SUMMARY
The Darwin region in northern Australia has experienced rapid population growth in recent years, and with it, an increased incidence of melioidosis. Previous studies in Darwin have associated the environmental presence of Burkholderia pseudomallei, the causative agent of melioidosis, with anthropogenic land usage and proximity to animals. In our study, we estimated the occurrence of B. pseudomallei and Burkholderia spp. relatives in faecal matter of wildlife, livestock and domestic animals in the Darwin region. A total of 357 faecal samples were collected and bacteria isolated through culture and direct DNA extraction after enrichment in selective media. Identification of B. pseudomallei, B. ubonensis, and other Burkholderia spp. was carried out using TTS1, Bu550, and recA BUR3–BUR4 quantitative PCR assays, respectively. B. pseudomallei was detected in seven faecal samples from wallabies and a chicken. B. cepacia complex spp. and Pandoraea spp. were cultured from wallaby faecal samples, and B. cenocepacia and B. cepacia were also isolated from livestock animals. Various bacteria isolated in this study represent opportunistic human pathogens, raising the possibility that faecal shedding contributes to the expanding geographical distribution of not just B. pseudomallei but other Burkholderiaceae that can cause human disease.
Key words: Cystic fibrosis, emerging infections, melioidosis, opportunist infections, veterinary epidemiology and bacteriology
INTRODUCTION
Australia has experienced rapid and marked ecological changes in the short time period following European colonization in 1788. These profound changes have concomitantly altered the landscape of infectious disease in this country. The greater Darwin region of the Northern Territory, where European colonization was long hindered by pervasive malaria [1], is now the second fastest growing capital region in the country [2]. Although malaria has been eradicated in Australia, melioidosis is one of the emerging infectious diseases which have accompanied the population expansion of Australia's tropical north [1].
Melioidosis is caused by Burkholderia pseudomallei, a non-spore-forming, Gram-negative, saprophytic bacterium that naturally inhabits tropical soil, the rhizosphere and water sources [3–6]. B. pseudomallei is endemic throughout Southeast Asia and northern Australia, and increased awareness and diagnostic capabilities in recent years have greatly expanded the known distribution of this bacterium [7]. Disease presentation, incubation period, and the potential for reactivation are highly variable and dependent on host immune status and the route of inoculation. B. pseudomallei is a dangerous organism with infection often being fatal without treatment, and as such is classified as a Tier 1 select agent by the U.S. Federal Select Agent Program (http://www.selectagents.gov/). Although culture isolation remains the gold standard of detection, the development of molecular assays have allowed for more sensitive and specific identification of B. pseudomallei in the environment [8–10].
The arsenal of adaptive mechanisms that allow B. pseudomallei and the closely related B. cepacia complex (Bcc) to survive in the competitive rhizosphere likely contributes to their success as opportunistic pathogens of humans and animals [6]. The Bcc comprises at least 18 different species [11], many of which are found in soil in northern Australia, with B. ubonensis being particularly prevalent in the Darwin region [12]. The metabolic versatility of Bcc spp. enables them to ward off plant pathogens and to degrade environmental pollutants; however, their promising agricultural and bioremediation potential has been overshadowed by the risks to human health [6]. B. pseudomallei and in particular Bcc spp. are increasingly significant pathogens of cystic fibrosis (CF) patients [3, 6]. Colonization of the CF lung with Bcc spp. can result in the potentially fatal ‘cepacia syndrome’ [6], and person-to-person transmission is possible with particular ‘epidemic’ Bcc strains [13].
Melioidosis cases in animals are well documented and encompass a wide array of species, with considerable variability in susceptibility seen between species [14, 15]. Reports of melioidosis in marsupials are extremely rare [16]; however, fatal cases in captive or highly stressed animals have occurred [17]. Melioidosis cases in birds [18], primates [19], and reptiles [20], and outbreaks associated with zoos [21] and intensive livestock agriculture [22], support the notion that stressful conditions may trigger the onset of disease.
Although there is no evidence of a sustained zoonotic transmission cycle for Burkholderia spp., indirect transmission from animals and animal products poses a tangible risk to human health, particularly to those with melioidosis risk factors [14]. Animal migration and transport may also contribute indirectly to human melioidosis by assisting with the dissemination of B. pseudomallei in the environment [23]; animals may also be an important vehicle for B. pseudomallei to expand its presence beyond traditional endemic regions. For example, bird migration has been proposed as a possible mechanism for Australian B. pseudomallei dissemination into the Pacific Islands [18]. In addition, localized epizootic outbreaks have occurred well beyond endemic regions following animal importation, likely as a result of subclinical infections in animals ‘which, because of travelling, climate or husbandry, start to excrete bacteria along with faeces or through abscesses’ [23, p. 306]. Experimental infection with B. pseudomallei demonstrates that the absence of overt symptoms in animals does not exclude the possibility for gastrointestinal colonization and bacterial shedding [24].
As therapy for melioidosis is complicated and prolonged and post-exposure prophylaxis is of uncertain efficacy [3], understanding the factors that contribute to B. pseudomallei distribution in the local environment is important in guiding preventative health strategies. Previous studies have established an association between the presence of B. pseudomallei in soil and proximity to animals, suggesting a possible animal role in bacterial amplification [25]. Local macropod species, particularly Macropus antilopinus (antilopine kangaroo) and M. agilis (agile wallaby), are known to consume invasive grass species preferentially colonized by B. pseudomallei [26]. As highly mobile animals with a virtual absence of documented clinical melioidosis despite routine exposure [16], wallabies are considered a likely candidate for the dispersal of B. pseudomallei through subclinical bacterial shedding in faeces. The aim of this study was to evaluate the prevalence of B. pseudomallei and the near-neighbour Bcc spp. in faecal matter of wildlife, livestock, and domestic animals in the Darwin region, with an emphasis on macropods. Faecal shedding of B. pseudomallei by animals is a suspected mechanism in several epizootic outbreaks [23] and was confirmed in laboratory settings [24]. In the 1970s, the isolation of B. pseudomallei from faecal matter was reported in captive horses [27] and native Australian rodents [28]; however, to our knowledge the faecal shedding of B. pseudomallei by captive or wild animals has not been demonstrated since these early studies.
METHODS
Animal ethics
Two methods were utilized for animal faecal sample collection: faecal swabs were collected by veterinary personnel, and faecal samples were obtained from wildlife and livestock collected in situ. Approval for faecal swab collection was granted by the Charles Darwin University Animal Ethics Committee (A 13 024) and Menzies School of Health Research Animal Research License (no. 055). Animal ethics approval was not required for non-invasive faecal sample collection.
Faecal swab collection
Thirty-six faecal swabs samples were collected by veterinarians from domestic and wildlife animals brought to two Darwin area animal hospitals for care between February and July 2014. Ten additional faecal swab samples were collected in March 2014 by a veterinary organization carrying out dog health programmes in Indigenous communities. Swabs were immediately stored in sterile containers with modified Ashdown's broth [29]. Containers were kept in biohazard bags with information on animal species, age, sex, location and health status, and stored at room temperature until laboratory processing. All samples were processed within 7 days of collection.
Faecal sample collection
There were three rounds of faecal sample collections. Between 2010 and 2012, wallaby scats were collected on an opportunistic basis with 14 scats collected near Darwin and Katherine (April, July 2010) and another two scats near Darwin in November 2012. From March to July 2014, the main study was conducted with a total of 228 in situ faecal samples collected from various animal species in the Darwin area (Table 1). In September 2014, an additional 67 wallaby scats were collected near Katherine (a small rural township 270 km southeast of Darwin). Faecal samples were collected with sterile disposable gloves or a trowel which was cleaned with 70% ethanol after each collection. Animal sources were determined by visual confirmation or based on identification with a scat manual [30]. Samples were stored in biohazard bags kept at room temperature and were processed within 7 days.
Table 1.
Animal faecal sources from the Darwin and Katherine regions of the Northern Territory, Australia, and molecular screening of samples for Burkholderia spp.
| Animal source | No. total samples | No. faecal swabs | No. faecal samples | No. positive samples (TTS1 qPCR) | No. positive samples (Bu550 qPCR) | No. positive samples (recA qPCR) | No. collection sites |
|---|---|---|---|---|---|---|---|
| Macropods* | 263 | 6 | 257 | 6† | 0 | 50 | 17, vets A, B |
| Possums | 3 | 3 | 0 | 0 | 0 | 0 | Vets A, B |
| Dogs/cats | 37 | 36 | 1 | 0 | 0 | 1 | Vets A, B, C |
| Cattle/buffalo | 15 | 0 | 15 | 0 | 0 | 2 | 4 |
| Goats/sheep | 11 | 0 | 11 | 0 | 0 | 4 | 3 |
| Horses | 5 | 0 | 5 | 0 | 0 | 1 | 1 |
| Pigs | 4 | 0 | 4 | 0 | 0 | 2 | 1 |
| Birds | 17 | 0 | 17 | 1 | 0 | 1 | 4, vet A |
| Other | 2 | 1 | 1 | 0 | 0 | 0 | Vet A |
| Total | 357 | 46 | 228 | 2 | 0 | 61 | 22, vets A, B, C |
83/263 macropod faecal samples were tested only with the TTS1 quantitative polymerase chain reaction (qPCR) assay (sampling prior to 2014 and in September 2014), 180/263 with TTS1, Bu550 and recA qPCR assays (main study March–July 2014).
2/6 TTS1 qPCR-positive samples were also positive by culture.
Animals listed as ‘Other’ were a black flying fox (Pteropus alecto) and a frilled-neck lizard (Chlamydosaurus kingii). All samples were screened with the TTS1 [31], Bu550 [32] and recA BUR3–BUR3 [33] qPCR assays with the exception of 83 macropod samples only screened with the TTS1 assay. ‘No. of collection sites’ represents the number of sampling sites per animal source. ‘Vet’ indicates samples collected by one of three veterinary professionals (A, B and C) rather than from a field site.
Culture and DNA extraction
Faecal swabs were incubated at 37 °C in modified Ashdown's broth with 50 mg/l colistin. After 2 and 7 days, 100 μl and 10 μl of supernatant were cultured onto modified Ashdown's agar (ASA) containing 8 mg/l gentamicin (ASA) [29]. For initial polymerase chain reaction (PCR) screening, DNA was extracted from sweeps of bacteria on ASA after days 2 and 7 using 10% Chelex 100 resin (Bio-Rad, Australia), and B. pseudomallei was detected using the TTS1 quantitative PCR (qPCR) assay (see ‘Identification of Burkholderia spp.’ Section below). Final identification of subcultured B. pseudomallei was through the TTS1 qPCR assay and latex agglutination.
DNA extraction from faecal samples
DNA was extracted from faecal samples as previously described from soil samples [8]. Briefly, 20 g faecal matter was mixed in 20 ml modified Ashdown's broth. Samples were shaken and incubated for 42 h at 37 °C and the pellet of the faecal matter processed with the PowerSoil DNA Isolation kit (MoBio Laboratories, USA) as described previously [8].
Identification of Burkholderia spp.
DNA extracted from samples were screened by the B. pseudomallei specific quantitative PCR assay TTS1 [31]. This assay detects a 115 bp segment of orf2, an open reading frame within the type III secretion system gene cluster that is unique to B. pseudomallei [8, 31]. Samples collected during February to July 2014 were also screened by qPCR for other members of the Burkholderia genus using two additional assays; the Bu550 and recA assays [32, 33]. Non-acetylated bovine serum albumin at a final concentration of 400 ng/μl was added to all PCRs to bind potential PCR inhibitors [8]. B. ubonensis, a common inhabitant of soil in northern Australia that is phenotypically indistinguishable from B. pseudomallei, was identified using the Bu550 assay as described previously [32]. DNA samples extracted using Chelex 100 resin were diluted 1:100; MoBio DNA samples were run undiluted. All qPCRs were conducted on the AB7900HT platform (Applied Biosystems, Australia). DNA of TTS1- and Bu550-negative samples was amplified with the recA assay with primers BUR3 and BUR4 [33], which target members of the Burkholderia genus and the closely related Ralstonia, Cupriavidus, Delftia, and Pandoraea genera.
B. pseudomallei inoculation in faecal samples
To verify that our soil DNA extraction protocol [8] was effective in retrieving B. pseudomallei DNA from faecal matter, a controlled spiking experiment was conducted. A mixture of faecal samples from cattle, wallaby and chicken confirmed to be B. pseudomallei-negative according to TTS1 qPCR were used for the spiking experiment. MSHR2817, an environmental strain collected in the Darwin rural region, was used for inoculation of the faecal samples; a negative control (no B. pseudomallei added) was also included. MSHR2817 was enriched in Ashdown's broth for 18 h at 37 °C. The optical density of the inoculated broth was measured at 600 nm and the broth was diluted to three different concentrations and added in duplicate to 4 ml of 20 g animal faeces in water. The seven samples were incubated in Ashdown's broth for 42 h at 37 °C and DNA was directly extracted as described above. Subsequent viable plate counts of the three MSHR2817 dilutions showed that 120, 9000 and 80 000 colony-forming units (c.f.u.) were added to the animal faeces.
recA sequencing
DNA from 16 culture isolates derived from 16 faecal samples negative for the TTS1 and Bu550 assays was sent for recA nucleotide sequencing [33] at Macrogen Inc. (Republic of Korea). Thirteen nucleotide sequences of sufficient quality were trimmed and aligned using ChromasPro 1·7·6 (Technelysium Pty Ltd, Australia). Consensus sequences were examined using the NCBI Basic Local Alignment Search Tool (www.ncbi.nlm.nih.gov). BLAST searches were conducted with both the complete and draft nucleotide databases using a Burkholderiales filter (taxid: 80 840) for computational efficiency to identify probable species. Sequence identities ⩾99% were considered to be a robust species match.
Nucleotide sequence accession numbers
Ten recA nucleotide sequences were submitted to GenBank under the following accession numbers: KT445904–KT445913.
RESULTS
Opportunistic sampling of wallaby scats was conducted during 2010 and 2012 in the Darwin and Katherine regions. In April and July 2010, 14 wallaby faecal samples were collected and tested with the TTS1 qPCR assay, of which three (21%) were positive for the presence of B. pseudomallei following direct DNA extraction methods. An additional two samples (2/2, 100%) collected in November 2012 near Darwin were culture positive for B. pseudomallei. Subsequent to these preliminary findings, more intensive sampling was undertaken between February and July 2014. Two-hundred and seventy-four faecal samples were collected, of which 228 samples consisted of faecal matter collected from 20 field sites in the Darwin area, and 46 samples were faecal swabs collected from animals cared for at three veterinary practices and organizations in the Darwin area (Table 1). Based on the 2010–2012 results, macropods were the primary focus of sampling in 2014, representing 65% (180/274) of all faecal samples. Additional animal sources included dogs, cats, livestock herbivores, and birds (Table 1). B. pseudomallei was detected by faecal DNA extraction and TTS1 qPCR assay in two faecal samples, one from a wallaby and one from a chicken (2/274, 0·7%) (Table 2) at sites 6·7 km apart (Fig. 1). No clinical disease was reported in the chicken. In September 2014, an additional 67 wallaby faecal samples were collected from various locations in the Katherine area where B. pseudomallei had been detected in wallaby scats in 2010. All 67 samples were negative for B. pseudomallei.
Table 2.
Identification of bacterial species using the TTS1 quantitative polymerase chain reaction (qPCR) assay and NCBI BLAST of recA sequences
| Bacterial species | Sample/isolate no. | Animal source | DNA extraction from | Identification method | Collection date (month/year) |
|---|---|---|---|---|---|
| Burkholderia pseudomallei | LRS 225 | Wallaby | Direct – faecal | TTS1 qPCR assay | Apr. 2010 |
| ICE 73 | Wallaby | Direct – faecal | TTS1 qPCR assay | July 2010 | |
| LRS 83 | Wallaby | Direct – faecal | TTS1 qPCR assay | July 2010 | |
| LRS 687 | Wallaby | Culture | TTS1 qPCR assay, latex agglutination | Nov. 2012 | |
| LRS 697 | Wallaby | Culture | TTS1 qPCR assay, latex agglutination | Nov. 2012 | |
| SCAT 58 | Wallaby | Direct – faecal | TTS1 qPCR assay | May 2014 | |
| SCAT 179 | Chicken | Direct – faecal | TTS1 qPCR assay | July 2014 | |
| B. multivorans | MSMB2372 | Wallaby | Culture | 99% NCBI BLASTn | May 2014 |
| MSMB2368 | Wallaby | Culture | 99% NCBI BLASTn | May 2014 | |
| B. cenocepacia | MSMB2427 | Wallaby | Culture | 99% NCBI BLASTn | July 2014 |
| MSMB2424 | Wallaby | Culture | 99% NCBI BLASTn | July 2014 | |
| MSMB2400 | Horse | Culture | 99% NCBI BLASTn | July 2014 | |
| B. cepacia | MSMB2428 | Wallaby | Culture | 99% NCBI BLASTn | July 2014 |
| MSMB2365 | Water buffalo | Culture | 99% NCBI BLASTn | Apr. 2014 | |
| Pandoraea faecigallinarum | MSMB2432 | Wallaby | Culture | 99% NCBI BLASTn | May 2014 |
| Pandoraea spp. | MSMB2376 | Wallaby | Culture | 97% NCBI BLASTn | Apr. 2014 |
| MSMB2369 | Wallaby | Culture | 97% NCBI BLASTn | May 2014 |
B. pseudomallei was detected following direct DNA extraction from wallaby and chicken faecal samples. Two wallaby scats were positive for B. pseudomallei by culture. Bcc spp. and Pandoraea spp. were identified with the NCBI BLASTn program using results of ⩾99% sequence identity to recA database entries. Two samples were determined to likely be members of Pandoraea based on 97% sequence identity to recA database entries.
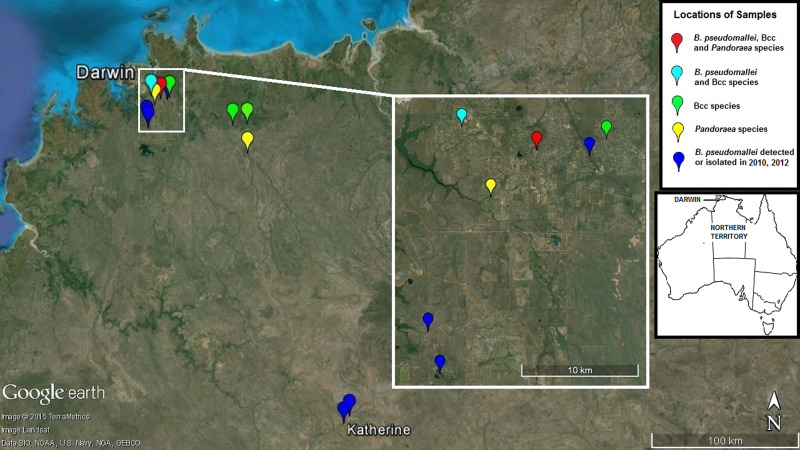
Fig. 1.
Collection sites of faecal samples in the Darwin and Katherine regions in which Burkholderia pseudomallei and B. cepacia complex (Bcc) spp. were detected. Most samples in this study were collected at hobby farms outside Darwin in a previously rural area which has undergone significant population growth in recent years. One site elicited B. pseudomallei, Bcc spp. and Pandoraea spp. in different wallaby scats, and B. pseudomallei and B. cepacia were identified at a nearby site in wallaby and horse faeces. Opportunistic sampling in 2010 and 2012 (dark blue) identified B. pseudomallei in five wallaby scats collected near Darwin and Katherine. The inset map shows the location of Darwin in Australia. (Source: commons.wikimedia.org.)
In a spiking experiment, B. pseudomallei was detected by TTS1 qPCR in all six faecal samples inoculated with 120, 9000, and 80 000 c.f.u. of MSHR2817, whereas the negative control with no B. pseudomallei added was negative. All samples screened for B. ubonensis using the Bu550 assay were negative (Table 1). A total of 61/274 (22%) faecal DNA extracts from 2014 had a positive recA assay, and 26/60 (43%) also elicited cultivable bacteria on selective Ashdown's media. DNA from 16 recA-positive faecal samples were sent for recA sequencing; 13 resulted in sequences of sufficient quality and ten were identified using NCBI BLAST. B. multivorans was isolated from two wallaby scats, B. cenocepacia was found in two wallaby scats and one horse faecal sample (Fig. 1), and B. cepacia was isolated from a wallaby scat and from one water buffalo faecal sample. No clinical disease was reported in livestock animals with Bcc-positive faecal matter. Pandoraea faecigallinarum was isolated from a wallaby scat, and two other isolates from wallaby scats likely belong to the genus Pandoraea based on 97% recA sequence identity.
DISCUSSION
This study revealed the presence of the opportunistic human pathogens, B. pseudomallei, Bcc spp. and Pandoraea spp., in the faecal matter of wildlife and livestock animals in the Darwin region. In 2014, B. pseudomallei was detected through direct DNA extraction methods in two faecal specimens (2/274, 0·7%); one from a chicken and the other from a wallaby. In contrast, opportunistic sampling of wallaby faecal samples in 2010 and 2012 revealed considerably higher detection rates of B. pseudomallei using molecular methods (3/14, 21%) with two scats from November 2012 also eliciting cultivable bacteria. The higher rate of B. pseudomallei detection in 2010 may have been due to in situ faecal collection during and after the particularly intense monsoon season, which saw an unprecedented number of human melioidosis cases presenting to Royal Darwin Hospital [34]. Furthermore, the collection of samples in November 2012 coincided with the onset of the northern Australian wet season (October–May), and this particular month saw 79 mm of rainfall with the sampling occurring shortly after the first major rainfall of the season (Bureau of Meteorology, Australia, http://www.bom.gov.au) which may have stimulated the dissemination of B. pseudomallei in soils and greater macropod access to grass shoots [26]. In contrast, faecal sample collection between February and July 2014 occurred during and after a below average wet season compared to previous wet seasons from years 1981 to 2010 (Bureau of Meteorology, Australia, www.bom.gov.au).
As B. pseudomallei was primarily detected through direct DNA extraction rather than culture isolation, it is possible that qPCR-positive but culture-negative samples represent the passage of ingested soil, water, or vegetation contaminated with B. pseudomallei rather than a colonizing infection. However, B. ubonensis, a common inhabitant of soil and water sources in the Darwin region [12], was not detected by culture or molecular methods, supporting that these positive samples do not represent contamination of ingested material nor contamination of faecal samples from contact with soil. Another explanation for the predominance of non-cultivable B. pseudomallei detected by molecular methods is that these infections represent bacteria in a viable but non-culturable (VBNC) state. In this state, metabolic activity is minimized and bacteria fail to grow on routine culture media. VBNC is the suspected mechanism of latency in tuberculosis infections [35], and may explain the melioidosis reactivation in some human cases [3, 5], as well as discrepancies between qPCR and culture detection of B. pseudomallei in environmental surveys [4, 8, 10].
The identification of B. multivorans, B. cenocepacia and B. cepacia and recA-positive culture isolates in faecal samples suggests that gastrointestinal colonization and faecal shedding of Bcc bacteria may be relatively common in the Darwin area. Documented animal infections with Bcc spp. are rare; however, Bcc spp. have been implicated in epizootic outbreaks in sheep, cattle, and horses [36] and were isolated from abscesses following the death of a domestic parrot [37]. As with B. pseudomallei, it is possible that stress related to captivity and livestock agriculture may contribute to the onset of disease in otherwise subclinical infections. Two isolates from wallaby faecal samples are most likely members of the Pandoraea genus. Pandoraea spp. are also commonly isolated from northern Australian soil [12], and this genus is often associated with polluted environmental sites featuring persistent organic pollutants and hydrocarbons, with some Pandoraea spp. associated with opportunistic infections in CF patients. An additional wallaby scat sample was determined to be P. faecigallinarum, a species first identified in the faecal matter of a chicken [38].
B. multivorans and B. cenocepacia are the predominant Bcc spp. impacting CF patients worldwide. Despite rigorous infection control strategies, diverse and novel strains increasingly present at CF centres, prompting urgent calls to identify environmental sources of these emerging pathogens [13]. Based on the isolates found in this study, exposure to animal faecal matter, directly or indirectly, may serve as a source of Bcc and Pandoraea infections. B. ubonensis was not detected in any faecal samples despite being overwhelmingly prevalent in environmental samples from the Darwin area [12], adding to a mounting body of evidence that this bacterium, unlike other Bcc members, may not be pathogenic in humans or animals due to its inability to colonize such hosts.
Previous studies have established the association between the presence of B. pseudomallei and human-modified landscapes [25]. The spread of invasive grass species [26], loss of woodland ecosystems [39], and environmental presence of organophosphorus compounds [40], hydrocarbons, and heavy metals [41] appear to favour the selection of Burkholderia and Pandoraea spp. Exposure to antibiotics is thought to have contributed to an epizootic outbreak of B. cenocepacia [36], and studies in Australia have detected antibiotic-resistance gene profiles in wallaby microbiota identical to those found in livestock and human pathogens [42]. Wallabies, which straddle rural and peri-urban ecotones, may serve as vehicles of dissemination, thereby altering the presence or density of microbes along the human–livestock–wildlife interface. As the isolates in this study represent adept opportunistic pathogens, it is not unreasonable to question whether simultaneous exposure to anthropogenic selection pressures and various animal hosts alter the pathogenicity profile of these bacteria. The Darwin region represents an area that is rapidly being altered by anthropogenic impacts, with concomitant effects at a microbial level. Fragmentation of natural bushland creates more areas in which humans, livestock, and wildlife interact, and the expanding population of Australia's north will increase the likelihood of exposure to B. pseudomallei and Bcc spp. in the future. The finding that wildlife and livestock animals shed B. pseudomallei and related bacteria in faecal matter is not in itself surprising as rectal swabs from humans diagnosed with melioidosis often elicit B. pseudomallei [43]. Furthermore, B. pseudomallei was previously isolated from horse faeces [27] following an epizootic outbreak of melioidosis at a zoo in Paris in 1975. This outbreak and the subsequent spread of B. pseudomallei to other animal facilities, also resulting in human fatalities [23], serves as an example that environmental contamination and zoonotic exposure can indeed occur following the importation of animals. As bacterial faecal shedding is possible in the absence of clinical disease [24] animal and human movements cannot be ruled out as contributing factors to the increasing geographical range of B. pseudomallei [3, 7] and the global distribution of Bcc strains [13]. Zoonotic cases of melioidosis have been described in Australia [14] and thus both medical and veterinary practitioners within and beyond endemic regions should consider animals and animal faecal matter as potential sources of exposure. Further studies are required to determine the extent to and conditions under which animal carriage and faecal shedding play a role in the epidemiology of these emerging pathogens.
ACKNOWLEDGEMENTS
The authors thank Dr Ilana Hoffmann, Dr Jan Allen and the Animal Management in Rural & Remote Indigenous Communities organization, as well as the very accommodating property owners in the rural Darwin area, for assisting with sample collection.
This project was funded by the Australian Research Council Linkage Grant LP110100691 and grant 1 046 812 from the Australian National Health and Medical Research Council.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.McFarlane RA, Sleigh AC, McMichael AJ. Land-use change and emerging infectious disease on an island continent. International Journal of Environmental Research and Public Health 2013; 10: 2699–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Australian Bureau of Statistics. Regional population growth, Australia, 2012–2013. 2015.
- 3.Currie BJ. Melioidosis: evolving concepts in epidemiology, pathogenesis, and treatment. Seminars in Respiratory and Critical Care Medicine 2015; 36: 111–125. [DOI] [PubMed] [Google Scholar]
- 4.Limmathurotsakul D, et al. Burkholderia pseudomallei is spatially distributed in soil in northeast Thailand. PLoS Neglected Tropical Diseases 2010; 4: e694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inglis TJJ, Sagripanti JL. Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Applied and Environmental Microbiology 2006; 72: 6865–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environmental Microbiology 2005; 7: 1673–1685. [DOI] [PubMed] [Google Scholar]
- 7.Dance DAB. Melioidosis in Puerto Rico: the iceberg slowly emerges. Clinical Infectious Diseases 2015; 60: 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaestli M, et al. Sensitive and specific molecular detection of Burkholderia pseudomallei, the causative agent of melioidosis, in the soil of tropical northern Australia. Applied and Environmental Microbiology 2007; 73: 6891–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trung TT, et al. Highly sensitive direct detection and quantification of Burkholderia pseudomallei bacteria in environmental soil samples by using real-time PCR. Applied Environmental Microbiology 2011; 77: 6486–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau SKP, et al. Burkholderia pseudomallei in soil samples from an oceanarium in Hong Kong detected using a sensitive PCR assay. Emerging Microbes & Infections 2014; 3: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim JM, et al. Survival and susceptibility of Burkholderia cepacia complex in chlorhexidine gluconate and benzalkonium chloride 2015. Journal of Industrial Microbiology and Biotechnology 2015; 42: 905–913. [DOI] [PubMed] [Google Scholar]
- 12.Ginther JL, et al. Identification of Burkholderia pseudomallei near-neighbor species in the Northern Territory of Australia. PLoS Neglected Tropical Diseases 2015; 9: e0003892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baldwin A, et al. Environmental Burkholderia cepacia complex isolates from human infections. Emerging Infectious Diseases 2007; 13: 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Low Choy J, et al. Animal melioidosis in Australia. Acta Tropica 2000; 74: 153–158. [DOI] [PubMed] [Google Scholar]
- 15.Limmathurotsakul D, et al. Melioidosis in animals, Thailand, 2006–2010. Emerging Infectious Diseases 2012; 18: 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladds P. Pathology of Australian Native Wildlife. Collingwood: CSIRO Publishing, 2009. [Google Scholar]
- 17.Egerton JR. Melioidosis in a tree climbing kangaroo. Australian Veterinary Journal 1963; 39: 243–244. [Google Scholar]
- 18.Hampton V, et al. Melioidosis in birds and Burkholderia pseudomallei dispersal, Australia. Emerging Infectious Diseases 2011; 17: 1310–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufmann AF, et al. Melioidosis in imported non-human primates. Journal of Wildlife Diseases 1970; 6: 211–219. [DOI] [PubMed] [Google Scholar]
- 20.Zehnder AM, et al. Burkholderia pseudomallei isolates in 2 pet iguanas, California, USA. Emerging Infectious Diseases 2014; 20: 304–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasantikul T, et al. Retrospective study on fatal melioidosis in captive zoo animals in Thailand. Transboundary and Emerging Diseases. Published online: 13 January 2015. doi: 10.1111/tbed.12315. [DOI] [PubMed]
- 22.Ketterer PJ, et al. Melioidosis in intensive piggeries in south eastern Queensland. Australian Veterinary Journal 1986; 63: 146–149. [DOI] [PubMed] [Google Scholar]
- 23.Sprague LD, Neubauer H. Melioidosis in animals: a review on epizootiology, diagnosis and clinical presentation. Journal of Veterinary Medicine 2004; 51: 305–320. [DOI] [PubMed] [Google Scholar]
- 24.Goodyear A, et al. Persistent gastric colonization with Burkholderia pseudomallei and dissemination from the gastrointestinal tract following mucosal inoculation of mice. PLoS ONE 2012; 7: e37324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaestli M, et al. Landscape changes influence the occurrence of the melioidosis bacterium Burkholderia pseudomallei in soil in northern Australia. PLoS Neglected Tropical Diseases 2009; 3: e364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaestli M, et al. Out of the ground: aerial and exotic habitats of the melioidosis bacterium Burkholderia pseudomallei in grasses in Australia. Environmental Microbiology 2012; 14: 2058–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galimand M, Dodin A. Update on melioidosis in the world [in French]. Bulletin de la Société de pathologie exotique et de ses filiales 1982; 75: 375–383. [PubMed] [Google Scholar]
- 28.Glazebrook JS, et al. Rodent zoonoses in north Queensland: the occurrence and distribution of zoonotic infections in north Queensland rodents. Australian Journal of Experimental Biology and Medical Science 1978; 56: 147–156. [PubMed] [Google Scholar]
- 29.Ashdown LR, Clarke SG. Evaluation of culture techniques for isolation of Pseudomonas pseudomallei from soil. Applied Environmental Microbiology 1992; 58: 4011–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Triggs B. Tracks, Scats and Other Traces: a Field Guide to Australian Mammals, 4th edn. South Melbourne: Oxford University Press, 2004. [Google Scholar]
- 31.Novak RT, et al. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. Journal of Clinical Microbiology 2006; 44: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price EP, et al. Accurate and rapid identification of the Burkholderia pseudomallei near-neighbour, Burkholderia ubonensis, using realtime PCR. PLoS ONE 2013; 8: e71647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Payne GW, et al. Development of a recA gene-based identification approach for the entire Burkholderia genus. Applied Environmental Microbiology 2005; 71: 3917–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parameswaran U, et al. Melioidosis at Royal Darwin Hospital in the big 2009–2010 wet season: comparison with the preceding 20 years. Medical Journal of Australia 2012; 196: 345–348. [DOI] [PubMed] [Google Scholar]
- 35.Oliver JD. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiology Reviews 2010; 34: 415–425. [DOI] [PubMed] [Google Scholar]
- 36.Berriatua E, et al. Outbreak of subclinical mastitis in a flock of dairy sheep associated with Burkholderia cepacia complex infection. Journal of Clinical Microbiology 2001; 39: 990–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akkoc A, et al. Burkholderia cepacia and Aeromonas hydrophila septicemia in an African grey parrot (Psittacus erithacus erithacus). Turkish Journal of Veterinary and Animal Sciences 2008; 32: 233–236. [Google Scholar]
- 38.Sahin N, et al. Pandoraea oxalativorans sp. nov., Pandoraea faecigallinarum sp. nov. and Pandoraea vervacti sp. nov., isolated from oxalate-enriched culture. Journal of Systematic and Evolutionary Biology 2011; 61: 2247–2253. [DOI] [PubMed] [Google Scholar]
- 39.Nüsslein K, Tiedje JM. Soil bacterial community shift correlated with change from forest to pasture vegetation in a tropical soil. Applied Environmental Microbiology 1999; 65: 3622–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itoh H, et al. Bacterial population succession and adaptation affected by insecticide application and soil spraying history. Frontiers in Microbiology 2014; 5: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy A, et al. Characterization of culturable bacterial communities in petroleum hydrocarbon contaminated sludge of oil refineries and oil exploration sites. Journal of Environmental Research and Development 2014; 8: 451–458. [Google Scholar]
- 42.Power ML, Emery S, Gillings MR. Into the wild: dissemination of antibiotic resistance determinants via a species recovery program. PLoS ONE 2013; 8: e63017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wuthiekanun V, et al. The use of selective media for the isolation of Pseudomonas pseudomallei in clinical practice. Journal of Medical Microbiology 1990; 33: 121–126. [DOI] [PubMed] [Google Scholar]