1 plants. To check the possibility of functional redundancy between CSA1 and CSA2 proteins, we generated csa1csa2 double-knockout by CRISPR/Cas9 targeting of Csa2 gene in csa1 background (Figure S1). Then, we generated genome-wide CPD repair map of csa1csa2 plants following the same XR-seq approach applied for csa1 and csa2 plants. The nucleotide content and size distribution of excision products in csa1csa2 showed similarity with WT, csa1, and csa2 plants (Fig. 3). We detected TCR activity in csa1csa2 plants, but it was lower than csa1, and the TCR level difference between csa1csa2 and csa1 is similar to the difference between csa2 and WT (Fig. 4). Our XR-seq analyses show that efficient TCR requires Arabidopsis CSA1 protein and CSA2 has little effect on TCR in spite of its high sequence similarity to CSA1 protein.
DISCUSSION
The genome-wide repair map of CPDs shows that TCR is prominent in plants and maintains gene expression, thereby biological processes, under DNA-damaging UV irradiation (3). Although how TCR works is well-understood in bacteria and mammalian cells, the molecular mechanism of TCR in plants is yet to be clarified. XR-seq method is a novel and highly efficient approach to screen TCR dynamics throughout the genome and enables the study of TCR when combined with reverse genetics. Using XR-seq, we examined whether Arabidopsis TCR pathway requires CSA homologs, which show 92% protein sequence similarity, interact with each other and make plants more tolerant to UV exposure. Our analysis demonstrates that CSA1 is required for an efficient TCR, while CSA2 has a minor contribution to TCR.
In csa2 plants, we detected a little decrease in TCR compared with WT, suggesting that CSA2 protein is not crucial for TCR although its binding to CSA1 has been shown before (8). The fact that TCR level difference between WT and csa2 and between csa1 and csa1csa2 was similar shows that TCR decrease in csa2 is real. TCR is proportional to transcription rate that makes XR-seq a highly useful tool to detect the dynamics of transcription genome-wide and an alternative to available NGS methods used to map transcription. In addition to excision repair activity on transcribed strands of genes, we detect the transcription level of the genes by XR-seq. Therefore, the TCR reduction in csa2 plants might result from the decrease in transcription levels, that is currently under investigation. CSA2 might indirectly affect TCR activity by altering the transcription rates of genes instead of being involved in the detection of DNA damage and these genes might be involved in UV protection which can explain the CSA2’s role in UV tolerance. The other reason for minor contribution of CSA2 to TCR could be the low transcription level of CSA2 in 10-day old seedlings.
Even though TCR in csa1csa2 is nearly lost, we still could detect a little amount of activity that apparently does not result from functional redundancy between CSA1 and CSA2 proteins. This suggests that TCR can occur in the absence of CSA proteins in Arabidopsis. In mammalian cells, CSA and CSB detect damage-stalled RNA polymerase II and are required for TCR to begin. Arabidopsis possess two CSB homologs: CHR8 and CHR24. Whether CSB homologs are the reason for the TCR activity in the absence of CSA proteins, and whether these CSB homologs are involved in Arabidopsis TCR have to be tested in future approaches.
Using XR-seq, we determined that CSA1, but not CSA2, play an essential role for TCR to function efficiently and maintain transcription under UV stress. Our study contributes to the understanding of nucleotide excision repair at molecular level that is required for generating crops resistant to DNA-damage causing environmental stress whose level is increasing by the influence of global warming.
Supplementary Material
Figure S1. Genotyping of csa1 and csa2 plants and generation of csa1csa2 plants.
Figure S2. Genome coverage profiles of XR-seq datasets.
Figure S3. Transcription levels of CSA1 and CSA2 genes based on XR-seq data.
Figure S4. Repair profiles on Araport11 protein-coding genes.
Figure S5. Repair profiles on TSS and TES regions of Araport11 protein-coding genes.
Acknowledgements
This paper is dedicated to Professor Jean Cadet for his outstanding scientific contributions to the field of DNA damage and mutagenesis by UV light and ionizing radiation, for his dedication to the promotion of the science of DNA damage chemistry for his service at many levels to the promotion of the fields of DNA damage, repair, and mutagenesis and for fostering international scientific collaboration.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article:
This article is part of a Special Issue celebrating the achievements of Dr. Jean Cadet.
DATA AVAILABILITY STATEMENT
The sequencing data have been deposited in the National Center for Biotechnology Information Short Read Archive (SRA) under the accession number PRJNA750873.
REFERENCES
- 1.Sancar A (2016) Mechanisms of DNA repair by photolyase and excision nuclease (Nobel lecture). Angew. Chem. Int. Ed 55, 8502–8527. [DOI] [PubMed] [Google Scholar]
- 2.Hanawalt PC and Spivak G (2008) Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol 9, 958–970. [DOI] [PubMed] [Google Scholar]
- 3.Oztas O, Selby CP, Sancar A and Adebali O (2018) Genomewide excision repair in Arabidopsis is coupled to transcription and reflects circadian gene expression patterns. Nat. Commun 9, 1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henning KA, Li L, Iyer N, McDaniel LD, Reagan MS, Legerski R, Schultz RA, Stefanini M, Lehmann AR, Mayne LV and Friedberg EC (1995) The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell 82, 555–564. [DOI] [PubMed] [Google Scholar]
- 5.Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K and Nakatani Y (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113, 357–367. [DOI] [PubMed] [Google Scholar]
- 6.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D and Hoeijmakers JH (1992) ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell 71, 939–953. [DOI] [PubMed] [Google Scholar]
- 7.Selby CP and Sancar A (1997) Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. USA 94, 11205–11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C, Guo H, Zhang J, Guo G, Schumaker KS and Guo Y (2010) Arabidopsis cockayne syndrome A-like proteins 1A and 1B form a complex with CULLIN4 and damage DNA binding protein 1A and regulate the response to UV irradiation. Plant Cell 22, 2353–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molinier J, Lechner E, Dumbliauskas E and Genschik P (2008) Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress. PLoS Genet. 4, e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC and Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- 11.Xie K, Zhang J and Yang Y (2014) Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol. Plant 7, 923–926. [DOI] [PubMed] [Google Scholar]
- 12.Peterson BA, Haak DC, Nishimura MT, Teixeira PJPL, James SR, Dangl JL and Nimchuk ZL (2016) Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PLoS One 11, e0162169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clough SJ and Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- 14.Hu J, Li W, Adebali O, Yang Y, Oztas O, Selby CP and Sancar A (2018) Genome-wide mapping of nucleotide excision repair with XR-seq. Nat. Protoc 14(1), 248–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. [Google Scholar]
- 16.Langmead B and Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat. Meth 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng C-Y, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S and Town CD (2017) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804. [DOI] [PubMed] [Google Scholar]
- 18.Quinlan AR (2014) BEDTools: The Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinform 47, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn RM, Haussler D and Kent WJ (2013) The UCSC genome browser and associated tools. Brief Bioinform. 14, 144–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G and Mesirov JP (2011) Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K and Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformaics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K and Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA and Caves LSD (2006) Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics 22, 2695–2696. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen L-T, Schmidt HA, von Haeseler A and Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol 32(1), 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, Adar S, Selby CP, Lieb JD and Sancar A (2015) Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 29, 948–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canturk F, Karaman M, Selby CP, Kemp MG, Kulaksiz-Erkmen G, Hu J, Li W, Lindsey-Boltz LA and Sancar A (2016) Nucleotide excision repair by dual incisions in plants. Proc. Natl. Acad. Sci. USA 113, 4706–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Genotyping of csa1 and csa2 plants and generation of csa1csa2 plants.
Figure S2. Genome coverage profiles of XR-seq datasets.
Figure S3. Transcription levels of CSA1 and CSA2 genes based on XR-seq data.
Figure S4. Repair profiles on Araport11 protein-coding genes.
Figure S5. Repair profiles on TSS and TES regions of Araport11 protein-coding genes.
Data Availability Statement
The sequencing data have been deposited in the National Center for Biotechnology Information Short Read Archive (SRA) under the accession number PRJNA750873.


