SUMMARY
Genotype G12 strains are now considered to be the sixth most prevalent human rotaviruses worldwide. In two Sicilian cities, Palermo and Messina, surveillance of rotavirus circulation performed since 1985 and 2009, respectively, did not detect G12 strains until 2012. From 2012 to 2014 rotavirus infection was detected in 29·7% of 1647 stool samples collected from children admitted for acute gastroenteritis to three Sicilian hospitals in Palermo, Messina and Ragusa. In 2012, G12P[8] was first detected in Palermo and then in Messina where it represented the second most frequent genotype (20% prevalence) after G1P[8]. Thereafter, G12 strains continued to circulate in Sicily, showing a marked prevalence in Ragusa (27·8%) in 2013 and in Palermo (21%) and Messina (16·6%) in 2014. All but one of the Sicilian G12 strains carried a P[8] VP4 genotype, whereas the single non-P[8] rotavirus strain was genotyped as G12P[9]. Phylogenetic analysis of the VP7 and VP4 sequences allowed distinction of several genetic lineages and separation of the G12P[8] strains into three cluster combinations. These findings indicate independent introductions of G12 rotavirus strains in Sicily in recent years.
Key words: G12, rotavirus, Sicily
INTRODUCTION
Group A rotaviruses (RVAs) are a major cause of acute infantile diarrhoea worldwide, accounting for over half a million deaths and 2 million hospitalizations each year [1]. Since 2009, the WHO has recommended the introduction of infant rotavirus vaccination to reduce the burden of disease. RVAs are genetically and antigenically heterogeneous. On the basis of the two outer capsid proteins VP7 and VP4, both stimulating the production of neutralizing antibodies, RVAs are classified into 27 G (VP7 specificity) and 37 P (VP4 specificity) genotypes, affecting humans and animals [2, 3]. RVA genotype distribution varies seasonally and among countries and regions. Studies from European countries report G1P[8], G2P[4], G3P[8], G4P[8] and G9P[8] as the most frequently identified RVA genotypes [4]. However, over recent years a growing number of reports have signalled the detection of G12 strains in humans worldwide, both in developed and developing countries, either sporadically or with marked incidence [5–12]. G12 is now considered to be the sixth most prevalent human RVA VP7 genotype. G12 viruses are predominantly associated with either a P[6] or P[8] VP4 genotype and, less commonly, with P[4] and P[9] specificity [9–11, 13, 14].
In Sicily, South Italy, documented continuous RVA surveillance has been conducted in Palermo since 1985 and in Messina since 2009, and G12 strains were not detected until 2012 [15, 16].
The present study aims to describe the emergence and the prolonged circulation of rotavirus G12 in Sicily supplemented with the evaluation of the genetic diversity of these strains.
MATERIALS AND METHODS
In this study, a total of 1647 stool samples were collected from children (aged 0–14 years) admitted for acute gastroenteritis to three Sicilian hospitals. In particular, 1267 and 344 samples were collected respectively from the G. Di Cristina Children's Hospital of Palermo and the G. Martino University Hospital of Messina from January 2012 to December 2014. An additional 36 samples were obtained from the Civil Hospital of Ragusa from December 2012 to August 2013. The three hospitals included in the study host the most important paediatric clinics in the respective geographical area, serving 320659 of the 736079 people aged <15 years in Sicily (official population figures at 1 January 2015). The entire set of samples received from Messina and Ragusa was screened for rotavirus. The entire collection from Palermo was screened from January 2012 to June 2013, while thereafter (July 2013-December 2014) only about one-third of the samples were arbitrarily selected (one every three sequential numbers from each monthly set). Although the collection of faecal samples was part of the general process of diagnosis of the acute gastroenteritis affecting the minors/children, we obtained verbal informed consent from the family/caregivers and the consent was recorded in the patient's medical chart.
Rotavirus testing
Rotavirus screening was performed with commercial tests validated in each hospital laboratory, including immunochromatographic (Certest Biotec, Spain; Vikia RotaAdeno, bioMérieux, Italy) and immunoenzymatic (RIDASCREEN Rotavirus; R-Biopharm AG, Germany) methods. After rotavirus identification by local laboratories, positive stool samples were stored at −20 °C and shipped to the typing centre in Palermo, for strain characterization, maintaining the cold-chain until genotyping.
RNA extraction and reverse transcription–polymerase chain reaction (RT–PCR)
Rotavirus RNA was extracted from 10% faecal suspensions in RNase-free sterile water using a QIAamp viral RNA mini kit (Qiagen/Westburg, The Netherlands) according to the manufacturer's instructions. The extracted RNA was resuspended in RNase-free sterile water and used for RT–PCR with random primers [17]. To determine the G and P genotypes, specimens were analysed by hemi-nested RT–PCR using type-specific primers (G1–G4, G6, G8–G11 and G12 and P[4], P[6], P[8]–P[11] and P[14] for VP7 and VP4, respectively) [18–22].
Sequence alignment and phylogenetic analysis
The nearly full-length VP7 gene and VP8* fragment of the VP4 gene were amplified with consensus primers Beg9/End9 and Con3/Con2, respectively, and the sequences were determined by direct sequencing. Sequence alignment was performed with CLUSTAL W [23]. Phylogenetic analysis was performed using MEGA software v. 6.0 [24], using the Kimura two-parameter model as a method of substitution and the neighbour-joining method to construct phylogenetic trees from partial sequences of VP7 (997 nt) and VP8* (862 nt, including most of the 741 nt of VP8*). The statistical significance of the phylogenies inferred was estimated by bootstrap analysis with 1000 pseudoreplicate datasets.
The VP7 and VP8* nucleotide sequences of seven Sicilian G12 strains representative of the phylogenetic clusters obtained in this study (PA121/12, PA414/12, PA525/14, ME760/12, ME650/14, ME659/14, RG178/13) were submitted to GenBank database and the accession numbers are given in Figure 2.
Fig. 2.
Phylogenetic analysis of partial VP7 (997 nt) (a) and VP8* (862 nt) (b) nucleotide sequences of the Sicilian G12P[8] isolates detected in the study. The phylogenetic trees were built using the neighbor-joining method and Kimura's two-parameter model, and bootstrapped with 1000 repetitions. Bootstrap values ⩾75% are indicated.
RESULTS AND DISCUSSION
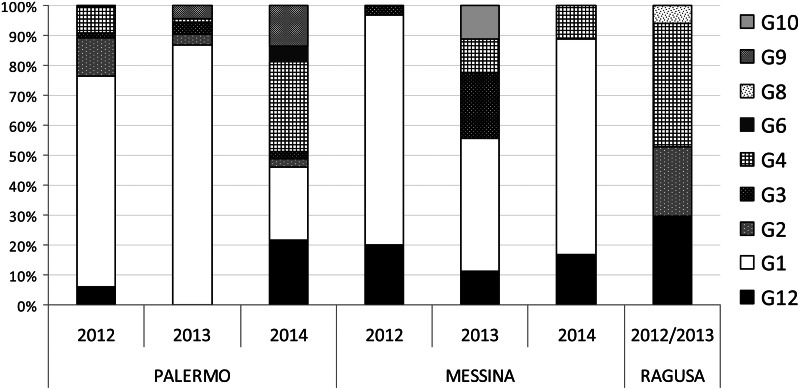
In the current study, the presence of rotavirus was investigated in stool samples collected from children admitted for acute gastroenteritis to three Sicilian hospitals in Palermo, Messina and Ragusa during three consecutive years (2012–2014). Rotavirus infection was detected in 29·7% (489/1647) of samples tested but the prevalence rates differed in the three years of the study and in the three sentinel hospitals. In Palermo, the yearly prevalence of rotavirus infection progressively decreased from 34·7% in 2012 to 24·2% in 2014, while in Messina it fluctuated, decreasing from 21·9% in 2012 to 10·9% in 2013 and then increasing to 25·3% in 2014 (Table 1). For most (79·8%) of the RVA-positive samples enough faecal material or viral nucleic acid was available for successful characterization. When considering the whole sampling, G1P[8] was the predominant strain, being detected in 75% of the specimens that could be typed. Interestingly, G12P[8] was the second most frequent genotype in 2012 in Messina (20%), in 2013 in Ragusa (27·8%) and in 2014 in both Palermo (23·7%) and Messina (16·6%) (Table 1, Fig. 1). The detection of rotaviruses with G12 specificity in this study represents the first report of such strain in Sicily. Continuous surveillance of RVA circulation has been conducted in Palermo since 1985 and, despite G12 specific primers being systematically used from January 2008 and G-untyped samples collected in 2006 and 2007 being retrospectively tested, G12 strains were not detected until 2012 [16]. Overall, since their introduction into Sicily, G12 RVAs have accounted for 8·7% of the strains characterized, showing a fluctuating circulation (Table 1).
Table 1.
Geographical and temporal distribution of G12 rotavirus strains detected in Sicily from 2012 to 2014
| Location | Year | No. of samples | No. RVA- positive samples (%) | No. RVA typed (%) | No. of G12 strains (%) | VP7 lineage and cluster of G12 strains (n) | VP8* lineage and cluster of G12 strains (n) | Temporal distribution of G12 strains (n) |
|---|---|---|---|---|---|---|---|---|
| Palermo | 2012 | 691 | 240 (34·7) | 185 (77) | 11 (5·9) | IIIb (5) IIIa (6) | P[8]-IIIb (5) P[8]-IIIa (3)‡ | February (4), March (1) September (6) |
| 2013 | 382 | 116 (30·4) | 92 (79) | 0 | − | − | − | |
| 2014 | 194 | 47 (24·2) | 38 (79) | 8 (21) | IIIa (8) | P[8]-IIIa (2) P[8]-IIIc (6) | August (2) October (1), November (5) | |
| Messina | 2012 | 173 | 38 (21·9) | 30 (79) | 6 (20) | IIIa (5) II (1) | P[8]-IIIa (5) P[9] (1) | July (3), August (2) August (1) |
| 2013 | 92 | 10 (10·9) | 9 (90) | 1 (9) | IIIa (1) | P[8]-IIIa (1) | May (1) | |
| 2014 | 79 | 20 (25·3) | 18 (90) | 3 (16·6) | IIIa (2) IIIb (1) | P[8]-IIIc (2) P[8]-IIIb (1) | May (3) | |
| Ragusa | 2012–2013† | 36 | 18 (50) | 18 (100) | 5 (27·8) | IIIa (5) | P[8]-IIIc (5) | March (5) |
| Total | 1647 | 489 (29·7) | 390 (79·8) | 34 (8·7) |
RVA, Group A rotavirus.
Samples from Ragusa were collected only from December 2012 to August 2013.
Only three of the six VP7 cluster IIIa rotavirus strains could be submitted to phylogenetic analysis for VP8* due to the limited amount of the original sample and amplified cDNA.
Fig. 1.
Percent distribution of rotavirus G types in Palermo, Messina and Ragusa from 2012 to 2014.
Historically, genotype G12 was first reported in humans in 1987 in the Philippines [25]. In the last decade rotavirus G12 strains have been increasingly identified in diarrhoeic children in Africa, Asia and South America [6, 26–30]. Similarly, sporadic detection of G12 strains has been documented in many European countries, first in UK in 2002 and then in Belgium (2003), Spain, Northern Italy, Sweden (2004–2005), Hungary (2005), France and Slovenia (2005–2006), Germany (2008) and Greece (2009–2010) [5, 9, 31–34]. According to the latest EuroRotaNet report, G12P[8] strains increased significantly from 2006 onwards and reached considerable prevalence rates in a few European countries [13]. For instance, in the 2010/2011 season, The Netherlands, Slovenia, Spain and the UK reported G12P[8] strain circulation affecting mostly single regions of these countries, which may represent localized outbreaks [35]. In Spain, rotavirus G12 was detected in the Basque Country during a 2010/2011 outbreak for seven consecutive months (September–March) [5]. In Italy, recently, Delogu et al. reported a G12 circulation in the central region of Umbria in 2012–2013 [36].
The genetic diversity within the VP7 and VP4 genes of the G12 strains detected in Sicily was investigated by phylogenetic analyses. In literature, genotype G12 strains have been subdivided into four VP7 lineages with lineage III containing the majority of the currently known G12 strains, which are associated with either P[6] or P[8] types [9, 37]. Upon phylogenetic analysis of VP7, Wangchuk et al. suggested that lineage III could be further divided into two clusters, with one cluster mostly comprising of G12P[8] strains (cluster IIIa) and the other mostly comprising of G12P[6] strains (cluster IIIb) [38]. In the present study, the majority of the Sicilian G12P[8] strains (28/34) segregated in cluster IIIa, while six strains were in cluster IIIb. A single strain from Messina clustered in the VP7 lineage II (Fig. 2a). According to the phylogenetic analysis, part of the Sicilian G12 strains belonging to cluster IIIa were most closely related (99·7% identity) to the Kenyan KDH651 strain, obtained from a rectal swab collected in Kiambu in 2010, while G12 strains detected in Bangladesh, Germany, USA, and Spain from 2002 to 2011 presented a lower correlation (Fig. 2a). The remaining Sicilian strains in cluster IIIa had a high sequence similarity to Italian G12 sequences detected in the Umbria region in 2013. The cluster IIIb sequences obtained in this study found their best match (99·0% identity) with the VP7 of a German G12P[6] strain from 2008, while no other sequence in GenBank showed >98·5% similarity. Sequence analysis of the VP8* fragment of the VP4 gene demonstrated that the Sicilian G12 strains belonging to VP7 lineage III showed a P[8] genotype, while the lineage II strain from Messina had a P[9] genotype [39]. The P[8] type is the VP4 genotype most frequently associated with G12 VP7 genotype in Europe, largely exceeding both the P[6] and P[4] genotypes [13]. In the VP8* tree (Fig. 2b), the Sicilian G12P[8] strains belonged to P[8] lineage III, i.e. a lineage different from the vaccine strains Rotarix-A41CB052A (P[8] lineage I) and RotaTeq-WI79-4 (P[8] lineage II). In addition, the P[8]-III strains segregated into three distinct clusters (herewith named cluster P[8]-IIIa, b and c, respectively). For both clusters P[8]-IIIb and P[8]-IIIc many G12 sequences with >99% similarity could be found in GenBank and are represented in Figure 2b by the US strain Vanderbilt VU-08-09-6 and the Kenyan strain KDH651, respectively. Differently, the Sicilian VP8* sequences belonging to P[8]-IIIa cluster had close similarity (99·1% identity) to a single G12P[8] sequence in GenBank, German strain GER126-08, while the remaining closest (>99% identity) BLAST hits were from G1, G3, G4 or G9P[8] strains circulating worldwide from 1999 to 2013 and are represented in Figure 2 by the KJ919669 G1P[8] sequence. According to the GenBank file, the GER126-08 G12 sequence was obtained from a stool sample of a diarrhoeic 10-year-old boy suffering from a G3 + G12P[8] co-infection. Furthermore, P[8] sequences of G1 strains circulating in 2012–2014 in Sicily also showed >99% identity to Sicilian cluster P[8]-IIIa G12 sequences (Fig. 2b).
Strains in VP7 cluster IIIb segregated in the VP8* cluster P[8]-IIIb, while strains in VP7 cluster IIIa segregated partly in VP8* cluster P[8]-IIIa and partly in VP8* cluster P[8]-IIIc. Therefore, among the Sicilian G12P[8] strains three VP7/VP8* cluster combinations could be defined: VP7/VP8* G12-IIIa/P[8]-IIIa (a/a), G12-IIIa/P[8]-IIIc (a/c) and G12-IIIb/P[8]-IIIb (b/b). The temporal and geographical distribution of the three cluster combinations showed peculiar patterns (Table 1). VP7/VP8* cluster b/b was the first to be detected in Sicily (Palermo), in February–March 2012, soon followed by cluster a/a in July–September 2012 in Messina and Palermo. The VP7/VP8* cluster a/c initially appeared in Ragusa in March 2013 and remained the major Sicilian G12 clone in 2014, circulating first in Messina (May) and then in Palermo (October–November). The detection of three different clusters of G12P[8] strains and of a G12P[9] strain indicates that the G12 strains circulating in Sicily are different RVA clones and were probably introduced by independent events. The circulation of many G12 clones in the same study period has been documented also in other settled populations, such as in Buthan, Central Myanmar and in the Kiambu area, Kenya [28, 38, 40]. However, generally a single epidemic clone has been introduced in European countries, where it was able to rapidly spread [5, 31]. G12 strains might have been transported across the globe by the increasing mobility of humans and animals. The peculiar epidemiological pattern observed in Sicily could be accounted for by the strong ongoing immigration pressure in the island. In fact, Sicily is the first shore of the sea route to Europe for migrants from the Middle East and from the African continent. The finding that some of the Sicilian G12P[8] strains (cluster a/a) possessed a VP8* sequence very similar to contemporary non-G12 strains detected in Sicily and worldwide would suggest that these G12P[8] strains may have resulted from the introduction of a VP7 gene into a common Wa-like rotavirus strain through gene reassortment. Whether the reassortment event took place in an autochthonous strain or elsewhere is not easy to determine but the detection in GenBank of a VP8* sequence of cluster P[8]-IIIa in a G12 + G3 co-infected child suggests that events potentially able to generate such hybrid genomes are not rare.
G12 strain circulation has been significantly correlated to the rural populations in Europe, possibly due to their zoonotic origin [35]. In our study most of the G12 strains were isolated from patients living in urban areas. Interestingly, for the single unusual G12P[9] strain detected in Messina retrospective epidemiological information recovered by online social network interaction with the parents allowed us to ascertain that the affected child stayed at a farmhouse for a few days before the onset of the diarrhoeic illness. The G12P[9] strain ME848/12 exhibited a complex genome constellation with at least three segments (VP6, NSP3, NSP5) closely related to those of animal rotaviruses, suggesting an animal origin for the infection of the child [39].
The emergence of new rotavirus genotypes poses a threat to vaccine efficacy but recent studies indicate that both Rotarix and RotaTeq vaccines should be effective also against G12 RVA [41, 42]. Universal rotavirus vaccination with Rotarix (GlaxoSmithKline, USA) was introduced in Sicily starting from January 2013. Regional vaccine coverage increased from 5·8% of newborns in 2012 to 32·2% in 2013 and reached 45% in 2014. However, large fluctuations in vaccine coverage were observed in the nine districts of the Sicilian island, with Palermo having high coverage (44·6% in 2013 and 62·4% in 2014) and Messina and Ragusa having lower immunization rates (respectively, 6·5% and 16% in 2013, rising to 23·6% and 29·2% in 2014). Interestingly, none of the children with G12 RVA-associated gastroenteritis had been vaccinated against rotavirus. This finding would exclude failures in vaccine-induced immunity. The age of the infected patients ranged from 2 to 99 months, but only two cases were aged >4 years and the median age was 19 months. Most cases were hospitalized for symptoms of enteritis and signs of dehydration but four developed diarrhoea >2 days after hospitalization for respiratory illness (mononucleosis or pneumonia) and therefore were considered nosocomial infections. The symptoms always included diarrhoea (>3 watery stools per day in 55·2% of cases) but also vomiting was a common sign (89·7%), while abdominal pain was less frequent (37·9%). In half (51·7%) of the cases the temperature exceeded 38 °C. There is, thus far, no evidence to suggest a difference in the virulence of the different rotavirus genotypes. However, the introduction of a new strain into a naive population should result in extensive rotaviral disease. In a prospective community-based study reported by Linhares et al., G9P[8] rotavirus, which at that time was an emerging human genotype, was associated with more severe disease in Latin America [43]. Conversely, no difference in clinical presentation and severity was found in genotypes in a 4-year prospective surveillance in France [44]. In our study, strain surveillance was hospital-based and the number of rotavirus infections caused by the newly introduced G12 strains was small. Strains identified at presentation to the hospital are likely to be associated with more severe disease; however, no significant difference was found when severity of Sicilian G12 cases was compared to the most common G1P[8] genotype (data not shown).
CONCLUSIONS
The incidence of G12 rotaviruses has shown a stable increasing tendency over recent years across Europe that justifies the inclusion of this genotype among those considered to be the most common human genotypes and suggests that it may become a predominant genotype in the future [35]. However, seasonal epidemics were generally reported in European countries, and local outbreaks failed to sustain a continuous or prolonged circulation. In Sicily, three different clusters of G12P[8] strains have been detected (VP7/VP8* cluster combinations a/a, a/c and b/b). Apparently, the Sicilian G12P[8] clusters were able to spread across the whole region through transmission among humans although at limited prevalence in some settings. Whether the introduction of rotavirus vaccination facilitated the introduction and persistent circulation at significant rates of G12P[8] rotavirus in Sicily remains to be clarified. In fact, globalization, increased travel and population movements are more likely to influence the transmission of genotypes across countries. In recent years Sicily, due to its peculiar geographical position at the border between Europe and Africa, has served as a gateway to the European Union for migrant populations from Africa and the Middle East. Meanwhile, increasing tourist flows, both national and foreign, have been attracted to the island. Finally, the animal reservoir should also be considered as a source of rotavirus infections or of gene segments for human–animal gene reassortment.
Continuous surveillance is necessary to monitor the circulation of emergent and re-emergent RVA strains, chiefly in the era of rotavirus vaccines. Whole genome analyses will be useful to investigate the pathways of evolution of G12 RVAs and to understand if the different VP7/VP8* cluster combinations we detected differ in their genomic backbone.
ACKNOWLEDGEMENTS
This study was supported, in part, by the Hungarian Scientific Research Fund (OTKA, T100727) and the Momentum program.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Parashar UD, et al. Global mortality associated with rotavirus disease among children in 2004. Journal of Infectious Diseases 2009; 200 (Suppl. 1): S9–S15. [DOI] [PubMed] [Google Scholar]
- 2.Estes MK, Kapikian AZ. Rotaviruses, 5th edn. Philadelphia: Lippincott, Williams and Wilkins, 2007, pp. 1917–1974. [Google Scholar]
- 3.Papp H, et al. Global distribution of group A rotavirus strains in horses: a systematic review. Vaccine 2013; 31: 5627–5633. [DOI] [PubMed] [Google Scholar]
- 4.Banyai K, et al. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine 2012; 30 (Suppl. 1): A122–130. [DOI] [PubMed] [Google Scholar]
- 5.Cilla G, et al. Rotavirus genotypes in children in the Basque Country (North of Spain): rapid and intense emergence of the G12[P8] genotype. Epidemiology and Infection 2013; 141: 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das S, et al. Emergence of novel human group A rotavirus G12 strains in India. Journal of Clinical Microbiology 2003; 41: 2760–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez MM, et al. Distinct evolutionary origins of G12P[8] and G12P[9] group A rotavirus strains circulating in Brazil. Infection Genetics and Evolution 2014; 28: 385–388. [DOI] [PubMed] [Google Scholar]
- 8.Pongsuwanna Y, et al. Detection of a human rotavirus with G12 and P[9] specificity in Thailand. Journal of Clinical Microbiology 2002; 40: 1390–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman M, et al. Evolutionary history and global spread of the emerging G12 human rotaviruses. Journal of Virology 2007; 81: 2382–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samajdar S, et al. Changing pattern of human group A rotaviruses: emergence of G12 as an important pathogen among children in eastern India. Journal of Clinical Virology 2006; 36: 183–188. [DOI] [PubMed] [Google Scholar]
- 11.Seheri M, et al. Update of rotavirus strains circulating in Africa from 2007 through 2011. Pediatric Infectious Diseases Journal 2014; 33 (Suppl. 1): S76–84. [DOI] [PubMed] [Google Scholar]
- 12.Shinozaki K, et al. Characterization of human rotavirus strains with G12 and P[9] detected in Japan. Journal of Medical Virology 2004; 73: 612–616. [DOI] [PubMed] [Google Scholar]
- 13.Iturriza-Gomara M, et al. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiology and Infection 2011; 139: 895–909. [DOI] [PubMed] [Google Scholar]
- 14.Mwenda JM, et al. Burden and epidemiology of rotavirus diarrhea in selected African countries: preliminary results from the African Rotavirus Surveillance Network. Journal of Infectious Diseases 2010; 202 (Suppl.): S5–S11. [DOI] [PubMed] [Google Scholar]
- 15.Colomba C, et al. Viral gastroenteritis in children hospitalised in Sicily, Italy. European Journal of Clinical Microbiology and Infectious Diseases 2006; 25: 570–575. [DOI] [PubMed] [Google Scholar]
- 16.De Grazia S, et al. Data mining from a 27-years rotavirus surveillance in Palermo, Italy. Infection Genetics and Evolution 2014; 28: 377–384. [DOI] [PubMed] [Google Scholar]
- 17.Iturriza-Gomara M, et al. Comparison of specific and random priming in the reverse transcriptase polymerase chain reaction for genotyping group A rotaviruses. Journal of Virological Methods 1999; 78: 93–103. [DOI] [PubMed] [Google Scholar]
- 18.Arista S, et al. Identification of human rotavirus strains with the P[14] genotype by PCR. Journal of Clinical Microbiology 1999; 37: 2706–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee I, et al. Modification of rotavirus multiplex RT-PCR for the detection of G12 strains based on characterization of emerging G12 rotavirus strains from South India. Journal of Medical Virology 2007; 79: 1413–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouvea V, et al. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. Journal of Clinical Microbiology 1990; 28: 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gouvea V, Santos N, Timenetsky Mdo C. Identification of bovine and porcine rotavirus G types by PCR. Journal of Clinical Microbiology 1994; 32: 1338–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iturriza-Gomara M, Kang G, Gray J. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. Journal of Clinical Virology 2004; 31: 259–265. [DOI] [PubMed] [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 1994; 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, et al. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 2013; 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taniguchi K, et al. Nucleotide sequence of the VP7 gene of a bovine rotavirus (strain 61A) with different serotype specificity from serotype 6. Nucleic Acids Research 1990; 18: 4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunliffe NA, et al. Serotype G12 rotaviruses, Lilongwe, Malawi. Emerging Infectious Diseases 2009; 15: 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espejo PW, et al. Diarrhoea caused by rotavirus in a regional Peruvian hospital: determination of circulating genotypes. Transactions of the Royal Society of Tropical Medicine and Hygiene 2014; 108: 425–430. [DOI] [PubMed] [Google Scholar]
- 28.Komoto S, et al. Whole genomic analysis of human G12P[6] and G12P[8] rotavirus strains that have emerged in Kenya: identification of porcine-like NSP4 genes. Infection Genetics and Evolution 2014; 27: 277–293. [DOI] [PubMed] [Google Scholar]
- 29.Mandile MG, et al. Surveillance of group A Rotavirus in Buenos Aires 2008–2011, long lasting circulation of G2P[4] strains possibly linked to massive monovalent vaccination in the region. Journal of Clinical Virology 2014; 60: 282–289. [DOI] [PubMed] [Google Scholar]
- 30.Ndze VN, et al. One year survey of human rotavirus strains suggests the emergence of genotype G12 in Cameroon. Journal of Medical Virology 2013; 85: 1485–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banyai K, et al. Emergence of serotype G12 rotaviruses, Hungary. Emerging Infectious Diseases 2007; 13: 916–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kokkinos PA, et al. Rare types of rotaviruses isolated from children with acute gastroenteritis in Patras, Greece. Intervirology 2013; 56: 237–241. [DOI] [PubMed] [Google Scholar]
- 33.Pietsch C, Liebert UG. Human infection with G12 rotaviruses, Germany. Emerging Infectious Diseases 2009; 15: 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steyer A, et al. Rotavirus genotypes in Slovenia: unexpected detection of G8P[8] and G12P[8] genotypes. Journal of Medical Virology 2007; 79: 626–632. [DOI] [PubMed] [Google Scholar]
- 35.Iturriza-Gomara M, Gray J, EuroRotanet. 5th Year EuroRotaNet Report; 2012.
- 36.Delogu R, et al. Unexpected spreading of G12P[8] rotavirus strains among young children in a small area of central Italy. Journal of Medical Virology 2015; 87: 1292–1302. [DOI] [PubMed] [Google Scholar]
- 37.Matthijnssens J, et al. Phylodynamic analyses of rotavirus genotypes G9 and G12 underscore their potential for swift global spread. Molecular Biology and Evolution 2010; 27: 2431–2436. [DOI] [PubMed] [Google Scholar]
- 38.Wangchuk S, et al. Dominance of emerging G9 and G12 genotypes and polymorphism of VP7 and VP4 of rotaviruses from Bhutanese children with severe diarrhea prior to the introduction of vaccine. PLoS ONE 2014; 9: e110795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Grazia S, et al. Identification of a multi-reassortant G12P[9] rotavirus with novel VP1, VP2, VP3 and NSP2 genotypes in a child with acute gastroenteritis. Infection Genetics and Evolution 2015; 35: 34–37. [DOI] [PubMed] [Google Scholar]
- 40.Ide T, et al. Whole genomic analysis of human G12P[6] and G12P[8] rotavirus strains that have emerged in Myanmar. PLoS ONE 2015; 10: e0124965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Payne DC, et al. Effectiveness of pentavalent and monovalent rotavirus vaccines in concurrent use among US children <5 years of age, 2009–2011. Clinical Infectious Diseases 2013; 57: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steele AD, et al. Human rotavirus vaccine Rotarix provides protection against diverse circulating rotavirus strains in African infants: a randomized controlled trial. BMC Infectious Diseases 2012; 12: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linhares AC, et al. Rotavirus serotype G9 is associated with more-severe disease in Latin America. Clinical Infectious Diseases 2006; 43: 312–314. [DOI] [PubMed] [Google Scholar]
- 44.de Rougemont A, et al. Molecular and clinical characterization of rotavirus from diarrheal infants admitted to pediatric emergency units in France. Pediatric Infectious Diseases Journal 2011; 30: 118–124. [DOI] [PubMed] [Google Scholar]