SUMMARY
ST131 Escherichia coli is an emergent clonal group that has achieved successful worldwide spread through a combination of virulence and antimicrobial resistance. Our aim was to develop a mathematical model, based on current knowledge of the epidemiology of ESBL-producing and non-ESBL-producing ST131 E. coli, to provide a framework enabling a better understanding of its spread within the community, in hospitals and long-term care facilities, and the potential impact of specific interventions on the rates of infection. A model belonging to the SEIS (Susceptible–Exposed–Infected–Susceptible) class of compartmental models, with specific modifications, was developed. Quantification of the model is based on the law of mass preservation, which helps determine the relationships between flows of individuals and different compartments. Quantification is deterministic or probabilistic depending on subpopulation size. The assumptions for the model are based on several developed epidemiological studies. Based on the assumptions of the model, an intervention capable of sustaining a 25% reduction in person-to-person transmission shows a significant reduction in the rate of infections caused by ST131; the impact is higher for non-ESBL-producing ST131 isolates than for ESBL producers. On the other hand, an isolated intervention reducing exposure to antimicrobial agents has much more limited impact on the rate of ST131 infection. Our results suggest that interventions achieving a continuous reduction in the transmission of ST131 in households, nursing homes and hospitals offer the best chance of reducing the burden of the infections caused by these isolates.
Key words: Epidemiology, Escherichia coli, ST131, extended-spectrum β-lactamases, mathematical model, transmission
INTRODUCTION
A clonal group of Escherichia coli isolates belonging to phylogroup B2, namely O25b:H4/ST131, recently emerged as an important cause of human infections worldwide [1, 2]. This clone has been found to be the predominant clonal group associated with antibiotic-resistant E. coli in different areas [1–4]. Although it was initially found in relation to extended-spectrum β-lactamase (ESBL) production, recent studies show that most ST131 isolates do not produce ESBLs [3, 5]. Fluoroquinolone resistance has been significantly associated with the most common lineages within this clone [4]; antimicrobial resistance and virulence factors are thought to have been important in the spread of this clone [1, 2]. However, the interactions between the epidemiological determinants of ST131 are not well-characterized.
The application of mathematical models to study the dissemination of infectious diseases is now common practice [6–8]. Some models have been developed on the dissemination of E. coli O157 [9]; or cephalosporin-resistant E. coli in hospitalized patients [10]. However, to the best of our knowledge, no mathematical model has yet been proposed to analyse the dissemination of the ST131 clonal complex.
The objective of this study was to construct an epidemiological framework and mathematical model to explain the spread of ESBL-producing and non-ESBL-producing E. coli ST131 isolates using a population-based perspective and the potential impact of control interventions.
Study design and data collection
To construct the model, we used recent data from five different prospective studies: (1) A 30-week prospective study in Seville, Spain, was performed to estimate the actual prevalence of ST131 isolates [5]. Briefly, 4308 E. coli isolated were screened by PCR for ST131. (2) Faecal carriage of ST131 isolates was evaluated using rectal swabs from patients diagnosed with E. coli ST131 infection, their contacts (household members for community cases and room-mates for hospitalized cases) [11, 12]. Mean duration of ST131 intestinal carriage was analysed in a subset of the participants. (3) Individual risk factors for infection due to E. coli ST131 were investigated using a case-control study [13]. (4) The risk factors and prognosis for bloodstream infections caused by ST131 were studied, using a case-control design (data on file). (5) Faecal carriage data from 54 cases of community-acquired urinary tract infection (UTI) caused by ESBL-producing E. coli; 105 relatives and 54 controls were used to provide information about spread of ESBL-producing E. coli in households [14]. The studies were approved by the Institutional Review Board at the University Hospital Virgen Macarena (Seville, Spain), which waived the need for consent except for the study of faecal carriers for whom written informed consent was obtained.
Model methodology
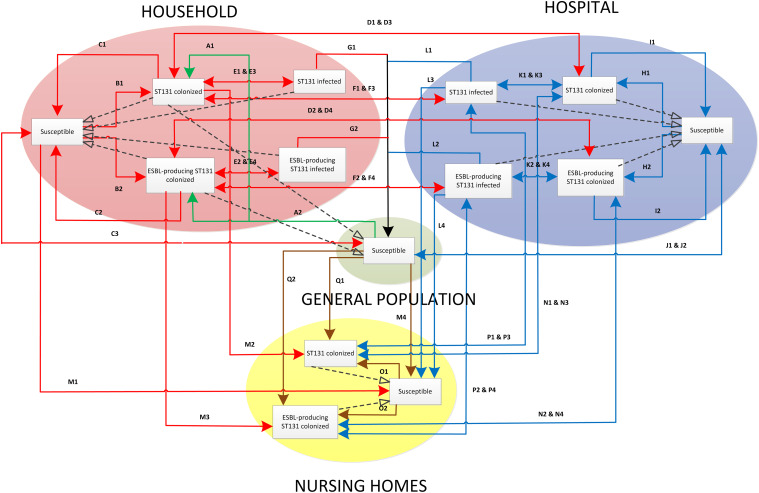
A structural model was proposed. The exposed individuals were divided into separate subpopulation groups with specific transmission environments (households, nursing homes, hospitals). Epidemiological models in the literature often analyse these environments separately; by contrast, this work aims to emphasize the interactions among different populations liable to suffer contagion by ESBL-producing and non-ESBL-producing ST131.
Epidemiological states considered included susceptible, colonized and infected. The model was developed using the following stepwise objectives: (1) To analyse possible interactions between the different variables, taking cause–effect relationships into account, and to describe these relationships via mathematical equations, leading to a mathematical model of the system. (2) To implement the mathematical model using a high-level programming language. (3) To validate the model with the dynamics observed. (4) To exploit the model by simulating different scenarios to help analyse the effects of possible strategies for the prevention and control of bacterial transmission.
Model description and assumptions
We considered the epidemiological background of ST131 [1, 2], data from ad-hoc studies (see above) [5, 11–14], and the various models available that address the transmission of infectious diseases [6–10]. The proposed structural model falls within the SEIS class (Susceptible–Exposed–Infected–Susceptible) of compartmental models, in which we used the term ‘colonized’ instead of ‘exposed’.
The following assumptions according to available data were established:
The main mechanism of transmission was considered to be person-to-person. ST131 E. coli has been isolated only occasionally from food and companion animals [1, 15–18].
From available evidence and clinical studies, there is no interaction among colonized or infected individuals by non-ESBL-producing ST131 and those colonized or infected by ESBL-producing ST131. ESBL-producing ST131 formed distinct clusters separated from ESBL-negative isolate [19].
The following data were used for estimations: prevalence of colonized persons with ST131 in the whole population, 2%; with ESBL-producing ST131, <1%; 93% of infections caused by ST131 infections are caused by non-ESBL-producers; 78% of infections are community onset [5]; 18% of members of households of a colonized person are colonized with ST131 and 6% with ESBL-producing ST131; the remainder (76%) are susceptible individuals [11]. Regarding hospitalized individuals, 15% are colonized by ST131, 8% of these with an ESBL-producing isolate [11, 12]. Fifty per cent of infected patients are still colonized after 1 week [14]. Re-colonization was considered possible.
The majority of the total population considered is included in the ‘susceptible’ group of the ‘general population’ compartment. These subjects are those not sharing a household with a colonized patient and therefore are at lower risk of acquiring E. coli ST131.
Households considered were those with at least one colonized member. The ‘susceptible’ members may acquire ST131 E. coli mainly from the colonized household member [11]. The initial proportion of susceptible (non-colonized) to colonized persons in households was estimated to be 3:1, and the average number of persons in a household to be n = 4 [11, 12].
In acute healthcare institutions (hospitals) the proportion of susceptible/colonized individuals was considered to be 5:1 [11]. The estimated prevalence of patients colonized by E. coli ST131 at admission was 2% [11]; of these, 10% are colonized by an ST131 ESBL-producing E. coli [5]. Because available data suggest that transmission of E. coli ST131 within acute-care hospitals is low [11, 20], we assume that 2% of discharged patients harbour this microorganism.
In nursing homes, the input of colonized individuals is derived predominantly from households, with outputs being those who die or are transferred to acute healthcare institutions, which is particularly frequent in the group developing infection [11].
Previous antibiotic use (mainly fluoroquinolones and β-lactam/β-lactamase inhibitors) is a risk factor for infection caused by ST131 and for those caused by ESBL-producing ST131 [13] but not for colonization as extrapolated from a study on ESBL-producing isolates [14].
The relationships between the birth, death, immigration and emigration rates are ignored because the simulation length covers a period of only 1 year.
Additional flows between compartments were included in order to adapt the SEIS model to the conditions of our study. For example, a new flow was added regarding individuals who are admitted in the hospital suffering a pathology not related to E. coli infection (i.e. they flow from the susceptible group of general population to the susceptible group of the hospital). Later, they may either return to be susceptible in the general population compartment (once they are recovered) or be colonized by E. coli and then infected. Nevertheless, a major proportion of individuals recovered from hospital will return to the susceptible group of the general population compartment and the remainder to the susceptible group of the nursing-home compartment.
Figure 1 presents the structure of the proposed model and the cause–effect relationships between variables, based on the above-mentioned assumptions.
Fig. 1.
Model structure.
Quantification of the model was based on the law of mass preservation, with state variables corresponding to the populations under consideration in households, nursing homes, and hospitals, and enabled relationships in the flows of individuals between the different compartments to be determined. On the basis of this methodology, a set of first-level coupled differential equations was proposed representing the dynamic behaviours of population states. In general, state equations are expressed as:
| 1 |
where Dpi represents the population state of compartment i, and Fap (Fdp) expresses the flow of individuals/day entering (or leaving) that state. A detailed mathematical description can be found in the Supplementary material.
Model parameters were determined according to the subpopulation size. Thus, a probabilistic quantification was used for small-sized subpopulations (such as hospitals and nursing homes). Due to the insufficient amount of available data from the analysed population, it is not possible to perform a statistical study that provides an average parameter for each subpopulation. Therefore, as done in other works of epidemiological modelling, the contagious probability is proposed based on experience of clinical experts.
For large-sized subpopulations (such as households and general population), a deterministic quantification was used adjusted using the least squares method. According to data from previous studies [5, 11–14], the mean probability [21] for colonization was calculated for each compartment of the model through an adjustment by the least squares method from clinical data collected. Thus, the parameters γ affecting flows A1, A2, B1, and B2 mean the probability that a person has to be colonized by (ESBL- or non-ESBL-producing) ST131 on a day in the general population and household.
The initial conditions and parameter values of the model are shown in Supplementary Tables S1, S2 and S3. Initial conditions and parameters can ease the use of the global model or a simplified version of it for other population or bacteria.
We developed different simulations with the aim of analysing the dynamics of the different populations from the variation of the parameters and initial conditions in the model rather than providing precise estimates on the number of persons colonized.
Sensitivity analysis
All the parameters were analysed to determine their influence in the number of colonized individuals. Results showed that the most dominant parameters were: (1) those related to colonization acquisition, (2) those affecting susceptible individuals in the general population (γrST131 and γrESBL), (3) the infection parameter, and (4) the initial populations of colonized individuals. For the sensitivity analysis of initial populations, variations were considered by using a multivariate technique, in which different parameters are considered at the same time.
The remaining set of parameters has a lower relevance since they influence smaller population niches. For these parameters, the sensitivity analysis was performed using a univariate technique, in which the model is analysed considering variations in one of these parameters [22–25].
RESULTS
Model validation
In order to assess the validity of the model, we estimated the number of persons colonized with E. coli ST131 based on the initial conditions. To do this, the complete set of individuals with infections caused by non-ESBL-producing and ESBL-producing E. coli ST131 in 1 year was considered. The size of the population in each compartment in 1 year was estimated based on local demographic data and was 800000, 70000 and 7000 for the general population, the hospital population, and the nursing home population, respectively. The estimated cumulative numbers of persons infected with ESBL-producing and ESBL-non-producing ST131 E. coli over 2 years are shown in Supplementary Figure S1. The results obtained are in agreement with the epidemiological data.
Sensitivity analysis of the parameters and starting conditions in the model
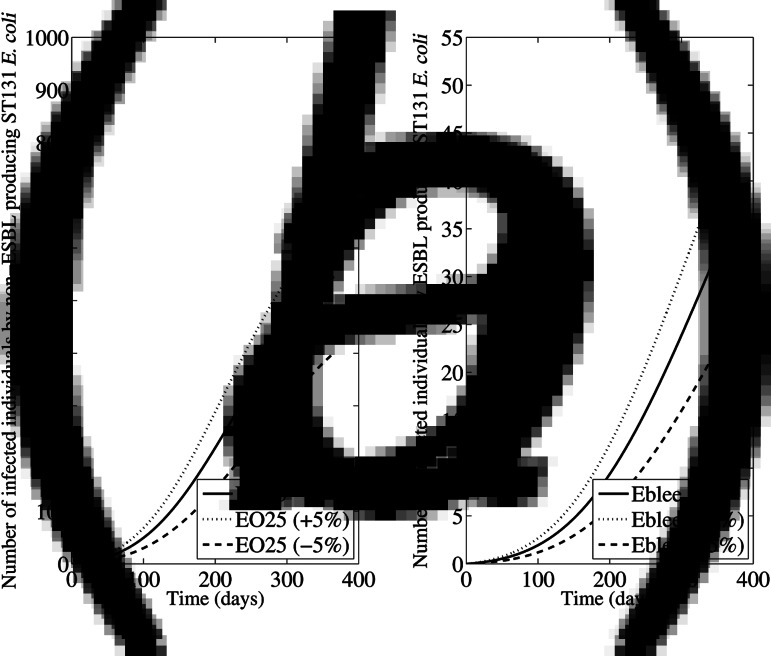
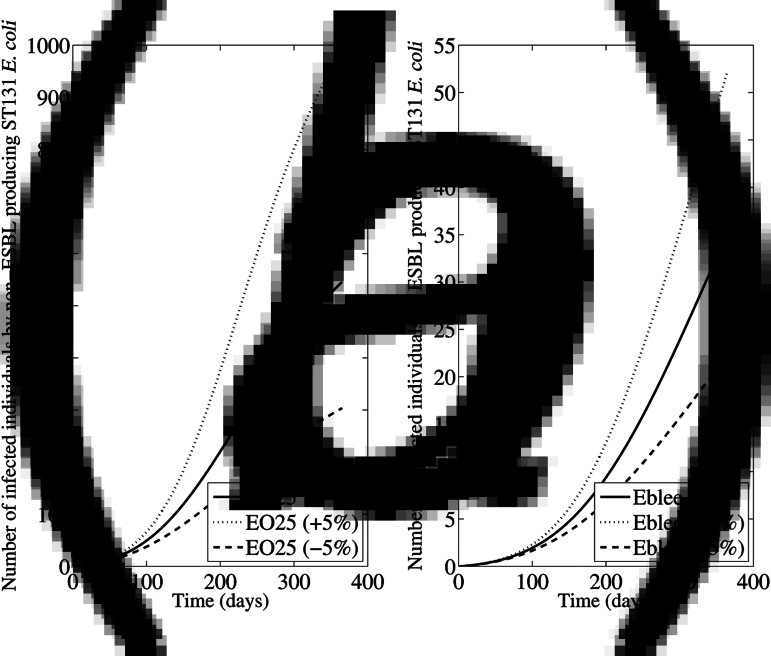
The first simulation considered a variation in ±50% of the initial colonized and infected persons in households, nursing homes and hospitals. The changes in the total number of persons infected with ESBL-producing and ESBL-non-producing ST131 E. coli are shown in Figure 2; the change was more pronounced when the initial proportions were reduced in 50% than when they were increased in 50%. The second simulation was performed considering a ±5% variation in the rate of acquisition of colonization in all compartments; as shown in Figure 3, such variations have a very important impact in the number of persons infected with ESBL-producing and ESBL-non-producing ST131 E. coli, being the most sensitive parameter in the model. The changes were bigger when the acquisition rate was 5% higher than in initial estimations. The third simulation (Supplementary Fig. S2) was performed considering a ±5% variation in the proportion of infected patients in all compartments. In this case, the impact was much lower than in previous simulations.
Fig. 2.
Sensitivity analysis considering a variation in ±50% of the initial colonized and infected persons in households, nursing homes and hospitals. The data represent the number of persons infected due to (a) non-ESBL-producing ST131 E. coli (EO25) and (b) ESBL-producing ST131 E. coli (Eblee) over 1 year.
Fig. 3.
Sensitivity analysis considering a ±5% variation in the rate of acquisition in all compartments. The data represent the number of persons infected due to (a) non-ESBL-producing ST131 E. coli (EO25), and (b) ESBL-producing ST131 E. coli (Eblee) over 1 year.
Estimation of the impact of specific interventions
We estimated the impact of an intervention (e.g. improving adherence to hand hygiene) achieving a 10% reduction in the acquisition rate implemented in the sixth month. As shown in Figure 4, such a reduction in transmission had a large impact, mainly in reducing the number of new infections due to non-ESBL-producing E. coli ST131.
Fig. 4.
Comparison of the estimated numbers of individuals predicted to become infected with non-ESBL-producing ST131 E. coli (EO25) and ESBL-producing ST131 E. coli (Eblee) with and without the implementation of an intervention achieving a 50% reduction in the person-to-person transmission rate, implemented at month 6.
Finally, we examined the impact of changes in exposure to antimicrobials on the generation of new cases of infection; for this, a reduction from 5% to 0% in the exposure to fluoroquinolones and cephalosporins was considered in the already colonized population. As shown in Figure 5, the impact was much more limited compared to the previous intervention.
Fig. 5.
Comparison of the estimated numbers of individuals predicted to become infected with non-ESBL-producing ST131 E. coli (EO25) and ESBL-producing ST131 E. coli (Eblee), with and without the implementation of an intervention reducing exposure to fluoroquinolones and cephalosporins from 5% to 0% in the colonized population.
DISCUSSION
We constructed a mathematical model to predict the occurrence of new cases of infection caused by ESBL-producing and ESBL-non-producing E. coli ST131, based on epidemiological data mostly developed for the model. The model suggests that implementing sustained activity capable of reducing person-to-person transmission in households and healthcare centres would significantly reduce the incidence of new cases of infection caused by ST131; the impact would be less for ST131 isolates that produce ESBLs. The impact of reducing exposure to antimicrobials in already colonized populations would be lower.
Previous mathematical models developed to investigate the spread of antibiotic-resistant bacteria were mainly performed for nosocomial transmission of antibiotic-resistant Staphylococcus aureus and Enterococcus spp. [6]. However, designing mathematical models is much more complex when population-based community transmission is studied. Recently, a dynamic transmission model was developed for methicillin-resistant S. aureus in the community [26]. To the best of our knowledge, ours is the first attempt to develop a model of the transmission dynamics of ST131 considering both the community and healthcare centre settings. Some particular aspects of the epidemiology of E. coli should be considered. First, E. coli is part of the normal flora of all humans. Second, humans can be colonized by different E. coli phylogroups and clones [27], meaning that one person can be colonized by several clones of E. coli. Third, the mechanism of transmission of E. coli has not been systematically studied in all settings, and specific phylogroups or clones may show differences in main reservoirs, virulence features and competing influences; in fact, some may be preferentially acquired through the food chain or via contact with animals [27, 28], while others, including ST131, seem to be more frequently transmitted by direct person-to-person contact [1, 15, 19, 29]. The fact that ST131 is causing an increasing number of infections is thought to be facilitated both by its virulence profile as an extraintestinal pathogenic E. coli (typical of the B2 phylogroup) and its antibiotic resistance profile [1, 2, 4, 14, 29].
The results of the sensitivity analysis showed that the structure of the model is robust and stable, and the validation showed that the estimations in the population dynamics are in accord with available epidemiological data. The simulations performed showed a greater decrease in the number of newly infected individuals when the interventions reducing person-to-person transmissions were implemented in a sustainable manner. It is well known that transmission of multidrug-resistant pathogens can be reduced by hand hygiene in healthcare centres; in households, hand hygiene is also important to avoid transmitting pathogens [30, 31]. We recognize that more data are needed to definitively assess that colonized persons in households are actually the main source for ST131; there is a strong rationale supporting an important role for hand hygiene in the prevention of transmission.
Antibiotic use is always an important variable in the epidemiology of antibiotic-resistant organisms. Exposure to fluoroquinolones and to cephalosporins are risk factors for infections caused by multidrug-resistant E. coli, including ST131 [13, 29], although such exposure has not been found to increase the risk of colonization with ESBL-producing E. coli [14]. Although more data are needed to investigate the effect of exposure on ST131 acquisition in different settings, our results suggest that reducing transmission would have a more significant impact than reducing the potential selective pressure caused by exposure to antibiotics.
Our study has several limitations. Models are simplifications of reality and are based on assumptions. Clearly, the model parameters would need to be changed if future studies show that some of the assumptions made are incorrect. Moreover, our estimates would only be applicable in areas with a similar epidemiology. The epidemiology behaviour of ST131 E. coli may vary according to social and epidemiological factors including international travel and migration, hand hygiene habits, etc. Finally, subclones may show specific epidemiological features [2].
In conclusion, we built a population-based model of the transmission dynamics of E. coli ST131; our results suggest that interventions achieving a sustained reduction in the transmission of these isolates in households, nursing homes and hospitals are most likely to reduce the burden of infections that they cause.
ACKNOWLEDGEMENTS
This study was funded by the Ministerio de Economía y Competitividad, Instituto de Salud Carlos III – co-financed by European Development Regional Fund ‘A way to achieve Europe’ ERDF, Spanish Network for the Research in Infectious Diseases (REIPI RD12/0015), Fondo de Investigación Sanitaria (grant nos. 070190, 10/02021, 10/01955, and 10/00795), and Junta de Andalucía (grant nos. 0048/2008, and CTS-5259 and CTS210). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
DECLARATION OF INTEREST
A.P. has been a consultant for Merck and Pfizer, has served as speaker for AstraZeneca, Merck, and Pfizer and received research support from Merck and Pfizer. J.R.B. has been a scientific advisor for Merck and AstraZeneca, and has served as speaker for Merck, Pfizer, AstraZeneca and Astellas. All other authors declare no conflict of interest.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268816000030.
click here to view supplementary material
REFERENCES
- 1.Nicolas-Chanoine MH, et al. Clinical Microbiology Reviews 2014; 27: 543–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee R, Johnson JR. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrobial Agents and Chemotherapy 2014; 58: 4997–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco J, et al. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. Journal of Antimicrobial Chemotherapy 2012; 66: 2011–2021. [DOI] [PubMed] [Google Scholar]
- 4.Johnson JR, et al. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clinical Infectious Diseases 2010; 51: 286–294. [DOI] [PubMed] [Google Scholar]
- 5.López-Cerero L, et al. Escherichia coli O25b:H4/ST131 are prevalent in Spain and are often not associated with ESBL or quinolone resistance. Enfermedades Infecciosas y Microbiología Clínica 2013; 31: 385–388. [DOI] [PubMed] [Google Scholar]
- 6.van Kleef E, et al. Modelling the transmission of healthcare associated infections: a systematic review. BMC Infectious Diseases 2013; 13: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grassly NC, Fraser C. Mathematical models of infectious disease transmission. Nature Reviews in Microbiology 2008; 6: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riley S. Large-scale spatial-transmission models of infectious diseases. Science 2007; 316: 1298–1301. [DOI] [PubMed] [Google Scholar]
- 9.Seto EYW, Soller JA, Colford JM. Strategies to reduce person-to-person transmission during widespread Escherichia coli O157:H7 outbreak. Emerging Infectious Diseases 2007; 13: 860–866. [DOI] [PubMed] [Google Scholar]
- 10.Bootsma MC, et al. An algorithm to estimate the importance of bacterial acquisition routes in hospital settings. American Journal of Epidemiology 2007; 166: 841–851. [DOI] [PubMed] [Google Scholar]
- 11.Torres E, et al. Faecal carriage of Escherichia coli O25b:h4/ST131: preliminary results of a survey in nosocomial and community settings. In: 22nd European Congress of Clinical Microbiology and Infectious Diseases (ESCMID), 31 March–3 April 2012. London, UK. Abstract P1297.
- 12.Torres E, et al. Prospective longitudinal carriage study of Escherichia coli O25B:H4/ST131 in nosocomial and community settings. In: 23rd European Congress of Clinical Microbiology and Infectious Diseases (ESCMID), 27–30 April 2013. Berlin, Germany. Abstract P1214.
- 13.López-Cerero L, et al. Escherichia coli belonging to the worldwide emerging epidemic clonal group O25b/ST131: risk factors and clinical implications. Journal of Antimicrobial Chemotherapy 2014; 69: 809–814. [DOI] [PubMed] [Google Scholar]
- 14.Rodríguez-Baño J, et al. Faecal carriage of extended-spectrum beta-lactamase-producing Escherichia coli: prevalence, risk factors and molecular epidemiology. Journal of Antimicrobial Chemotherapy 2008; 62: 1142–1149. [DOI] [PubMed] [Google Scholar]
- 15.Johnson JR, et al. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967–2009. Emerging Infectious Diseases 2012; 18: 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhanji H, et al. Cephalosporin resistance mechanisms in Escherichia coli isolated from raw chicken imported into the UK. Journal of Antimicrobial Chemotherapy 2010; 65: 2534–2537. [DOI] [PubMed] [Google Scholar]
- 17.Egea P, et al. Increased raw poultry meat colonization by extended spectrum beta-lactamase-producing Escherichia coli in the south of Spain. International Journal of Food Microbiology 2012; 159: 69–73. [DOI] [PubMed] [Google Scholar]
- 18.Pomba C, et al. Within-lineage variability of Escherichia coli ST131 isolates from humans and pets in the South of Europe. Journal of Antimicrobial Chemotherapy 2014; 69: 271–273. [DOI] [PubMed] [Google Scholar]
- 19.Price LB, et al. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 2013; 4: e00377–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilty M, et al. Transmission dynamics of extended-spectrum β-lactamase-producing Enterobacteriaceae in the tertiary care hospital and the household setting. Clinical Infectious Diseases 2012; 55: 967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyman JM, Li J. An intuitive formulation for the reproductive number for the spread of diseases in heterogeneous populations. Mathematical Biosciences 2000; 167: 65–86. [DOI] [PubMed] [Google Scholar]
- 22.Iman RL, Helton JC. An investigation of uncertainty and sensitivity analysis techniques for computer models. Risk Analysis 1988; 8: 71–90. [Google Scholar]
- 23.Perez L, Dragicevic S. An agent-based approach for modeling dynamics of contagious disease spread. International Journal of Health Geographics 2009; 5: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kardas-Sloma L, et al. Impact of antibiotic exposure patterns on selection of community-associated methicillin-resistant Staphylococcus aureus in hospital settings. Antimicrobial Agents and Chemotherapy 2011; 55: 4888–4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nsoesie EO, Beckman RJ, Marathe MV. Sensitivity analysis of an individual-based model for simulation of influenza epidemics. PLoS ONE 2012; 7: e45414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogea C, van Effelterre T, Acosta CJ. A basic dynamic transmission model of Staphylococcus aureus in the US population. Epidemiology and Infection 2014; 142: 468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson JR, Clabots C, Kuskowski MA. Multiple-host sharing, long-term persistence, and virulence of Escherichia coli clones from human and animal household members. Journal of Clinical Microbiology 2008; 46: 4078–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kluytmans JA, et al. Extended-spectrum β-lactamase-producing Escherichia coli from retail chicken meat and humans: comparison of strains, plasmids, resistance genes, and virulence factors. Clinical Infectious Diseases 2013; 56: 478–487. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee R, et al. Escherichia coli sequence type 131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infection Control and Hospital Epidemiology 2013; 34: 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren-Gash C, Fragaszy E, Hayward AC. Hand hygiene to reduce community transmission of influenza and acute respiratory tract infection: a systematic review. Influenza and Other Respiratory Viruses 2013; 7: 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nerby JM, et al. Risk factors for household transmission of community-associated methicillin-resistant Staphylococcus aureus. Pediatric Infectious Diseases Journal 2011; 30: 927–932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268816000030.
click here to view supplementary material