SUMMARY
After the introduction of conjugate vaccines, a strong rearrangement of pneumococcal serotypes was observed globally. Probably most concerning was the emergence of serotype 19A, which has not only high invasive disease potential, but also high antibiotic resistance. In the current study we focused on the increased prevalence of serotype 19A after the PCV vaccination rate became widely used in Hungary. A total of 2262 children aged 3–6 years were screened for pneumococcus carriage using nasal swabs. Children were divided into two groups according to the vaccination rates, low level (group 1) vs. high level (group 2). While the carriage rate did not change over time (average 32·9%), the serotype distribution differed greatly in the two groups. The prevalence of serotype 19A increased >eightfold. Almost all 19A isolates had high-level macrolide resistance and elevated penicillin minimum inhibitory concentrations. Genotyping methods revealed that these new 19A isolates are different from the previously frequent Hungary19A−6 PMEN clone. Both the carriage rate and the overall penicillin and macrolide resistance remained stable over time, but while several serotypes were represented in group 1, serotype 19A alone was clearly dominant in group 2.
Key words: Pneumococcus, resistance, serotypes, vaccination
INTRODUCTION
Streptococcus pneumoniae is one of the leading human pathogens in all age groups, but especially in young children and the elderly. It is the causative agent of community-acquired pneumonia (CAP), sinusitis, otitis media, and invasive pneumococcal diseases (IPD) such as sepsis or meningitis [1], and it is still the cause of death of half a million children aged <5 years worldwide [2]. The infection often derives from symptomless carriers, usually small children, as this bacterium repeatedly colonizes the nasopharynx [3, 4].
To prevent pneumococcal infections, two types of vaccines were developed, polysaccharide and conjugate vaccines. The latter were designed for children aged <2 years, since they are not able to develop a T-cell-dependent immune response against polysaccharides. The 13-valent PCV13 (Prevenar) contains serotypes 1, 3, 4, 5, 6A, 6B, 7F, 9V, 14, 18C, 19A, 19F, and 23F [5].
In Hungary, PCV7 was introduced in 2005, but in the first 3 years the uptake was low, due to its high cost and a low awareness. In October 2008, PCV7 was made available free of charge for children aged <2 years in the framework of a pneumococcus surveillance programme, and in April 2009 it was included in the national immunization programme (NIP) as a recommended, but not mandatory vaccine. Within 1 year, according to data of the National Centre for Epidemiology, the vaccination rate increased to >80% and it has reached nearly 95% for children born in 2012 [6–8]. In September 2010, PCV13 replaced PCV7 in Hungary, and PCV13 was integrated in the NIP in the 2 + 1 schedule from 1 July 2014 as a mandatory vaccine. PCV10 does not currently belong to the supported vaccinations in Hungary, and none of the children in this study received PCV10.
Due to vaccination with PCV7, a strong reduction in the serotypes included in the vaccine (VTs, vaccine types) was observed worldwide, especially in the incidence of IPDs in young children [9, 10]. On the other hand, however, increased prevalence of certain non-vaccine types (NVTs), such as 1, 3, 5, 6A, 7F or 19A was observed [9]. For that reason, the above-mentioned six new serotypes were added to the higher-valency conjugate vaccines, to target the residual burden of both invasive and non-invasive pneumococcal infections [11]. Serotype 19A is distinguished among these serotypes, not only because of its high invasive disease potential, but also because of its antibiotic resistance capacity. Many studies have demonstrated that after PCV7 vaccination there was a strongly increased rate of serotype 19A in IPD, in pneumonia, and in acute otitis media [11].
Before the PCV era, similarly to other countries, the old, successful ‘paediatric’ clones (serotypes 14, 19F, 23F, 6A, 6B) dominated in Hungary [12]. However, a marked rearrangement of serotypes among carried strains has been observed recently [13], with serotype 14 disappearing, and others, such as 19A predominating. In that paper [13], the screening results of 1022 children attending 18 day-care centres was reported; here we publish extended data about 2262 children attending 40 nurseries.
Furthermore, the current paper specifically focuses on changes in the colonization rate of serotype 19A, 3 years after widespread PCV7 vaccination started in Hungary, because it showed the greatest increase in non-PCV7 serotypes. We also investigated whether these 19A strains are similar to the well-known, highly resistant PMEN clone, Hungary19A−6 [14].
METHODS
Study population
In total, 2262 healthy children, aged 3–6 years, from 40 nurseries in 17 cities or villages in different parts of Hungary were screened between March 2009 and June 2012. Informed consent obtained from the parents was a condition for enrolment. In the form, data were collected on the children's vaccination status, siblings, history of recurrent otitis media or other severe infections, and exposure to passive smoking. For statistical analysis, where appropriate, the χ2 test was used.
Ethical permission (TUKEB 4-3/2009) was issued by the Regional and Institutional Committee of Science and Research Ethics of Semmelweis University.
Specimen collection and identification of pneumococci
Nasal samples were taken from both nostrils with sterile swab (Transwab, Medical Wire & Equipment, UK), transported to the laboratory within a few hours and inoculated onto Columbia blood agar plates. After incubating the cultures overnight at 37 °C, in the presence of 5% CO2, suspected colonies showing typical colony morphology of Streptococcus pneumoniae (α-haemolysis and umbilicus colonies) were subcultured and tested for optochin sensitivity (5 µg discs, Mast Diagnostica, UK). The identity of the strains was confirmed by detecting the lytA (autolysin) gene by polymerase chain reaction (PCR) in every case [15] and confirmed strains were stored at −80 °C on cryobeads (Mast Diagnostica).
Serotyping
To determine the serotypes of the isolates, the Pneumotest Latex kit (Statens Serum Institut, Denmark) was used. Factor determination was done either by PCR using primers published by CDC [16], or others [17]; or by the Quellung method at the Hungarian National Pneumococcal Reference Centre and the German National Reference Centre for Streptococci (GNRCS).
Genotyping
To demonstrate the clonality of the serotype 19A strains, pulsed-field gelelectrophoresis (PFGE) was used, using SmaI restriction digestion, as described previously [18]. Multilocus sequence typing (MLST) was also performed for ten isolates, selected from the PFGE dendrogram. Well-defined sections of seven housekeeping genes (aroe, gdh, gki, recP, spi, xpt, ddl) were amplified by PCR, using the primers provided on the MLST website [19], except for recP, where the recP reverse primer was 5′-TGCCAACCGTAGGCATTGTAAC-3′. The products were purified by the QIAquick PCR purification kit (Qiagen, Germany) and sent for sequencing to BIOMI Ltd, Gödöllő, Hungary. The allele sequences were compared to the MLST database and the sequence types identified.
In the case of two strains (L477, L23), MLST results were extracted from next-generation sequencing data. The following briefly introduces the applied method. Sequencing-compatible genomic DNA was isolated (Wizard Genomic DNA Purification kit, Promega, USA; Lysozyme, Roche, Switzerland) from the specified strains. The library was prepared using enzymatic fragmentation (Ion Xpress Plus Fragment Library kit, Ion Xpress Barcode Adapters 1–16 kit) and sequenced on an Ion Proton semiconductor sequencer (Ion PI Template OT2 200 kit v. 3, Ion PI Sequencing 200 kit v. 3, Ion PI Chip kit v. 2, Life Technolgies, USA). The sequenced reads were aligned (Torrent Suit software, Life Technolgies) to S. pneumoniae reference genome (NCBI Reference Sequence: NC_010380.1). S. pneumoniae MLST gene-specific primers were aligned on the contigs by BLAST [20]. Proprietary software (Matlab, www.mathworks.com) was used to identify the MLST gene sequences and determine the sequence types.
Antibiotic susceptibility testing
The antibiotic sensitivity of the strains to penicillin, cefotaxime, erythromycin, clindamycin, levofloxacin and moxifloxacin was determined by the agar dilution method using an A400 multipoint inoculator (AQS Manufacturing Ltd, UK), and by minimum inhibitory concentration (MIC) test strips (Liofilchem, Italy), on Mueller–Hinton blood agar plates, supplemented with horse blood and 20 mg/l nicotinamide adenine dinucleotide. Incubation was performed at 37 °C in 5% CO2 and ATCC 49 619 was used as the control strain. The susceptibility and resistance rates were determined using the breakpoints suggested by EUCAST guidelines [21]. As our collection consisted of carried strains (i.e. non-meningitis cases), we used the ⩽0·06/>2 breakpoints for penicillin. Inducible clindamycin resistance was tested with the double-disc method, using 15 µg erythromycin and 2 µg clindamycin discs, as described by EUCAST. The strains were tested for the presence of ermB and mef genes [22, 23], and the distinction between mefA and mefE was carried out by BamHI digestion, which generates two fragments in mefA, but none in mefE, as described previously [22, 24].
RESULTS
Serotype arrangement
Nurseries were divided into two groups, according to time of screening and vaccination percentage. Group 1 (GR1) comprised of nurseries from the earlier part of the study (March 2009–April 2010), when the vaccination rate was low (average 16·4%, range 0–33·3%); while children attending nurseries with a higher level of vaccination (average 48·0%, range 19·6–74·4%), screened in the two subsequent winter seasons (October 2010–June 2012) made up group 2 (GR2). Although PCV7 became a free vaccine in Hungary in 2008, widespread vaccination only started in April 2009, upon its inclusion in the NIP. Children start nursery at age 3 years in Hungary, so those who attended nursery in 2011–2012, were vaccinated with increased rates. All children in the study received PCV7.
In GR1, 634 children (attending 15 nurseries) were screened and 216 carried S. pneumoniae, this corresponds to a carriage rate of 34·1%. In GR2, 1628 children (attending 25 nurseries) were screened, and 529 carried pneumococcus, resulting in a carriage rate of 32·5%. On three occasions double carriage was identified, therefore the number of pneumococcal isolates was 218 in GR1 and 530 in GR2.
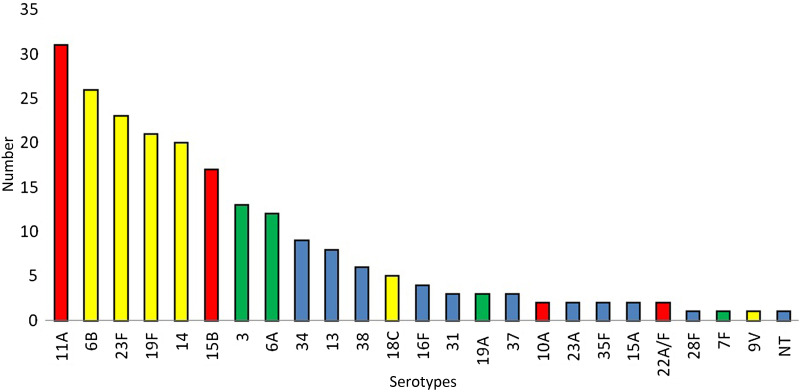
The serotype distribution in GR1 (Fig. 1) reflected the high prevalence of PCV7 serotypes (44·0%), which also indicated the need for vaccination. The calculated PCV13 coverage was 57·3% for GR1, which was particularly due to the relative high frequency of serotypes 3 and 6A (6·0% and 5·5%, respectively). Among the NVTs, 11A and 15B combined accounted for 22·0% of the isolates, but other NVTs were rare.
Fig. 1.
Serotype distribution in group 1 (n = 218 pneumococci). Yellow columns, PCV7 serotypes; green columns, additional serotypes in PCV13; red columns, PPV23 serotypes; blue columns, non-PCV serotypes; NT, non-typable.
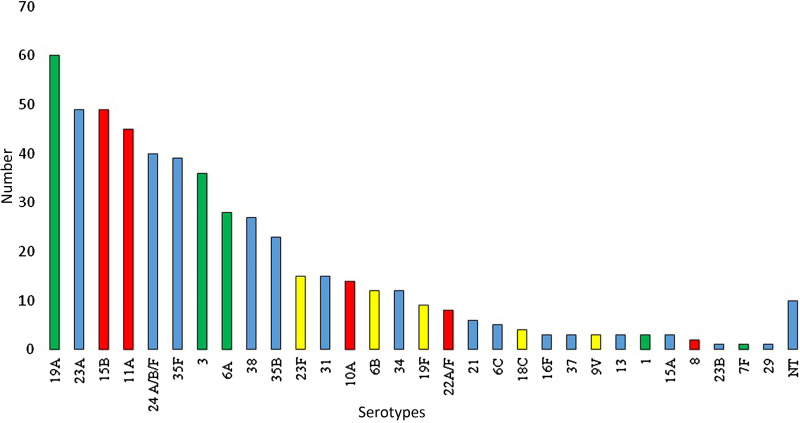
The serotype distribution in GR2 (Fig. 2) showed a marked rearrangement. The rate of PCV7 types decreased markedly (to 8·1%), e.g. serotype 14 vanished completely from the isolates. On the other hand, the prevalence of PCV13/non-PCV7 types increased considerably (being responsible for 24·5% of all strains in GR2). Serotypes 19A, 3 and 6A were represented in high numbers, while serotype 5 was completely absent throughout the study and there were only four strains of serotypes 1 or 7F. Five strains of serotype 6C were detected in GR2, while none were found in GR1. Furthermore, many previously very rare or absent serotypes, such as 23A, 24F, 35F or 38 emerged. One of the most marked differences in the serotype distribution occurred with serotype 19A: while there were only three isolates in GR1 (3/218 = 1·4% prevalence), its contribution increased to 11·5% in GR2 (61/530). This corresponds to a >eightfold increase in frequency. Two serotypes maintained a nearly constant rate: 6A and 3. A comparison of the prevalence of vaccine types is shown in Table 1.
Fig. 2.
Serotype distribution in group 2 (n = 530 pneumococci). Yellow columns, PCV7 serotypes; green columns, additional serotypes in PCV13; red columns, PPV23 serotypes; blue columns, non-PCV serotypes; NT, non-typable.
Table 1.
Prevalence of vaccine serotypes in groups 1 and 2
| Prevalence in group 1 (N = 218) | Prevalence in group 2 (N = 530) | |
|---|---|---|
| Serotype | n (%) | n (%) |
| PCV7 serotypes: 96 (44·0%) | PCV7 serotypes: 43 (8·1%) | |
| 4 | 0 (0·0) | 0 (0·0) |
| 6B | 26 (11·9) | 12 (2·3) |
| 9V | 1 (0·5) | 3 (0·6) |
| 14 | 20 (9·2) | 0 (0·0) |
| 18C | 5 (2·3) | 4 (0·8) |
| 19F | 21 (9·6) | 9 (1·7) |
| 23F | 23 (10·6) | 15 (2·8) |
| PCV13/non-PCV7 serotypes: 29 (13·3%) | PCV13/non-PCV7 serotypes: 129 (24·3%) | |
| 1 | 0 (0·0) | 3 (0·6) |
| 5 | 0 (0·0) | 0 (0·0) |
| 7F | 1 (0·5) | 1 (0·2) |
| 3 | 13 (6·0) | 36 (6·8) |
| 6A | 12 (5·5) | 28 (5·3) |
| 19A | 3 (1·4) | 61 (11·5) |
Antibiotic susceptibility
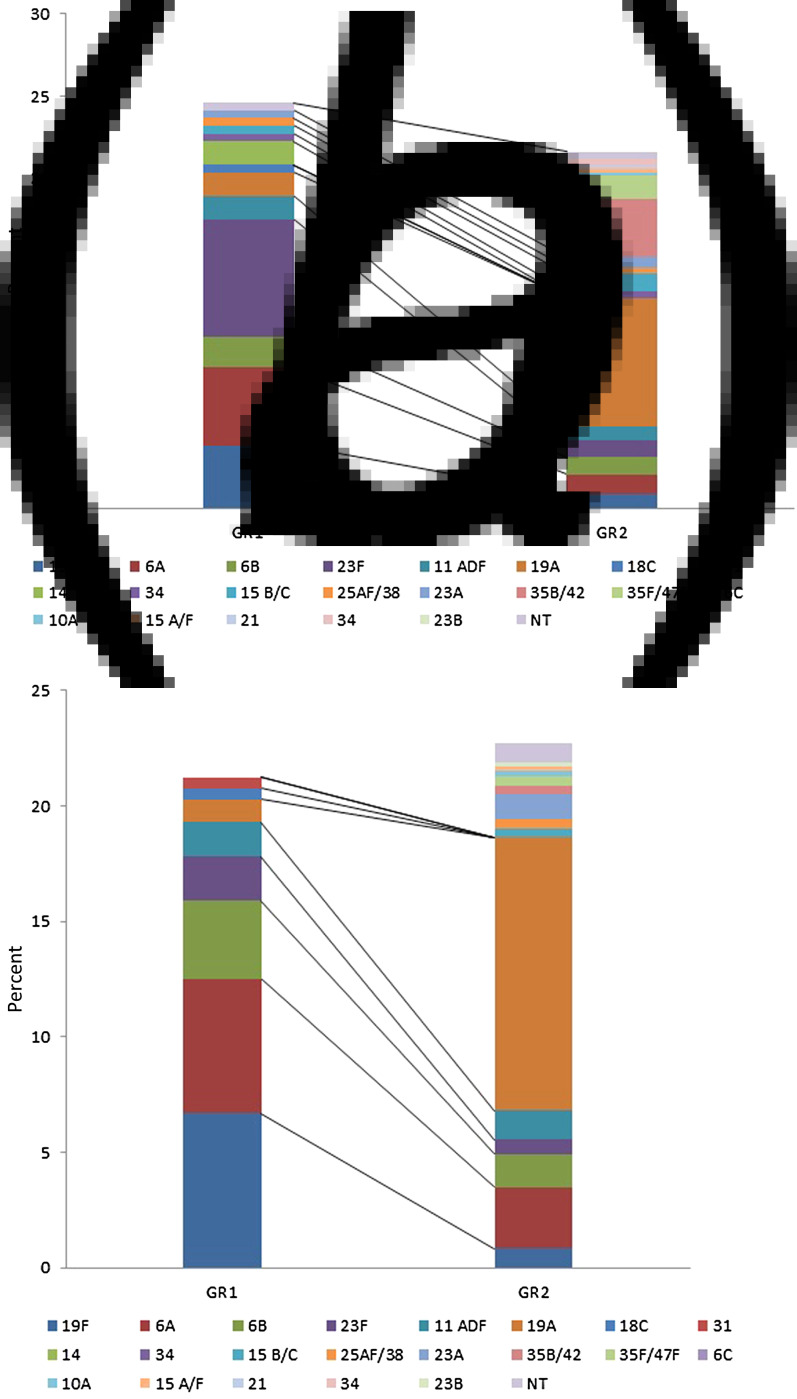
The antibiotic susceptibility rates in GR1 and GR2 did not change significantly, but the serotypes contributing to resistance showed great differences. The ratio of penicillin intermediate resistance (penI, MIC = 0·125–2 mg/l) was 24·6% and 21·6% in GR1 and GR2, respectively. While the old ‘paediatric’ serotypes (23F, 19F, 6A, 6B, 14) provided >70% of these isolates in GR1 (serotype 23F in the largest proportion, Fig. 3a), serotype 19A alone was completely dominant in GR2 (36·3% of penI isolates). Regarding erythromycin resistance (eryR), the overall rate increased slightly from 21·3% in GR1 to 22·7% in GR2. While in GR1 ~85% of the eryR isolates belonged to the paediatric serotypes (19F and 6A contributing equally, Fig. 3b), in GR2 serotype 19A was responsible for 52·1% of eryR and all other serotypes were represented only in small proportions.
Fig. 3.
Contribution of serotypes to penicillin intermediate resistance and erythromycin resistance, in groups 1 and 2 (GR1 and GR2). (a) Number of penicillin intermediate isolates were 54/218 vs. 115/530 in GR1 and GR2, respectively. (b) Number of erythromycin-resistant isolates were 46/218 vs. 120/530 in GR1 and GR2, respectively.
Serotype 19A strains
A total of 61 isolates of serotype 19A (three in GR1 and 58 in GR2) were detected. Regarding their antibiotic susceptibility (Table 2), only one isolate was fully resistant to penicillin (MIC = 4 mg/l), but 2/3 of them (67·2%, n = 41) fell into the intermediate category. Of concern, almost all serotype 19A isolates (n = 56) were highly resistant to both erythromycin and clindamycin (MIC >256 mg/l). All of these isolates carried the ermB gene, and one possessed ermB + mefE in combination. Three additional isolates had high-level erythromycin MICs, but were phenotypically sensitive to clindamycin, these had inducible clindamycin resistance; and surprisingly these were also double carriers of ermB + mefE. Furthermore, these four ermB/mefE positive isolates had elevated penicillin MICs (1 or 2 mg/l) and unusually high cefotaxime MICs (1–3 mg/l). Two isolates were fully sensitive to both erythromycin and clindamycin. All 61 serotype 19A isolates were sensitive to levofloxacin and moxifloxacin.
Table 2.
Antibiotic susceptibility results of the 61 serotype 19A isolates
| MIC range | S (%) | I (%) | R (%) | |
|---|---|---|---|---|
| Penicillin | 0·016–4 | 31·3 | 67·2 | 1·6 |
| Cefotaxime | 0·008–3 | 93·4 | 4·9 | 1·6 |
| Erythromycin | 0·064–>256 | 3·3 | — | 96·7 |
| Clindamycin | 0·032–>256 | 8·2 | — | 91·8 |
| Levofloxacin | 0·5–2 | 100 | — | — |
| Moxifloxacin | 0·032–0·5 | 100 | — | — |
MIC, Minimum inhibitory concentration.
Based on the information extracted from the questionnaires, two important risk factors could be identified for 19A carriage, i.e. male gender and passive exposure to smoking; however, none of them were statistically significant (using the χ2 test). Of the 61 children who carried 19A pneumococcus, 39 were males (63·9%), while among the non-19A carriers, gender was equally represented (52·3% males, P = 0·081). Regarding the other factor, 49·2% of the 19A carrier children were exposed to smoking in the household; this figure is 39·2% in the general carrier population (P = 0·139).
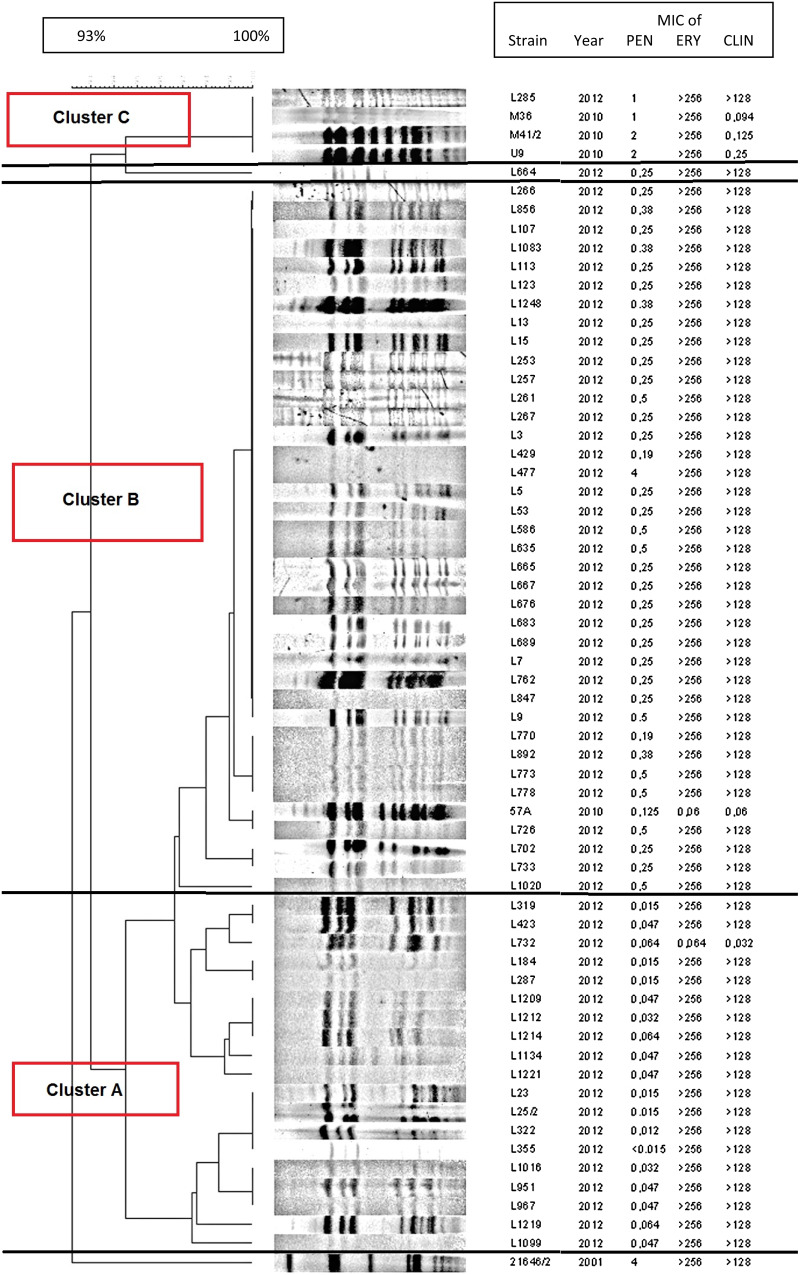
The clonality among serotype 19A isolates was examined with PFGE. The digested genome patterns (Fig. 4) showed that although all isolates were related to one another, several lineages could be identified. These PFGE clusters shared similar penicillin susceptibility rates. For instance, the isolates fully susceptible to penicillin formed a distinct group (cluster A). Another larger cluster comprised of isolates with intermediate penicillin MICs (cluster B). Interestingly, the single penicillin-resistant strain in the collection (L477) had an identical PFGE pattern to these isolates. Additionally, the four isolates possessing ermB + mefE in combination, were clustered into a separate small PFGE clone (cluster C). In order to compare the 19A strains of the current study to the previously frequent Hungary19A−6 PMEN clone [14, 18], which typically was resistant to penicillin, we have included one representative isolate of that clone (isolate 21646, isolated in 2001) in the PFGE analysis. As is obvious from Figure 4, the new 19A isolates differ greatly from this older clone.
Fig. 4.
PFGE patterns of the 61 serotype 19A isolates in the present study, compared to the Hungary19A−6 PMEN clone. The isolate 21 646/2 (last strain in the dendrogram) represents the Hungary19A−6 PMEN clone.
For MLST analysis, a few isolates were selected from each PFGE cluster. The MLST was determined for five isolates within PFGE cluster A (L23, L1214, L1134, L319 and L732, all penicillin sensitive, Fig. 4), and four of these isolates had identical sequence types (ST8430). The fifth isolate had a completely different sequence type (ST97), differing in six loci, but this isolate was sensitive to macrolides, unlike the rest (Table 3). The MLST was determined also for five isolates within PFGE cluster B (L477, L770, L762, 57A, L1020), and again, four belonged to the same sequence type (ST319), but the only macrolide-sensitive isolate (57A) was different (ST1611), in this case differing in three alleles. Regarding PFGE cluster C, MLST was done for one representative isolate (M41), and this proved to be ST320. Additionally, isolate L664, which showed a dissimilar PFGE pattern, proved to have an unrelated sequence type (ST1757).
Table 3.
Data of the serotype 19A strains with known sequence types
| Isolate | PFGE cluster | Penicillin MIC | Erythromycin MIC | Clindamycin MIC | Sequence type |
|---|---|---|---|---|---|
| L23 | A | 0·016 | >256 | >128 | ST8430 |
| L1214 | A | 0·064 | >256 | >128 | |
| L1134 | A | 0·064 | >256 | >128 | |
| L319 | A | 0·016 | >256 | >128 | |
| L732 | A | 0·064 | 0·064 | 0·032 | ST97 |
| L477 | B | 4 | >256 | >128 | ST319 |
| L770 | B | 0·25 | >256 | >128 | |
| L762 | B | 0·25 | >256 | >128 | |
| L1020 | B | 0·5 | >256 | >128 | |
| 57A | B | 0·125 | 0·06 | 0·06 | ST1611 |
| L664 | Misc. | 0·25 | >256 | >128 | ST1757 |
| M41/2 | C | 2 | >256 | 0·125 (Ri) | ST320 |
| 21 646 | PMEN | 4 | >256 | >128 | ST268 |
PFGE, Pulsed-field gel electrophoresis; MIC, minimum inhibitory concentration.
Based on the MLST data, we were able to confirm that the current isolates are different from the Hungary19A−6 PMEN clone. The latter belongs to the ST268 clone [25], while our isolates had different sequence types.
DISCUSSION
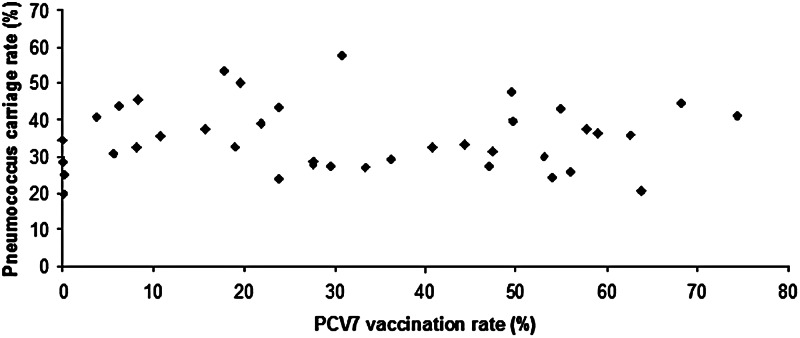
Comparing the two groups of children with low-level (GR1) and high-level (GR2) vaccination rates (both groups involving several hundreds of children), it was clear that, although global carriage rate hardly decreased (34·1% vs. 32·5%), the serotype distribution almost completely changed. This stable carriage rate is reflected in Figure 5, where the pneumococcal carriage rate is shown in relation to the vaccination rate of each nursery group.
Fig. 5.
Pneumococcus carriage rates in the individual nursery groups, in relation to the vaccination rate of each group.
Former dominant serotypes were nearly eliminated (such as serotype 14), and the vacated niche was replaced by circulating new types (e.g. 23A, 24F). Among the PCV13/non-PCV7 serotypes, the frequency of serotypes 3 and 6A remained stable, while the number of serotype 19A strains increased to a great extent. Unfortunately these isolates are fully resistant to erythromycin and clindamycin and have elevated MICs to penicillin.
The newest available European surveillance (ECDC) data published in February 2015 [26] also reflect the importance of serotype 19A: it was the leading serotype in IPD of children aged <1 year, the third most frequent of IPD in all age groups, and the second most frequent cause of fatal cases (after serotype 3) in a total of 520 reported deaths in 2012. Serotype 19A also ranked first in IPD of children in the United States after 7–8 years post-PCV7 [27]. Based on UK nationwide data, serotype 19A was shown to be associated with high mortality in the 5–64 years age group [28]. Nonetheless, this emergence of serotype 19A might not solely be the aftermath of vaccination with PCV7. Other factors, such as selective pressure of antibiotic usage or simply the natural fluctuations in seroepidemiology observed over larger time-frames could also play an important role [29–31]. However, independent of the driving force, serotype 19A is increasing in frequency and is a problem that must be dealt with.
As is well known, nasopharyngeal colonization precedes infection in most cases, and the conjugate vaccines have been proven to have an impact not only on pneumococcal diseases, but also on nasopharyngeal carriage [5, 32]. Therefore, several studies surveyed the rearrangement of serotype distribution in colonization after PCV7 vaccination [31, 33, 34] and also reported the emergence of 19A. In a randomized controlled trial in The Netherlands involving nearly 1000 children, van Gils et al. found that acquisition of serotype 19A was significantly higher in children who completed the 2 + 1 vaccination schedule with PCV7, compared to unvaccinated children [31]. In a study by Sleeman et al. [35], the duration of carriage of the individual serotypes was investigated, and an inverse relationship between attack rate and duration was found. They concluded that the disease-causing potential of the serotypes should be evaluated using the pneumococcal acquisition rates rather than the simple carriage rates. Serotype 19A belonged to the serotypes with high attack rates (>20 IPD cases/100 000 acquisitions), with an average duration of carriage of 12·6 weeks.
It was interesting to observe that the overall resistance rate remained stable over time; however, while several serotypes were represented in GR1, the absolute dominance of serotype 19A in GR2 was clear, with regard to both penicillin and erythromycin non-susceptibility.
To compare our results to disease-causing Hungarian pneumococci, data of invasive isolates, provided by the National Center for Epidemiology (NCE), are available [36]. Between 2008 and 2012, out of 545 IPD isolates, serotype 19A was stable around 6–6·6%, serotype 3 was the leading type (~27%) and serotype 14 disappeared. A more detailed comparison has been published previously [13].
The 19A strains were clonal, but PFGE allowed identification of some smaller clusters, which showed strong relation to penicillin sensitivity levels. It is clear from the PFGE patterns and MLST results that none of them are identical to the previously dominant, highly resistant and virulent Hungarian 19A clone. We were able to detect the presence of two major sequence types, ST319 and ST8430, both of which showed very good correlation with PFGE clustering.
In the MLST database [29], nine hits were found for ST319 (serotype 19A, n = 6; serotype 19F, n = 3), five strains from Germany, two from Poland, and one each from Greece and Spain. Five of these derived from invasive infections and two from the nasopharynx. They were all penicillin intermediate, like our isolates.
In the case of ST8430, only one strain was found in the MLST database (serotype 19A), also isolated in Germany and sensitive to penicillin.
Among our isolates in PFGE cluster C, showing elevated penicillin MICs, high-level erythromycin resistance, inducible clindamycin resistance, and possessing the ermB + mefE genes in combination, we identified ST320, which is a well-known multi-resistant pneumococcal clone circulating throughout the world. For instance, ST320 was first detected in 2005 in Barcelona, 4 years after the introduction of PCV7, and 5–6 years later it characterized nearly 3/4 of the serotype 19A isolates [37]. In Alaska, ST320 was also first detected only 6–7 years after the introduction of PCV7, but soon represented 13% of all 19A isolates [38]. All these ST320 isolates also carried the ermB + mefE gene combination and were multi-resistant [39], suggesting that ST320 probably developed from its ancestor Taiwan19F−14 PMEN clone (ST236) through a recombination event between a strain belonging to this clone and a serotype 19A strain [39], under the pressure of the use of PCV7. The isolates of this new ST320 were shown to have outcompeted Taiwan19F−14 (ST236) [40]. Interestingly, isolates belonging to the Taiwan19F−14 clone were found in clinical isolates before the PCV era in Hungary [22], so either a genetic recombination occurred also in Hungary leading to the development of ST320, or members of the ST320 clone have spread to Hungary. Based on the 12 isolates with known sequence types – with only one exception – PFGE and MLST showed a strong correlation.
Although not much time has passed since the introduction of PCV13, most recent international data suggest that following the worldwide emergence of non-PCV7 serotypes, a slowing in the rate of increase has been observed in the prevalence of these types, including 19A [11]. We hope this tendency will continue and the emergence of serotype 19A will be followed by a rapid decline also in Hungary now that PCV13 is a mandatory vaccine.
ACKNOWLEDGEMENTS
Part of this work was presented at the 23rd ECCMID (O397, Berlin, 2013). We acknowledge the use of the pneumococcal MLST database which is located at Imperial College London and is funded by the Wellcome Trust.
This project was funded by the National Research, Development and Innovation Office/Hungarian Scientific Research Fund (grant no. K108631).
DECLARATION OF INTEREST
M.vd.L. has been a member of advisory boards for, and has received research grants and speakers' honorary fees from, Pfizer, GSK, Merck and Sanofi-PasteurMSD. The remaining authors have nothing to declare.
REFERENCES
- 1.Scott JA. The preventable burden of pneumococcal disease in the developing world. Vaccine 2007; 25: 2398–2405. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Pneumococcal conjugate vaccine for childhood immunization – WHO position paper. Weekly Epidemiological Record 2012; 87: 129–144.24340399 [Google Scholar]
- 3.Bogaert D, De Groot R, Hermans PW. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infectious Diseases 2004; 4: 144–154. [DOI] [PubMed] [Google Scholar]
- 4.Dagan R, et al. Nasopharyngeal colonization in southern Israel with antibiotic-resistant pneumococci during the first 2 years of life: relation to serotypes likely to be included in pneumococcal conjugate vaccines. Journal of Infectous Diseases 1996; 174: 1352–1355. [DOI] [PubMed] [Google Scholar]
- 5.Ledwith M. Pneumococcal conjugate vaccine. Current Opinion in Pediatrics 2001; 13: 70–74. [DOI] [PubMed] [Google Scholar]
- 6.National Center for Epidemiology. Pneumococcal vaccination data in Hungary for those born in 2009 and 2010 [in Hungarian]. Epinfo 2012; 19: 262–264. [Google Scholar]
- 7.National Center for Epidemiology. Pneumococcal vaccination data in Hungary for those born in 2011 [in Hungarian]. Epinfo 2013; 20: 331–334. [Google Scholar]
- 8.National Center for Epidemiology. Pneumococcal vaccination data in Hungary for those born in 2012 [in Hungarian]. Epinfo 2014; 21: 362–365. [Google Scholar]
- 9.Isaacman DJ, McIntosh ED, Reinert RR. Burden of invasive pneumococcal disease and serotype distribution among Streptococcus pneumoniae isolates in young children in Europe: impact of the 7-valent pneumococcal conjugate vaccine and considerations for future conjugate vaccines. International Journal of Infectious Diseases 2010; 14: e197–209. [DOI] [PubMed] [Google Scholar]
- 10.Whitney CG, et al. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. New England Journal of Medicine 2003; 348: 1737–1746. [DOI] [PubMed] [Google Scholar]
- 11.Weil-Olivier C, et al. Prevention of pneumococcal diseases in the post-seven valent vaccine era: a European perspective. BMC Infectious Diseases 2012; 12: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tóthpál A, et al. Nasal carriage of Streptococcus pneumoniae among Hungarian children before the wide use of the conjugate vaccine. Acta Microbiologica et Immunologica Hungarica 2012; 59: 107–118. [DOI] [PubMed] [Google Scholar]
- 13.Tóthpál A, et al. Radical serotype rearrangement of carried pneumococci in the first 3 years after intensive vaccination started in Hungary. European Journal of Pediatrics 2015; 174: 373–381. [DOI] [PubMed] [Google Scholar]
- 14.McGee L, et al. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. Journal of Clinical Microbiology 2001; 39: 2565–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagai K, et al. Evaluation of PCR primers to screen for Streptococcus pneumoniae isolates and beta-lactam resistance, and to detect common macrolide resistance determinants. Journal of Antimicrobial Chemotherapy 2001; 48: 915–918. [DOI] [PubMed] [Google Scholar]
- 16.CDC List of oligonucleotide primers used for pneumococcal serotype deduction by conventional multiplex PCR (http://www.cdc.gov/streplab/downloads/pcr-oligonucleotide-primers.pdf). Accesed 13 March 2015.
- 17.Brito DA, Ramirez M, de Lencastre H. Serotyping Streptococcus pneumoniae by multiplex PCR. Journal of Clinical Microbiology 2003; 41: 2378–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobay O, et al. The relationship between serotypes and PFGE genotypes in isolates of Streptococcus pneumoniae from Hungary. Clinical Microbiology and Infection 2005; 11: 673–676. [DOI] [PubMed] [Google Scholar]
- 19.Streptococcus pneumoniae MLST website. (http://pubmlst.org/spneumoniae/). Accessed 13 March 2015.
- 20.BLAST. (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Accessed 13 March 2015.
- 21.EUCAST. The European Committee on Antimicrobial Susceptibility Testing (http://www.eucast.org/clinical_breakpoints/). Accesed 13 March 2015.
- 22.Dobay O, Rozgonyi F, Amyes SG. Molecular characterisation of Hungarian macrolide-resistant Streptococcus pneumoniae isolates, including three highly resistant strains with the mef gene. International Journal of Antimicrobial Agents 2005; 25: 488–495. [DOI] [PubMed] [Google Scholar]
- 23.Sutcliffe J, et al. Detection of erythromycin-resistant determinants by PCR. Antimicrobial Agents and Chemotherapy 1996; 40: 2562–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oster P, et al. Patterns of macrolide resistance determinants among community-acquired Streptococcus pneumoniae isolates over a 5-year period of decreased macrolide susceptibility rates. Antimicrobial Agents and Chemotherapy 1999; 43: 2510–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MLST databases. Streptococcus pneumoniae PMEN CLONE SET Hungary 19A-6 ST-268 (http://spneumoniae.mlst.net/sql/pmensts.asp?clone=Hungary%2019A-6&st=268). Accessed 13 March 2015.
- 26.European Centre for Disease Prevention and Control. Surveillance of invasive bacterial diseases in Europe, 2012. Stockholm: ECDC, 2015. [Google Scholar]
- 27.Kaplan SL, et al. Serotype 19A is the most common serotype causing invasive pneumococcal infections in children. Pediatrics 2010; 125: 429–436. [DOI] [PubMed] [Google Scholar]
- 28.van Hoek AJ, et al. Effect of serotype on focus and mortality of invasive pneumococcal disease: coverage of different vaccines and insight into non-vaccine serotypes. PLoS ONE 2012; 7: e39150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dagan R, et al. Introduction and proliferation of multidrug-resistant Streptococcus pneumoniae serotype 19A clones that cause acute otitis media in an unvaccinated population. International Journal of Infectious Diseases 2009; 199: 776–785. [DOI] [PubMed] [Google Scholar]
- 30.Reinert R, Jacobs MR, Kaplan SL. Pneumococcal disease caused by serotype 19A: review of the literature and implications for future vaccine development. Vaccine 2010; 28: 4249–4259. [DOI] [PubMed] [Google Scholar]
- 31.van Gils EJ, et al. Pneumococcal conjugate vaccination and nasopharyngeal acquisition of pneumococcal serotype 19A strains. Journal of the American Medical Association 2010; 304: 1099–1106. [DOI] [PubMed] [Google Scholar]
- 32.Simell B, et al. The fundamental link between pneumococcal carriage and disease. Expert Review of Vaccines 2012; 11: 841–855. [DOI] [PubMed] [Google Scholar]
- 33.Tocheva AS, et al. Distribution of carried pneumococcal clones in UK children following the introduction of the 7-valent pneumococcal conjugate vaccine: a 3-year cross-sectional population based analysis. Vaccine 2013; 31: 3187–3190. [DOI] [PubMed] [Google Scholar]
- 34.Sharma D, et al. Pneumococcal carriage and invasive disease in children before introduction of the 13-valent conjugate vaccine: comparison with the era before 7-valent conjugate vaccine. Pediatric Infectious Disease Journal 2013; 32: e45–53. [DOI] [PubMed] [Google Scholar]
- 35.Sleeman KL, et al. Capsular serotype-specific attack rates and duration of carriage of Streptococcus pneumoniae in a population of children. Journal of Infectous Diseases 2006; 194: 682–688. [DOI] [PubMed] [Google Scholar]
- 36.Tirczka T, Berta B. Results of the Streptococcus pneumoniae serotyping and antibiotic resistance surveillance, between 1 January 2008 and 31 August 2011 [in Hungarian]. Microbiology Circular 2012; 3: 3–17. [Google Scholar]
- 37.Gene A, et al. Pneumococcal serotypes causing acute otitis media among children in Barcelona (1992–2011): emergence of the multiresistant clone ST320 of serotype 19A. Pediatric Infectious Disease Journal 2013; 32: e128–133. [DOI] [PubMed] [Google Scholar]
- 38.Rudolph K, et al. Molecular epidemiology of serotype 19A Streptococcus pneumoniae among invasive isolates from Alaska, 1986–2010. International Journal of Circumpolar Health 2013; 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore MR, et al. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. Journal of Infectous Diseases 2008; 197: 1016–1027. [DOI] [PubMed] [Google Scholar]
- 40.Hsieh YC, et al. Expansion and evolution of Streptococcus pneumoniae serotype 19A ST320 clone as compared to its ancestral clone, Taiwan19F-14 (ST236). Journal of Infectious Diseases 2013; 208: 203–210. [DOI] [PubMed] [Google Scholar]