SUMMARY
Nebovirus is a new genus of viruses belonging to the Caliciviridae family recently characterized in cattle, and is associated with gastrointestinal disorders, such as diarrhoea, anorexia and intestinal lesions particularly in calves. The aim of this study was to investigate the prevalence of neboviruses in Brazilian cattle and analyse phylogenetically the virus strains detected. A prevalence of 4·8% of neboviruses in faecal samples from 62 head of cattle from different Brazilian states was detected. All positive animals were aged <20 days and had diarrhoea. Phylogenetic analysis clustered the virus sequences into the Newbury1 clade. There was >96·0% nt (100% aa) sequence identity between the virus sequences in this study and >88·8% nt (>94·4% aa) identity with Newbury1/UK. Our results indicate, for the first time, the occurrence of neboviruses in Brazil as well as in South America, and the first Newbury1-like nebovirus found outside the UK.
Key words: Caliciviridae, cattle diseases, diarrhoea, molecular epidemiology
Caliciviruses are single-stranded, positive-sense RNA viruses which infect a variety of animal hosts, causing gastrointestinal disorders and even cases of fatal haemorrhage [1, 2]. Of the genera belonging to the family Caliciviridae, the genus Nebovirus was recently characterized in cattle. Neboviruses are on average 7450 nucleotides (nt) long excluding the 3′-poly(A) tail, divided into two open reading frames (ORF); ORF-1 encodes a large non-structural polyprotein contiguous with the major capsid protein and ORF-2 encodes a small basic protein. Gnotobiotic calves experimentally infected with neboviruses showed changes in colour of stools, diarrhoea, anorexia and intestinal lesions [3, 4]. Although there is still no clear definition, the genotypes Nebraska (NB), Newbury1 (NA1) and Dijon have been suggested for this genus [5].
Despite the known relevance of viral agents causing enteric diseases in cattle, there are few studies on the prevalence of viral enteropathogens in Brazil, including predominantly rotavirus [6], coronavirus [7] and astrovirus [8]. We report the first molecular detection and characterization of a nebovirus in South America.
From 15 July 2012 to 25 May 2013, faecal samples were collected from the rectum of 62 cattle (beef and dairy) in rural areas of the states of São Paulo, Minas Gerais, Goiás and Rio Grande do Sul in Brazil. Of the studied animals, 27 (43·5%) had diarrhoea, 39 (62·9%) were aged <4 months, and the majority (51 animals, 82·2%) were female. Fifty-four (87·1%) were raised under the feedlot system and eight (12·9%) under free-range production conditions. Immediately after collection, the samples were kept in plastic bags at −4 °C until processing in the laboratory.
We extracted RNA using TRIzol™ Reagent (Invitrogen, USA). Reverse transcription was performed using ImProm-II™ Reverse Transcription System (Promega, USA) and random primers (Invitrogen), according to the manufacturer's instructions. Polymerase chain reaction (PCR) targeting the 3′ end of the polymerase gene of neboviruses [9] was done with GoTaq™ Colorless Master Mix (Promega), following manufacturer's protocol, using the bovine β-actin gene as internal control [10]. The primer sequences used were as follows: NBU-F 5′-TTTCTAACYTATGGGGAYGAYG-3′ and NBU-R 5′-GTCACTCATGTTTCCTTCTCTAAT-3′ (549-bp amplicon); actin-F 5′-ACCAGCCATCCAGACAAAAC-3′ and actin-R 5′-ATCTTCAGGGACTTTGGACG-3′ (360-bp amplicon). The amplified products were gel extracted and sequenced directly on both strands with the same primers used in the PCR, in an automated ABI 3730 DNA Analyser (Applied Biosystems, USA). Nucleotide sequence fragments were edited and assembled into consensus contigs using BioEdit v. 7.0.9 [11]. MEGA 6 [12] was used to perform the multiple sequence alignment and subsequently to build a neighbour-joining phylogenetic tree with bootstrap support. MatGAT v. 2.0 [13] was used for sequence identity calculations. Fourteen other nucleotide sequences representative of the three known genotypes for neboviruses were retrieved from GenBank.
Of the samples collected, three (4·8%) were positive for nebovirus by RT–PCR. All positive animals were aged <20 days and had diarrhoea, two positive calves were female, and two positive samples were from beef cattle. Two animals were from the state of Goiás and one from the state of São Paulo.
The amplified fragments were sequenced and deposited in GenBank under accession numbers KR136171-3 for virus strains 285/08/BRA, 283/BRA and 288/08/BRA, respectively.
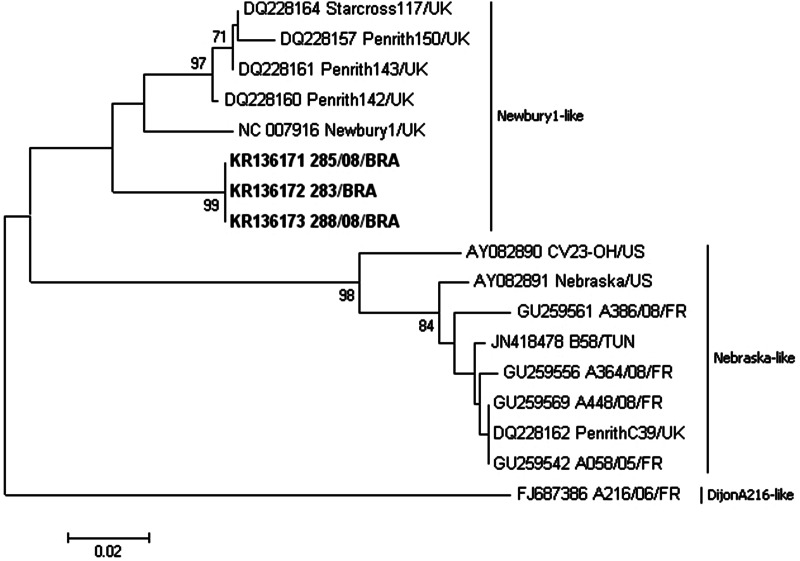
The samples sequenced in this study shared >96·0% nt (100% aa) sequence identity compared to each other. To reference virus strains of each nebovirus genotype, the viruses in the three samples shared 88·8–89·5% nt (94·4–94·8% aa) identity with Newbury1/UK, 80·0–81·8% nt (87·6–88·4% aa) identity with Nebraska/USA, and 80·8–81·4% nt (86·4–86·5% aa) identity with DijonA216/FR. Phylogenetic analysis clustered sequences described here into the Newbury1 clade (Fig. 1).
Fig. 1.
Cladogram representing a phylogenetic reconstruction using 162-aa-length sequences covering the 3′ end polymerase region of nebovirus. Bootstrap values higher than 70% for 1000 pseudo-replicates are showen at the nodes. The Brazilian nebovirus sequences determined in the present study are show in boldface. GenBank accession numbers are shown on the tree. The scale bar represents the phylogenetic distance between sequences.
The prevalence in our study was slightly higher than to that reported in Tunisia (3%) [14] and lower than the reported in the France [5] and the United States [9] (7% and 28%, respectively).
In summary, this is the first detection of neboviruses in Brazil as well as in South America, and the first Newbury1-like nebovirus found outside the UK. Nevertheless, a greater number of studies are needed to clarify the epidemiology of the infection by nebovirus in cattle, and evaluation of the prevalence and genetic variation of neboviruses in different countries.
A better understanding of the viruses that cause gastroenteritis in cattle is important for future prevention and control strategies in these animals.
ACKNOWLEDGEMENTS
This study was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, nos. 2006/52060–3 and 2012/18441-0) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, no. 472509/2010-1).
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Kapikian AZ, Estes MK, Chanock M. Norwalk group of viruses. In: Fields BN, et al. eds. Fields virology, 3rd edn, vol. 1. Philadelphia, PA: Lippincott-Raven Publishers, 1996, pp. 783–810. [Google Scholar]
- 2.Pedersen NC, et al. An isolated epizootic of hemorrhagic-like fever in cats caused by a novel and highly virulent strain of feline calicivirus. Veterinary Microbiology 2000; 73: 281–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smiley JR, et al. Characterization of anenteropathogenic bovine calicivirus representing a potentially new calicivirus genus. Journal of Virology 2002; 76: 10089–10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridger JC, Hall GA, Brown JF. Characterization of a calici-like virus (Newbury agent) found in association with astrovirus in bovine diarrhea. Infection and Immunity 1984; 43: 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplon J, et al. Possible novel nebovirus genotype in cattle, France. Emerging Infectious Diseases 2011; 17: 1120–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alfieri AA, et al. Frequency of group A rotavirus in diarrhoeic calves in Brazilian cattle herds, 1998–2002. Tropical and Animal Health Production 2006; 38: 521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stipp DT, et al. Frequency of BCoV detection by a semi-nested PCR assay in faeces of calves from Brazilian cattle herds. Tropical and Animal Health Production 2009; 41: 1563–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candido M, et al. Molecular detection and phylogenetic analysis of bovine astrovirus in Brazil. Archives of Virology 2015; 160: 1519–1525. [DOI] [PubMed] [Google Scholar]
- 9.Smiley JR, et al. Reverse transcription-PCR assays for detection of bovine enteric caliciviruses (BEC) and analysis of the genetic relationships among BEC and human caliciviruses. Journal of Clinical Microbiology 2003; 41: 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renshaw RW, Ray R, Dubovi EJ. Comparison of virus isolation and reverse transcription polymerase chain reaction assay for detection of bovine viral diarrhea virus in bulk milk tank samples. Journal of Veterinary Diagnostic Investigation 2000; 12: 184–186. [DOI] [PubMed] [Google Scholar]
- 11.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 1999; 41: 95–98. [Google Scholar]
- 12.Tamura K, et al. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 2013; 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campanella JJ, Bitincka L, Smalley J. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 2003; 4: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassine-Zaafrane M, et al. Molecular prevalence of bovine noroviruses and neboviruses detected in central-eastern Tunisia. Archives of Virology 2012; 157: 1599–1604. [DOI] [PubMed] [Google Scholar]