Abstract

The giant muscle protein titin plays important roles in heart function. Mutations in titin have emerged as a major cause of familial cardiomyopathy. Missense mutations have been identified in cardiomyopathy patients; however, it is challenging to distinguish disease-causing mutations from benign ones. Given the importance of titin mechanics in heart function, it is critically important to elucidate the mechano-phenotypes of cardiomyopathy-causing mutations found in the elastic I-band part of cardiac titin. Using single-molecule atomic force microscopy (AFM) and equilibrium chemical denaturation, we investigated the mechanical and thermodynamic effects of two missense mutations, R57C-I94 and S22P-I84, found in the elastic I-band part of cardiac titin that were predicted to be likely causing cardiomyopathy by bioinformatics analysis. Our AFM results showed that mutation R57C had a significant destabilization effect on the I94 module. R57C reduced the mechanical unfolding force of I94 by ∼30–40 pN, accelerated the unfolding kinetics, and decelerated the folding. These effects collectively increased the unfolding propensity of I94, likely resulting in altered titin elasticity. In comparison, S22P led to only modest destabilization of I84, with a decrease in unfolding force by ∼10 pN. It is unlikely that such a modest destabilization would lead to a change in titin elasticity. These results will serve as the first step toward elucidating mechano-phenotypes of cardiomyopathy-causing mutations in the elastic I-band.
Introduction
Titin is a giant muscle protein and spans half of the sarcomere from the Z-line to the M-line.1 Titin is composed of hundreds of tandem repeats of individually folded immunoglobulin (Ig), fibronectin modules, and unique unstructured sequences. The I-band part of titin is elastic and responsible for the passive tension and elastic properties of vertebrate striated muscle and thus plays critical roles in muscle function.
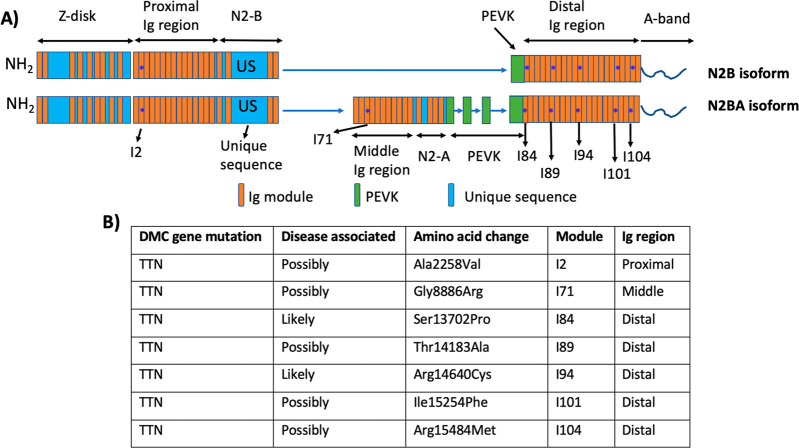
The composition of the I-band titin is regulated by alternative splicing, giving rise to muscle type-specific isoforms that determine the mechanical properties of specific muscles.2 The shortest titin isoform is the N2B isoform found in cardiac muscle and has a molecular weight of 2993 kDa. The elastic I-band of human cardiac N2B titin can be divided into four structurally distinct regions (Figure 1A): (1) a proximal Ig region containing 15 tandem Ig modules, (2) a middle unstructured N2B unique sequence (N2Bus) that contains 573 amino acid (aa) residues, (3) a largely unstructured, 186-amino acid residue PEVK sequence that is rich in proline (P), glutamate (E), valine (V), and lysine (K), and (4) a distal Ig region that contains 22 tandem repeats of Ig modules. N2BA is another titin isoform found in heart muscle. It contains additional Ig modules in the mid-Ig region and a longer PEVK sequence (Figure 1).2
Figure 1.
(A) Schematics of cardiac titin N2B and N2BA isoforms. Blue asterisks indicate the locations of the missense mutations identified in cardiac titin N2B and N2BA isoforms. (B) Missense mutations identified in titin N2B and N2BA isoforms that are possibly and likely causing cardiomyopathy.
The elasticity and extensibility of titin have been studied in detail both in situ(3−5) and at the single-molecule level.6−11 It is now well established that the I-band titin is a complex, multielement molecular spring. In addition to the entropic polymer elasticity of titin, mechanical unfolding of titin Ig modules also plays important roles in titin elasticity and extensibility when the muscle sarcomere is stretched close to or beyond the physiological range of sarcomere length.3,9,10,12−14 Titin elasticity is finely regulated by the length of titin elastic elements (i.e., the length of unstructured sequences and the number of folded Ig modules) as well as the mechanical stability of folded Ig modules. The mechanical properties of individual titin molecules can be scaled to explain the passive elastic properties of muscles, providing a detailed mechanistic understanding of muscle passive elasticity at the molecular level.9
Cardiac titin plays critical roles in heart function. The size of the stroke volume is determined by the passive diastolic tension of the cardiac muscle wall,1,15 which is directly regulated by titin. Given the importance of cardiac titin in regulating the passive elasticity of heart muscles, it has long been recognized that mutations found in titin that alter the structure and/or properties of cardiac titin could lead to cardiomyopathy.16−19 Despite the enormous size of the titin gene TTN (364 exons in total),2 early work revealed only a small number of mutations in TTN associated with cardiomyopathies.17,20−23 This was largely due to the challenges associated with sequencing such an enormous gene. Over the past few years, developments in next-generation high-throughput sequencing techniques have changed the field significantly, and sequencing TTN has become feasible at a reasonable cost. New experiments over the past decade have demonstrated that mutations in TTN have emerged as a major cause of familial cardiomyopathy.22,24−27
In a landmark study by Herman et al.,26 it was found that TTN truncation mutations are a common genetic cause of dilated cardiomyopathy (DCM), a major cause of heart failure and premature death. TTN-truncating mutations account for ∼25% of familial DCM cases and ∼18% of sporadic cases. Further studies have identified rare missense mutations that are linked to various cardiomyopathies, including DCM, hypertrophic cardiomyopathy (HCM), and arrhythmogenic right ventricular cardiomyopathy (ARVC).25
In a more recent study involving a cohort of 147 DCM index subjects, 348 rare missense variants were identified in cardiac titin N2B and N2BA isoforms.24 Using bioinformatics analysis, 44 “severe” rare variants were identified in 37 probands, and subsequent segregation analysis allowed the classification of these “severe” variants into five “likely”, five “unlikely”, and 34 “possibly”. The “likely” and “possibly” disease-causing variants are overrepresented in the A-band region of titin (23 in total). Because the A-band titin is inextensible and interacts with the myosin thick filament, it is likely that mutations found in A-band cardiac titin affect the protein–protein interactions between titin and the thick filament.
From the perspective of titin mechanics, it is very interesting to note that seven of the “likely” and “possibly” disease-causing variants were also found to be located in the elastic I-band of titin N2B and N2BA isoforms. All seven mutations were found in folded Ig modules, and none was in the unstructured N-2B unique sequence or PEVK sequence (Figure 1).
It is well established that point mutations could significantly destabilize I-band Ig modules (as shown for I91) so that they could potentially unfold under physiological conditions.19,28 It has been hypothesized that such mutations could alter titin-based passive stiffness and potentially cause cardiomyopathy.19 Recent studies on the T2850I variant of I10 lend supporting evidence to this hypothesis.29,30 T2850I is a missense mutation in the 10th Ig module of the I-band titin and has been confirmed to be linked to ARVC.20 Single-molecule atomic force microscopy (AFM) as well as equilibrium chemical denaturation studies showed that the T2850I mutation significantly destabilized I10, both thermodynamically and mechanically, and caused I10 to unfold at reduced forces29 and display a higher propensity for unfolding. These results suggested that T2850I-I10 may unfold under physiological conditions and be more susceptible to protein degradation, leading to altered titin passive elasticity.29,30
The discovery of the “likely” and “possibly” disease-causing variants in the Ig modules of the I-band part of cardiac titin provides a valuable opportunity to explore if these variants display mechano-phenotypes similar to that of T2850I-I10, and if mechanical effects caused by mutations can be used as screening tools for identifying cardiomyopathy-causing mutations. For these purposes, here we combined single-molecule AFM, equilibrium denaturation, and protein engineering techniques to investigate the mechanical and thermodynamic effects of two mutations, Ser13702Pro and Arg14640Cys, that are predicted to be likely to cause cardiomyopathy.
Experimental Section
Protein Engineering
Genes encoding wild type (wt) I94 and I84, carrying a 5′ BamHI, and 3′ BglII and KpnI restriction sites, of cardiac titin were custom synthesized (Gene-script). R57C-I94 and S22P-I84 were constructed by using the standard site-directed mutagenesis method. The genes encoding polyproteins Coh-(GB1-Ig)4 were constructed in the pUC57 vector following a well-established iterative protocol based on the identity of the sticky ends of digested BamHI and BglII restriction sites.31,32 The genes were then subcloned into the pQE80L expression vector. Overexpression of target proteins was carried out in Escherichia coli strain DH5α following well-established protocols. Briefly, after inoculation with 3 mL of the preculture, the cells were grown in 200 mL of Luria-Bertani medium containing 100 μg/mL ampicillin at 37 °C and 225 rpm. When the OD600 of the culture reached ∼0.7, protein overexpression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside and the protein expression continued for 4 h. Then, the cells were pelleted by centrifugation at 4000g for 10 min at 4 °C and resuspended in 10 mL of loading buffer [10 mM phosphate-buffered saline (PBS) (pH 7.4)]. After the addition of 10 μL of protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO), 100 μL of 50 mg/mL lysozyme, 1 mL of 10% (w/v) Triton X-100, and 50 μL of 1 mg/mL DNase I and RNase A to the loading buffer, cells were lysed for 40 min on ice. Cell debris was then removed by centrifugation at 22000g and 4 °C. Target proteins were purified by using Co2+ affinity chromatography (TALON Metal Affinity Resins, Clontech, Mountain View, CA). The purified protein sample had a concentration of ∼4 mg/mL.
Functionalization of AFM Cantilevers and Glass Coverslips
After air plasma cleaning for 30 min, cantilevers were salinized by incubating in 500 μL of (2-aminopropyl)-dimethyl-ethoxysilane (APDMES, ABCR, Karlsruhe, Germany) mixed with 0.5 μL of doubly distilled H2O (ddH2O) and 500 μL of ethanol for 2 min. The cantilevers were then rinsed with ethanol and water sequentially, followed by baking at 85 °C for 45 min. Salinized cantilevers were stored under argon and used within one month.
Glass coverslips were first washed by sonication in a 1:1 ddH2O/isopropanol solution for 10 min and then washed in a hot chromic acid solution for 20 min (Caution:Chromic acid is corrosive and a carcinogen and must be handled with extreme care!). Coverslips were then rinsed with ddH2O and ethanol and dried in a gentle stream of nitrogen. To functionalize the glass coverslips, coverslips were incubated in an APDMES/ethanol solution (88% ethanol, 10% ddH2O, and 2% APDMES in volume) on a tabletop shaker for 1 h. Then the coverslips were rinsed with ethanol and ddH2O and then baked at 85 °C for 45 min. The clean coverslips were then stored under argon and used within one month.
To functionalize the glass coverslip and AFM cantilevers with Cohesin and Dockrin fusion proteins, silanized coverslips and cantilevers were incubated in a solution of 5 kDa maleimide-PEG succinimidyl-NHS-acid ester (Creative PEGWorks, Durham, NC) dissolved in 50 mM HEPES (pH 7.4–7.6) at 25 mM (125 mg/mL) for 45 min. After being rinsed with ddH2O, the coverslips were incubated with the protein of interest at 6–10 μM [50 mM sodium phosphate (pH 7.2), 50 mM NaCl, and 10 mM EDTA buffer], and the cantilevers were incubated with 10 μM cys-X-dockerin [50 mM sodium phosphate (pH 7.2), 50 mM NaCl, and 10 mM EDTA buffer] for 1 h. After being rinsed with PBS buffer, the samples were used immediately in single-molecule AFM experiments. The pulling experiments were carried out in Tris buffer [25 mM Tris, 72 mM NaCl, and 1 mM CaCl2 (pH 7.2)].
Single-Molecule Force Spectroscopy (SMFS) Experiments
SMFS experiments were conducted on a home-built atomic force microscope, which was described previously.32 Before each SMFS experiment, the spring constant (∼40 pN/nm) of the silicon nitride cantilevers (MLCT, Bruker, Santa Barbara, CA) was calibrated by utilizing the thermal method in a PBS buffer solution (pH 7.4). In a typical constant-velocity SMFS experiment, the cantilever functionalized with dockerin was brought into contact (∼50 pN) with the surface functionalized with the target cohesin fusion proteins and then retracted at a constant velocity (e.g., 400 nm/s). The pulling was conducted in a semiautomatic fashion. The force–extension curves were then screened for single-molecule stretching events and analyzed using custom written codes in IgorPro (Wavemetrics, Lake Oswego, OR). The unfolding experiments were carried out in Tris buffer [25 mM Tris, 72 mM NaCl, and 1 mM CaCl2 (pH 7.2)]. To prevent endogenous cysteine residues in I94 from reacting with unreacted maleimide or forming a disulfide bond between them in the unfolded I94, the refolding experiments with I94 and R57C-I94 were carried out in the presence of 5 mM dithiothreitol.
Monte Carlo Simulations
Monte Carlo simulations were carried out following the well-established protocol, which was described in detail previously, to reproduce the experimentally determined force–extension curves. The mechanical unfolding of proteins was described using the Bell–Evans model, a(F) = a0 exp(FΔxu/kBT).33,34 The simulation results (unfolding force histograms and pulling speed dependence of the unfolding forces) were then compared with the experimental data to estimate the kinetic parameters characterizing the mechanical unfolding, the unfolding rate constant at zero force, α0, and the distance between the native and mechanical unfolding transition state, Δxu, performed to reproduce our experimental data to determine unfolding kinetics.
Chemical-Induced Denaturation
To determine the thermodynamic stability of I94, I84, and their mutants, equilibrium chemical denaturation experiments were performed on a Jasco-J810 spectropolarimeter using a micro fluorometer polystyrene plastic cuvette with a path length of 10 mm. Tryptophan fluorescence was used as the probe for protein unfolding. In the experiments, tryptophan fluorescence was excited at 280 nm, and the emission spectra were recorded from 300 to 500 nm. To measure the fluorescence spectra of proteins at different GdmCl concentrations, the protein concentration was kept the same at 0.2 mg/mL by mixing protein and GdmCl stock solutions. The prepared protein solutions were equilibrated at room temperature for at least 30 min before measurements. The experimentally determined tryptophan fluorescence as a function of GdmCl concentration was converted to the equilibrium denaturation curve. The denaturation curves were then fitted to a two-state model (eq 1) to estimate the m value, [D]1/2, and ΔGN–U
 |
1 |
where f is the fraction of folded proteins, R is the gas constant, and T is the absolute temperature.16ΔGN–U = m[D]1/2, where [D]1/2 is the GdmCl concentration at which the fraction of the folded protein is 50%.
Results
Ser13702Pro and Arg14640Cys Are Two Missense Mutations That Likely Cause Cardiomyopathy
Among the seven rare missense variants identified as “possibly” and “likely” variants in the I-band N2B/N2BA cardiac titin to cause DCM, Ser13702Pro and Arg14640Cys are two likely variants.24 Ser13702Pro is located in the I84 module, and Arg14640Cys is in the I94 module, both of which are homologous and in the distal Ig region of the I-band titin (Figure S1). Because all I-band Ig modules share a high degree of sequence homology among themselves, they are predicted to fold into similar three-dimensional structures.35,36 I1 and I91 modules are the only Ig modules in the I-band titin whose three-dimensional structures are available, and both of them fold into a β-sandwich structure.37,38 To gain insights into the structures of I84 and I94, we carried out homology modeling using SWISS-MODEL39 to predict their three-dimensional structures.
As expected, homology modeling revealed that both I94 and I84 fold into β-sandwich structures (Figure 2) that are similar to I91. I94 contains 90 residues. Residue Arg14640, which is Arg57 in the sequence of I94, is located at the beginning of β-strand E and solvent-exposed (Figure 2A). The Arg14640Cys mutation caused the change in a positively charged residue to a polar residue but did not cause significant structural changes to the β-sandwich structure of I94 (Figure 2B). In comparison, I84 contains 98 residues. Residue Ser13702, which is Ser22 in the sequence of I84, is located in the flexible loop connecting β strands A′ and B (Figure 2C). Although the Ser13702Pro mutation introduced a bend, this mutation was well-tolerated and did not cause significant structural changes to the β-sandwich structure (Figure 2D). These results indicated that the structural changes caused by these two mutations are likely small, and both mutant Ig modules are likely folded.
Figure 2.

Three-dimensional structures of (A) I94, (B) R57C-I94, (C) I84, and (D) S22P-I84 predicted by homology modeling. Both I94 and I84 modules fold into a β-sandwich structure, similar to that of I91. Homology modeling also predicted that mutations R57C and S22P do not alter the three-dimensional structure of I94 and I84 significantly.
R57C and S22P Destabilized I94 and I84 Thermodynamically
To investigate the thermodynamic effect of both mutations on I94 and I84, we first carried out equilibrium unfolding experiments to measure their thermodynamic stability. For the sake of convenience, Arg14640Cys-I94 is termed as R57C-I94 and Ser13702P-I84 as S22P-I84. Because both Ig modules contained a tryptophan residue in their hydrophobic core, we used tryptophan fluorescence as a probe to monitor their equilibrium unfolding as a function of the concentration of chemical denaturant guanidinium chloride (GdmCl). Figure 3 shows the equilibrium denaturation curves of both I94 and I84 modules as well as their mutants. Both I94 and I84 are thermodynamically stable, with [D]1/2 values, at which 50% of the protein is unfolded, of 1.17 ± 0.01 M [average (avg.) ± standard deviation (SD)] and 2.90 ± 0.01 M for I94 and I84, respectively. Fitting the experimental data to a two-state unfolding–folding model measured equilibrium free energies between the native and unfolded state (ΔGN–U) of 2.86 ± 0.10 kcal/mol for I9440 and 5.67 ± 0.18 kcal/mol for I84. Introducing mutations R57C and S22P into I94 and I84 resulted in a clear destabilization, as evidenced by the shift in [D]1/2 toward lower values. Fitting the data to the two-state model revealed ΔΔGN–U values of 1.09 kcal/mol for R57C-I94 and 0.87 kcal/mol for S22P-I84. Neither mutation significantly changed the m value, which is associated with the difference in the surface area between the native (N) and denatured (D) states,41 of both Ig modules. These results indicated that both mutations had a modest destabilization effect on I94 and I84, and both variants remained thermodynamically stable and folded under physiological conditions.
Figure 3.
Equilibrium denaturation curves of I94, (A) I94-R57C, and I84, (B) I84-S22P. Solid lines are fits of the experimental data to a two-state unfolding–folding model. The fitting parameters are listed in Table S1.
Single-Molecule AFM Experiments Revealed the Decreased Mechanical Stability of Both Variants
To examine the mechanical effect of the mutations on both Ig modules, we used single-molecule AFM to examine their mechanical unfolding. For this purpose, we constructed heteropolyproteins Coh-(GB1-I94)4 and Coh-(GB1-R57C-I94)4, where Coh represents the cohesin domain42 and GB1 represents the small protein B1 IgG binding domain of protein G. GB1, which has been studied in great detail32 and is characterized by an unfolding force ∼180 pN at a pulling speed of 400 nm/s and a contour length increment ΔLc of 18 nm, served as a fingerprint for identifying single-molecule stretching events as well as an internal force caliper for comparing the unfolding forces of I94 and its mutant I94-R57C.32
To stretch the heteropolyprotein in its entirely from its N- and C-termini, we used a well-established stretching strategy based on specific interactions between cohesin and dockerin (Figure 4A).42 Cohesin and dockerin are two interacting domains from Clostridium thermocellum and have been developed as a specific handle for stretching single proteins in AFM-based SMFS experiments.42,43 Bringing a dockerin-coated AFM tip into contact with Coh-(GB1-I94)4, which were chemically immobilized onto the glass substrate via the thiol-maleimide chemistry, allowed us to stretch Coh-(GB1-I94)4 from its N- and C-termini via the strong interaction between Coh-Doc. Figure 4B shows the representative force–extension curves of stretching (GB1-I94)4, which are of the characteristic sawtooth pattern-like appearance, with each sawtooth peak corresponding to the mechanical unfolding of one protein domain in the stretched polyprotein. Eight unfolding force peaks were observed in each force–extension curve, corresponding to the unfolding of the eight domains in the polyprotein, and the last peak corresponded to the rupture of the Coh-Doc complex. The unfolding events clearly showed two different ΔLc values. Fitting the data to the wormlike chain (WLC) model of polymer elasticity revealed a ΔLc1 of 18.1 ± 0.5 nm (avg. ± SD) and a ΔLc2 of 28.2 ± 0.6 nm. Clearly, the unfolding events with a ΔLc1 of 18 nm correspond to the unfolding of the fingerprint domain GB1, while those with 28 nm correspond to the unfolding of I94. It is worth noting that four unfolding force peaks of I94 were interspersed with four GB1 unfolding events, suggesting that I94 and GB1 unfolded at similar forces.
Figure 4.
Mechanical properties of I94 and R57C-I94. (A) Schematics of the single-molecule AFM pulling experiments using Coh-Doc interactions. Representative force–extension curves of (B) (GB1-I94)4 and (C) (GB1-R57C–I94)4. Two groups of unfolding force events were observed. Unfolding events colored black display a contour length increment ΔLc of 18 nm and correspond to the unfolding of GB1 domains, while unfolding events colored red display a contour length increment ΔLc of 28 nm and correspond to the unfolding of I94 or R57C-I94 domains. (D) Unfolding force histograms of I94 and R57C-I94 at a pulling speed of 400 nm/s. The average unfolding force is 187 ± 33 pN (avg. ± SD) for wt I94 (n = 208) or 145 ± 32 pN for R57C-I94 (n = 291). (E) Pulling speed dependence of the unfolding forces. Solid lines are Monte Carlo simulation results using a Δxu of 0.17 nm and α0 values of 0.025 s–1 for wt I94 and 0.08 s–1 for R57C-I94. The error bars are standard errors of the mean (sem).
Stretching (GB1-R57C-I94)4 resulted in force–extension curves similar to those of wild type (wt) (GB1-I94)4 (Figure 4C). The unfolding of I94-R57C displayed a ΔLc of 28 nm, identical to those of wt I94, suggesting that the R57C mutant is well folded. It is of note that, different from the wt protein, the four unfolding events of the mutant R57C often occurred prior to the four unfolding events of GB1, suggesting that the mechanical stability of R57C is lower than that of GB1 and thus lower than that of wt I94.
Quantitative analysis of the unfolding forces revealed that the average unfolding force of wt I94 was 187 ± 33 pN (avg. ± SD) at a pulling speed of 400 nm, while the unfolding force for R57C-I94 was 145 ± 32 pN, ∼40 pN lower than that of wt I94 (Figure 4D). Stretching (GB1-I94-R57C)4 at different pulling speeds revealed that the unfolding force of R57C was consistently lower than that of wt I94 by 30–40 pN (Figure 4E). The standard deviation of the unfolding forces is determined by kBT/Δxu, where kBT is the thermal energy and Δxu is the unfolding distance between the native state and mechanical unfolding transition state.33,34 The relatively large SD of the unfolding forces indicated a large Δxu for both I94 and R57-I94. It is worth noting that the standard error of the mean (sem) of the unfolding forces is small (∼2–3 pN), and a Student’s t test showed that the difference in unfolding forces of I94 and R57-I94 is statistically significant (p < 0.001). Hence, these results clearly indicated that I94 was weakened mechanically by the R57C mutation.
Using similar strategies, we characterized the mechanical unfolding of I84 and its variant S22P-I84 (Figure 5). The average unfolding forces at a pulling speed of 400 nm/s were 180 pN for wt I84 and 167 pN for S22P. Experiments at different pulling speeds revealed the same consistent trend. Clearly, S22P decreased the unfolding force of I84, but its weakening effect was much milder than that of R57C-I94. It is of note that although the weakening effect was small, the difference in the unfolding force of I84 and S22P-I84 is statistically significant (p < 0.001), while the difference in unfolding forces of GB1 in (GB1-I84)4 and (GB1-I84-S22P)4 is statistically not significant (p > 0.05).
Figure 5.
Mechanical properties of I84 and S22P-I84. Representative force–extension curves of (A) (GB1-I84)4 and (B) (GB1-S22P-I84)4. Two groups of unfolding force events were observed. Unfolding events colored black display a contour length increment ΔLc of 18 nm and correspond to the unfolding of GB1 domains, while unfolding events colored red display a contour length increment ΔLc of 28 nm and correspond to the unfolding of I84 or S22P-I84 domains. (C) Unfolding force histograms of I84 and S22P-I84 at a pulling speed of 400 nm/s. The average unfolding force is 180 ± 30 pN (avg. ± SD) for wt I84 (n = 276) or 167 ± 30 pN for S22P-I84 (n = 232). (D) Pulling speed dependence of the unfolding forces. Solid lines are Monte Carlo simulation results using a Δxu of 0.23 nm and α0 values of 0.002 s–1 for wt I84 and 0.004 s–1 for S22P-I84. Errors bars correspond to the sem.
To quantify the mechanical unfolding kinetics, we used the well-established Monte Carlo simulation protocols to reproduce the pulling speed dependence of the unfolding forces. Fitting the Monte Carlo simulation result to the experimental data allowed for the estimation of the key parameters characterizing the free energy profile of the mechanical unfolding of proteins: α0, the intrinsic unfolding rate constant at zero force, and Δxu, the distance between the native state and the mechanical unfolding transition state (⧧) (Figures 4E and 5D). Both mutations accelerate the unfolding kinetics of I94 and I84 but did not affect Δxu. The R57C mutation increased the α0 of I94 by ∼3-fold (from 0.025 to 0.080 s–1), while the S22P mutation increases the α0 of I84 by ∼2-fold (from 0.002 to 0.004 s–1).
It is worth noting that R57 is outside of the A-B and A′G patch, which are predicted to be responsible for the mechanical stability of titin Ig domains,12 yet R57C has an obvious mechanical destabilizing effect on I94. This result suggested that R57 may be involved in interactions with the A′G patch. To understand the molecular origin of this destabilization effect, more detailed analysis will be needed.
S22P and R57C Decelerated the Intrinsic Folding Kinetics of I94 and I84
Having characterized the mechanical effects of the two mutations on I94 and I84, we used single-molecule AFM to evaluate their effects on the folding kinetics of both proteins. For this, we used the double-pulse protocol19 to measure the folding kinetics of I94, I84, and their mutants. As shown in Figure 6A, in the first pulse, the full length polyprotein (GB1-I94)4 was stretched to unfold all of the protein domains (four GB1 domains and four I94 domains). Then the unfolded polypeptide chain was quickly relaxed to zero force and kept at zero force for a given time to allow the unfolded protein domains to refold. In the second pulse, the polyprotein was stretched again to count the number of folded I94 domains. By varying waiting time Δt, we measured the folding probability of I94 as a function of Δt. As shown in Figure 6B, wt I94 folded rapidly at zero force; within <1 s, almost 100% I94 domains managed to refold to regain their mechanical stability. Fitting the folding probability curve of I94 to a first-order rate equation yielded a folding rate constant β0 of 15 ± 5 s–1 at zero force (Figure 6C). In comparison, the folding of I94-R57C was clearly slowed by the R57C mutation, showing a folding rate constant β0 of 6.4 ± 1.1 s–1.
Figure 6.
Effects of mutations on the folding kinetics of I94 and I84. (A) Schematics of the double-pulse AFM protocol in measuring the folding kinetics of proteins. (B) Representative refolding curves of full length (GB1-I94)4. In the first pulse, four GB1 and four I94 unfolding events were observed. In the second pulse, four GB1 unfolding events were observed due to its fast-folding kinetics, while the number of refolded I94 depended on Δt. The refolding experiments were carried out in the presence of 5 mM dithiothreitol, which prevented endogenous cysteine residues in I94 from forming a disulfide bond or reacting with unreacted maleimide. Plots of folding probability vs Δt for (C) I94 and R57C-I94 and (D) I84 and S22P-I84. Solid lines are the fitting to the first-order rate law Pf = 1 – exp(−β0t).
Using the same strategy, we also examined the effect of S22P on the folding kinetics of I84 (Figure 6D). It is interesting to find that the folding of wt I84 is very slow, with a folding rate constant β0 of (1.8 ± 0.2) × 10–2 s–1 at zero force. Such a slow folding kinetics was observed in some distal Ig modules, such as I92 and I95.44 The folding of mutant S22P slowed the folding of I84 even further by ∼3.6-fold, leading to a β0 of (5.0 ± 0.3) × 10–3 s–1 at zero force for S22P-I84.
I94-R57C Displayed Mechano-phenotypes Similar to Those of ARVC-Causing Variant T1850I-I10
Although it has long been hypothesized that mutations in the elastic I-band titin that affect the mechanical properties of folded Ig modules may be a potential cause of cardiomyopathy,19 connecting rare missense mutations in titin with cardiomyopathy has been challenging. The first confirmed cardiomyopathy-causing missense mutation in the elastic I-band titin is T2850I, which is located in the I10 module and linked to ARVC.20,29,30 Single-molecule AFM studies revealed that the T1850I mutation destabilized I10 both thermodynamically and mechanically.29 The T1850I mutation resulted in a decrease in the mechanical unfolding force of I10 by ∼30 pN, a 4-fold increase in the unfolding rate. The mutation did not affect the folding kinetics of I10. Proteolysis analysis also revealed that the mutation increased the susceptibility of I10 to proteolysis. On the basis of these experimental data, it was concluded that the increased susceptibility of I10 to proteolysis, which is due to the increased unfolding kinetics and the disruption of the local structure, may be a key feature for this cardiomyopathy-causing variant. These mechano-phenotypes of a cardiomyopathy-causing mutation provided an invaluable starting point for further investigation of the mechanical effects of potential disease-causing mutations in the I-band titin. Our studies of R57C-I94 and S22P-I84 serve as an effort in this direction. Our results revealed different mechanical effects of these mutations on Ig modules.
The two missense variants I94-R57C and I84-S22P, identified from cardiomyopathy patients, were predicted to be likely to cause cardiomyopathy.24 Our single-molecule AFM studies revealed that the R57C mutation had mechano-effects similar to those of the cardiomyopathy-causing mutation T1850I-I10. I94-R57C decreased the mechanical unfolding force of I94 by ∼30–40 pN and accelerated the unfolding kinetics by 3-fold. Moreover, R57C slowed the folding kinetics of the I94 module. Both an increased unfolding rate constant and a decreased folding rate constant should increase the propensity of unfolding of the I94 module. A stretching force would further increase this propensity, as predicted by the Bell–Evans model.33,34 To quantitatively estimate the unfolding propensity of I94 in muscle sarcomeres, we assumed an average stretching force of 5 pN experienced by each individual titin molecule in sarcomeres.14 We found that under reduced conditions (which is the physiological condition), ∼2.3% of wt I94 would be unfolded at this force. However, the unfolded fraction of R57C-I94 increased to ∼15% under the same condition. This significant increase in the unfolded fraction of I94 suggested that the R57C mutation may render a significant fraction of I94 unfolded, thus making it more susceptible to proteolysis inside the cell. These effects are similar to those of T1850I-I10, suggesting that R57C may alter the elastic properties of I-band titin and make R57C a likely cardiomyopathy-causing mutation.
S22P Displayed a Small Effect on the Nanomechanical Properties of I84
For I84-S22P, the mutational effect appears to be more subtle. I84 is thermodynamically stable with a ΔGN–U of 5.67 kCal/mol measured from equilibrium denaturation experiments. A destabilization effect of 0.87 kcal/mol is rather modest. Single-molecule AFM experiments also revealed a small destabilization effect (a decrease of 10 pN in unfolding force).
Single-molecule AFM experiments revealed that I84 showed a slow folding kinetics, with a β0 of 0.018 s–1 at zero force. This slow folding kinetics and the measured mechanical unfolding kinetics would give rise to an apparent ΔGN–U of 1.38 kcal/mol, considerably smaller than the value measured from equilibrium chemical denaturation experiments (5.67 kcal/mol). A ΔGN–U of 1.38 kcal/mol would predict that ∼10% of I84 would be unfolded at equilibrium even in the absence of any stretching force. This is in sharp contrast with the predicted 0.01% unfolded fraction of I84 based on a ΔGN–U of 5.67 kcal/mol. To examine this discrepancy, we counted the number of folded and unfolded I84 modules in our single-molecule AFM pulling experiments on (GB1-I84)4. In our experiments, (GB1-I84)4 was stretched from its N- and C-termini. In principle, each force–extension curve should contain four GB1 and four I84 unfolding events. Missing an unfolding event, either GB1 or I84 would indicate that the missing domain was unfolded on the surface prior to stretching. We found that 99% of the force–extension curves contained four GB1 unfolding events and four I84 unfolding events, indicating that almost all I84 modules are properly folded in the experiments, in spite of the chemical immobilization onto the glass surface. This observation eliminated the possibility that 10% of I84 would be unfolded at equilibrium. Therefore, the observed slow folding rate constant of I84 in the AFM experiments was likely a result of kinetically trapping; i.e., the mechanically unfolded I84 was likely stretched into a kinetically trapped state. However, the origin of this kinetic trapping remains to be elucidated.
Therefore, on the basis of the ΔGN–U measured from equilibrium chemical denaturation experiments, the S22P mutation would not cause a significant change to the unfolding probability of I84 under a stretching force of 5 pN (0.1% for wt I84 vs 0.5% for S22P). In other words, the S22P mutation would have a very weak effect on the mechanical properties of I84. This result implied that from the viewpoint of titin elasticity, the S22P mutation would unlikely cause a significant change in the mechanical properties of I84, thus raising the question of whether S22P is a cardiomyopathy-causing variant. However, S22P does slow the refolding of I84. Because titin cyclically stretches during its life span, the reduced refolding rate could also lead to the change in titin elasticity.45 Thus, it remains an open question if the slower folding kinetics of S22P can lead to cardiomyopathy.
Challenges in Identifying Cardiomyopathy-Causing Mutations in I-Band Titin Ig Modules
Our single-molecule AFM experiments on the two missense mutants that were predicted by bioinformatics analysis to be likely causing cardiomyopathy24 revealed different mechanical effects. AFM results on R57C-I94 revealed clear mechanical signatures of the R57C mutant that are comparable to those found in a cardiomyopathy-causing mutation T1850I-I10,29 while S22P-I84 showed only minute changes to the mechanical properties of I84.
From a viewpoint of titin mechanics alone, the R57C mutation will likely alter the elasticity of cardiac titin and potentially lead to cardiomyopathy. However, S22P will unlikely alter titin elasticity. Although we cannot completely eliminate the possibility that S22P is linked to cardiomyopathy, our results do raise the question of whether S22P is indeed a cardiomyopathy-causing mutation.
The difference in the mechanical consequences of mutations in these two I-band Ig modules also points to the challenge in distinguishing disease-causing mutations from those benign ones. This challenge is inevitably linked to the very limited number of missense mutants that have been confirmed to be directly linked to cardiomyopathy. This situation is different from disease-causing mutations found in other sarcomeric proteins, such as myosin and myosin binding proteins, where cardiomyopathy has been found to be linked to missense mutations in these proteins.46 In the latter case, establishing mechano-phenotypes may become feasible. For example, single-molecule AFM has been used to try to establish mechano-phenotypes of disease-causing mutations found in myosin binding proteins.45 Therefore, developing innovative methods to help establish definitive connections between cardiomyopathy and missense variants in titin, especially I-band titin, will be a critical step toward establishing mechano-phenotypes for missense variants in elastic I-band titin. In this respect, the experimental efforts by Bogomolovas,30 in which transgenic knock-in mice containing the T1850I mutation were used to link DCM with T1850I, are critically important.
Conclusions
Using single-molecule AFM and equilibrium chemical denaturation, we investigated the mechanical and thermodynamic effects of two missense mutations found in the elastic I-band part of cardiac titin that were predicted to cause cardiomyopathy by using bioinformatics analysis. Our AFM results showed that R57C had a significant destabilization effect, both thermodynamic and mechanical, on the I94 module. R57C reduced the mechanical unfolding force of I94, accelerated the unfolding kinetics, and decelerated the folding kinetics. These effects collectively increased the unfolding propensity of I94, likely resulting in altered titin elasticity. In comparison, the S22P mutation led to only modest destabilization of I84, with a decrease in unfolding force by ∼10 pN. It is unlikely that such a modest destabilization would lead to a change in titin elasticity. These results will serve as the first step toward elucidating mechano-phenotypes of cardiomyopathy-causing mutations in the elastic I-band part of titin.
Acknowledgments
This work is supported by a Grant-in-Aid (G-19-0026357) from Hearts & Stroke Foundation Canada. The authors thank Prof. Algar for technical assistance in some experiments.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.langmuir.1c02006.
Sequence alignment of I84, I94, and I91 and a table of chemical denaturation parameters (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Labeit S.; Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science 1995, 270 (5234), 293–6. 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- Bang M. L.; Centner T.; Fornoff F.; Geach A. J.; Gotthardt M.; McNabb M.; Witt C. C.; Labeit D.; Gregorio C. C.; Granzier H.; Labeit S. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 2001, 89 (11), 1065–72. 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- Linke W. A.; Rudy D. E.; Centner T.; Gautel M.; Witt C.; Labeit S.; Gregorio C. C. I-band titin in cardiac muscle is a three-element molecular spring and is critical for maintaining thin filament structure. J. Cell Biol. 1999, 146 (3), 631–44. 10.1083/jcb.146.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzier H. L.; Labeit S. The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ. Res. 2004, 94 (3), 284–95. 10.1161/01.RES.0000117769.88862.F8. [DOI] [PubMed] [Google Scholar]
- Granzier H. L.; Irving T. C. Passive tension in cardiac muscle: contribution of collagen, titin, microtubules, and intermediate filaments. Biophys. J. 1995, 68 (3), 1027–44. 10.1016/S0006-3495(95)80278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermayer M. S.; Smith S. B.; Granzier H. L.; Bustamante C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science 1997, 276 (5315), 1112–6. 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- Rief M.; Gautel M.; Oesterhelt F.; Fernandez J. M.; Gaub H. E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 1997, 276 (5315), 1109–12. 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- Tskhovrebova L.; Trinick J.; Sleep J. A.; Simmons R. M. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature 1997, 387 (6630), 308–12. 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- Li H.; Linke W. A.; Oberhauser A. F.; Carrion-Vazquez M.; Kerkvliet J. G.; Lu H.; Marszalek P. E.; Fernandez J. M. Reverse engineering of the giant muscle protein titin. Nature 2002, 418 (6901), 998–1002. 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- Watanabe K.; Muhle-Goll C.; Kellermayer M. S.; Labeit S.; Granzier H. Different molecular mechanics displayed by titin’s constitutively and differentially expressed tandem Ig segments. J. Struct. Biol. 2002, 137 (1–2), 248–58. 10.1006/jsbi.2002.4458. [DOI] [PubMed] [Google Scholar]
- Marszalek P. E.; Lu H.; Li H.; Carrion-Vazquez M.; Oberhauser A. F.; Schulten K.; Fernandez J. M. Mechanical unfolding intermediates in titin modules. Nature 1999, 402 (6757), 100–3. 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- Linke W. A.; Grutzner A. Pulling single molecules of titin by AFM-recent advances and physiological implications. Pfluegers Arch. 2008, 456 (1), 101–15. 10.1007/s00424-007-0389-x. [DOI] [PubMed] [Google Scholar]
- Watanabe K.; Nair P.; Labeit D.; Kellermayer M. S.; Greaser M.; Labeit S.; Granzier H. Molecular mechanics of cardiac titin’s PEVK and N2B spring elements. J. Biol. Chem. 2002, 277 (13), 11549–58. 10.1074/jbc.M200356200. [DOI] [PubMed] [Google Scholar]
- Minajeva A.; Kulke M.; Fernandez J. M.; Linke W. A. Unfolding of titin domains explains the viscoelastic behavior of skeletal myofibrils. Biophys. J. 2001, 80 (3), 1442–51. 10.1016/S0006-3495(01)76116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady A. J. Mechanical properties of isolated cardiac myocytes. Physiol. Rev. 1991, 71 (2), 413–28. 10.1152/physrev.1991.71.2.413. [DOI] [PubMed] [Google Scholar]
- Itoh-Satoh M.; Hayashi T.; Nishi H.; Koga Y.; Arimura T.; Koyanagi T.; Takahashi M.; Hohda S.; Ueda K.; Nouchi T.; Hiroe M.; Marumo F.; Imaizumi T.; Yasunami M.; Kimura A. Titin mutations as the molecular basis for dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2002, 291 (2), 385–93. 10.1006/bbrc.2002.6448. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y.; Hayashi T.; Inagaki N.; Takahashi M.; Hiroi S.; Nakamura T.; Arimura T.; Nakamura K.; Ashizawa N.; Yasunami M.; Ohe T.; Yano K.; Kimura A. Functional analysis of titin/connectin N2-B mutations found in cardiomyopathy. J. Muscle Res. Cell Motil. 2006, 26 (6–8), 367–74. 10.1007/s10974-005-9018-5. [DOI] [PubMed] [Google Scholar]
- LeWinter M. M.; Granzier H. L. Cardiac titin and heart disease. J. Cardiovasc. Pharmacol. 2014, 63 (3), 207–12. 10.1097/FJC.0000000000000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Carrion-Vazquez M.; Oberhauser A. F.; Marszalek P. E.; Fernandez J. M. Point mutations alter the mechanical stability of immunoglobulin modules. Nat. Struct. Biol. 2000, 7 (12), 1117–20. 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- Taylor M.; Graw S.; Sinagra G.; Barnes C.; Slavov D.; Brun F.; Pinamonti B.; Salcedo E. E.; Sauer W.; Pyxaras S.; Anderson B.; Simon B.; Bogomolovas J.; Labeit S.; Granzier H.; Mestroni L. Genetic variation in titin in arrhythmogenic right ventricular cardiomyopathy-overlap syndromes. Circulation 2011, 124 (8), 876–85. 10.1161/CIRCULATIONAHA.110.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerull B.; Atherton J.; Geupel A.; Sasse-Klaassen S.; Heuser A.; Frenneaux M.; McNabb M.; Granzier H.; Labeit S.; Thierfelder L. Identification of a novel frameshift mutation in the giant muscle filament titin in a large Australian family with dilated cardiomyopathy. J. Mol. Med. (Heidelberg, Ger.) 2006, 84 (6), 478–83. 10.1007/s00109-006-0060-6. [DOI] [PubMed] [Google Scholar]
- Gerull B.; Gramlich M.; Atherton J.; McNabb M.; Trombitas K.; Sasse-Klaassen S.; Seidman J. G.; Seidman C.; Granzier H.; Labeit S.; Frenneaux M.; Thierfelder L. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 2002, 30 (2), 201–4. 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- Satoh M.; Takahashi M.; Sakamoto T.; Hiroe M.; Marumo F.; Kimura A. Structural analysis of the titin gene in hypertrophic cardiomyopathy: identification of a novel disease gene. Biochem. Biophys. Res. Commun. 1999, 262 (2), 411–7. 10.1006/bbrc.1999.1221. [DOI] [PubMed] [Google Scholar]
- Begay R. L.; Graw S.; Sinagra G.; Merlo M.; Slavov D.; Gowan K.; Jones K. L.; Barbati G.; Spezzacatene A.; Brun F.; Di Lenarda A.; Smith J. E.; Granzier H. L.; Mestroni L.; Taylor M.; Familial Cardiomyopathy R. Role of Titin Missense Variants in Dilated Cardiomyopathy. J. Am. Heart Assoc. 2015, 4 (11), e002645. 10.1161/JAHA.115.002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauveau C.; Rowell J.; Ferreiro A. A rising titan: TTN review and mutation update. Hum. Mutat. 2014, 35 (9), 1046–59. 10.1002/humu.22611. [DOI] [PubMed] [Google Scholar]
- Herman D. S.; Lam L.; Taylor M. R.; Wang L.; Teekakirikul P.; Christodoulou D.; Conner L.; DePalma S. R.; McDonough B.; Sparks E.; Teodorescu D. L.; Cirino A. L.; Banner N. R.; Pennell D. J.; Graw S.; Merlo M.; Di Lenarda A.; Sinagra G.; Bos J. M.; Ackerman M. J.; Mitchell R. N.; Murry C. E.; Lakdawala N. K.; Ho C. Y.; Barton P. J.; Cook S. A.; Mestroni L.; Seidman J. G.; Seidman C. E. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 2012, 366 (7), 619–28. 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansweijer J. A.; Nieuwhof K.; Russo F.; Hoorntje E. T.; Jongbloed J. D.; Lekanne Deprez R. H.; Postma A. V.; Bronk M.; van Rijsingen I. A.; de Haij S.; Biagini E.; van Haelst P. L.; van Wijngaarden J.; van den Berg M. P.; Wilde A. A.; Mannens M. M.; de Boer R. A.; van Spaendonck-Zwarts K. Y.; van Tintelen J. P.; Pinto Y. M. Truncating titin mutations are associated with a mild and treatable form of dilated cardiomyopathy. Eur. J. Heart Failure 2017, 19 (4), 512–21. 10.1002/ejhf.673. [DOI] [PubMed] [Google Scholar]
- Williams P. M.; Fowler S. B.; Best R. B.; Luis Toca-Herrera J.; Scott K. A.; Steward A.; Clarke J. Hidden complexity in the mechanical properties of titin. Nature 2003, 422 (6930), 446–9. 10.1038/nature01517. [DOI] [PubMed] [Google Scholar]
- Anderson B. R.; Bogomolovas J.; Labeit S.; Granzier H. Single molecule force spectroscopy on titin implicates immunoglobulin domain stability as a cardiac disease mechanism. J. Biol. Chem. 2013, 288 (8), 5303–15. 10.1074/jbc.M112.401372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolovas J.; Fleming J. R.; Anderson B. R.; Williams R.; Lange S.; Simon B.; Khan M. M.; Rudolf R.; Franke B.; Bullard B.; Rigden D. J.; Granzier H.; Labeit S.; Mayans O. Exploration of pathomechanisms triggered by a single-nucleotide polymorphism in titin’s I-band: the cardiomyopathy-linked mutation T2580I. Open Biol. 2016, 6 (9), 160114. 10.1098/rsob.160114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion-Vazquez M.; Oberhauser A. F.; Fowler S. B.; Marszalek P. E.; Broedel S. E.; Clarke J.; Fernandez J. M. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. U. S. A. 1999, 96 (7), 3694–9. 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.; Li H. Polyprotein of GB1 is an ideal artificial elastomeric protein. Nat. Mater. 2007, 6 (2), 109–14. 10.1038/nmat1825. [DOI] [PubMed] [Google Scholar]
- Bell G. I. Models for the specific adhesion of cells to cells. Science 1978, 200 (4342), 618–27. 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Evans E. Probing the relation between force--lifetime--and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 105–28. 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- Amodeo P.; Fraternali F.; Lesk A. M.; Pastore A. Modularity and homology: modelling of the titin type I modules and their interfaces. J. Mol. Biol. 2001, 311 (2), 283–96. 10.1006/jmbi.2001.4797. [DOI] [PubMed] [Google Scholar]
- Improta S.; Krueger J. K.; Gautel M.; Atkinson R. A.; Lefevre J. F.; Moulton S.; Trewhella J.; Pastore A. The assembly of immunoglobulin-like modules in titin: implications for muscle elasticity. J. Mol. Biol. 1998, 284 (3), 761–77. 10.1006/jmbi.1998.2028. [DOI] [PubMed] [Google Scholar]
- Improta S.; Politou A. S.; Pastore A. Immunoglobulin-like modules from titin I-band: extensible components of muscle elasticity. Structure 1996, 4 (3), 323–37. 10.1016/S0969-2126(96)00036-6. [DOI] [PubMed] [Google Scholar]
- Mayans O.; Wuerges J.; Canela S.; Gautel M.; Wilmanns M. Structural evidence for a possible role of reversible disulphide bridge formation in the elasticity of the muscle protein titin. Structure 2001, 9 (4), 331–40. 10.1016/S0969-2126(01)00591-3. [DOI] [PubMed] [Google Scholar]
- Waterhouse A.; Bertoni M.; Bienert S.; Studer G.; Tauriello G.; Gumienny R.; Heer F. T.; de Beer T. A. P.; Rempfer C.; Bordoli L.; Lepore R.; Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46 (W1), W296–W303. 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ΔGN–U reported here for wt I94 is smaller than that in ref (44). The difference is likely due to the difference in the experimental conditions. The measurements in ref (44) were performed in the presence of reducing agents, and our work was performed in the absence of reducing agents. It is likely that the two cysteines in I94 were oxidized into a disulfide bond by air oxygen in this work.
- Myers J. K.; Pace C. N.; Scholtz J. M. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995, 4 (10), 2138–48. 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl S. W.; Nash M. A.; Fried D. B.; Slutzki M.; Barak Y.; Bayer E. A.; Gaub H. E. Single-molecule dissection of the high-affinity cohesin-dockerin complex. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (50), 20431–6. 10.1073/pnas.1211929109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten M.; Ott W.; Jobst M. A.; Milles L. F.; Verdorfer T.; Pippig D. A.; Nash M. A.; Gaub H. E. From genes to protein mechanics on a chip. Nat. Methods 2014, 11 (11), 1127–30. 10.1038/nmeth.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. A.; Steward A.; Fowler S. B.; Clarke J. Titin; a multidomain protein that behaves as the sum of its parts. J. Mol. Biol. 2002, 315 (4), 819–29. 10.1006/jmbi.2001.5260. [DOI] [PubMed] [Google Scholar]
- Suay-Corredera C.; Pricolo M. R.; Velazquez-Carreras D.; Pathak D.; Nandwani N.; Pimenta-Lopes C.; Sanchez-Ortiz D.; Urrutia-Irazabal I.; Vilches S.; Dominguez F.; Frisso G.; Monserrat L.; Garcia-Pavia P.; de Sancho D.; Spudich J. A.; Ruppel K. M.; Herrero-Galan E.; Alegre-Cebollada J. Nanomechanical Phenotypes in Cardiac Myosin-Binding Protein C Mutants That Cause Hypertrophic Cardiomyopathy. ACS Nano 2021, 15 (6), 10203–16. 10.1021/acsnano.1c02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms A. S.; Thompson A. D.; Glazier A. A.; Hafeez N.; Kabani S.; Rodriguez J.; Yob J. M.; Woolcock H.; Mazzarotto F.; Lakdawala N. K.; Wittekind S. G.; Pereira A. C.; Jacoby D. L.; Colan S. D.; Ashley E. A.; Saberi S.; Ware J. S.; Ingles J.; Semsarian C.; Michels M.; Olivotto I.; Ho C. Y.; Day S. M. Spatial and Functional Distribution of MYBPC3 Pathogenic Variants and Clinical Outcomes in Patients With Hypertrophic Cardiomyopathy. Circ Genom Precis Med. 2020, 13 (5), 396–405. 10.1161/CIRCGEN.120.002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.