SUMMARY
Among the most prevalent Mycobacterium tuberculosis (Mtb) strains worldwide is the Beijing genotype, which has caused large outbreaks of tuberculosis (TB). Characteristics facilitating the dissemination of Beijing family strains remain unknown, but they are presumed to have been acquired through evolution of the lineage. To explore the genetic diversity of the Beijing family Mtb and explore the discriminatory ability of mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) loci in several regions of East Asia, a cross-sectional study was conducted with a total of 163 Beijing strains collected from registered TB patients between 1 June 2009 and 31 November 2010 in Funing County, China. The isolated strains were analysed by 15-MIRU-VNTR loci typing and compared with published MIRU-VNTR profiles of Beijing strains. Synonymous single nucleotide polymorphisms at 10 chromosomal positions were also analysed. The combination of SNP and MIRU-VNTR typing may be used to assess Mtb genotypes in areas dominated by Beijing strains. The modern subfamily in Shanghai overlapped with strains from other countries, whereas the ancient subfamily was genetically differentiated across several countries. Modern subfamilies, especially ST10, were prevalent. Qub11b and four other loci (MIRU 26, Mtub21, Qub26, Mtub04) could be used to discriminate Beijing strains.
Key words: Beijing family, MIRU, Mycobacterium tuberculosis, phylogeny, SNP
INTRODUCTION
Tuberculosis (TB) remains a major infectious and deadly disease in many parts of the world. Among the most prevalent Mycobacterium tuberculosis (Mtb) strains worldwide is the Beijing genotype, which has caused large outbreaks of TB in many countries [1–3] and been highly associated with drug resistance [4]. Currently, Beijing strains are present in >10% of patients with TB worldwide and are highly endemic throughout much of East and South East Asia, where they are present in >50% of TB patients [5, 6]. Mtb strains belonging to the Beijing genotype might have a selective advantage over other Mtb strains and be associated with hypervirulence and multidrug resistance [7–9].
Characteristics facilitating the dissemination of Beijing family strains remain unknown, but they are presumed to have been acquired through evolution of the lineage. Therefore, it is necessary to investigate the phylogeny of the sublineage and to elucidate the prevalence of these characteristics. The mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) may be phylogenetically informative for the Mtb Beijing family [10]. Phylogenetic application of MIRU-VNTR profiles may be assessed by a graphing method known as the minimum spanning tree (MST) [11–13]. The results of MIRU-VNTR genotyping exhibited concordance with the results of other genotyping techniques [14], including spoligotype clade assignment [6] and single nucleotide polymorphism (SNP) analyses [15, 16] in Mtb. Sublineages of the Beijing family could be classified using 10 synonymous SNPs [17, 18]. These SNPs were detected by comparison among the whole-genome sequences, including strain 210, a Beijing family strain belonging to the modern sublineage [15, 19].
The Beijing family was originally reported to be a genetically closely related genotype family in 1995 [20]. Later studies showed that these strains were prevalent in East Asia [21], the Former Soviet Union [22], and South Africa [23]. Whole genome analyses have indicated that the evolution of Mtb paralleled human migration and that Mtb expanded due to increases in human population density [24]. Several epidemiological studies have also indicated that Mtb genotype distribution is highly related to geography, human population and ethnicity [25–27].
This study utilized MIRU-VNTR genotyping and SNPs to analyse the genetic population structure and diversity of the Beijing family. These results were subsequently compared to published profiles of Beijing family Mtb strains isolated from patients in neighbouring geographical areas (e.g. other areas of China and Japan) to assess potential genetic linkages, which refer to phylogenetic relatedness. These analyses may promote a better understanding of the population genetic structure of the Beijing family and extend the information obtained from the MIRU-VNTR genotyping method.
MATERIALS AND METHODS
Study samples
A cross-sectional study was conducted with a total of 163 Beijing strains collected from registered TB patients between 1 June 2009 and 31 November 2010 in Funing County, China, a relatively closed and low socioeconomic county in eastern China with a population of 1·08 million, to explore the genetic diversity of the Beijing family in several regions of East Asia. After obtaining fresh isolates, all these cultures were stored and maintained at –80 °C until required at the molecular laboratory of the School of Public Health, Fudan University. The Beijing family strains assessed in this study were identified on the basis of an RD105 deletion. Further characterization of the modern and ancestral sublineages was performed following the presence of an RD181 deletion, as determined by polymerase chain reaction amplification, which was used to subclassify all of the Beijing family strains in our collection as ancient [RD181(+)] and modern [RD181(–)] types.
The 199 Mtb isolates obtained from the study site consisted of 182 Beijing family strains; of these, 19 were excluded because of insufficient of DNA samples. The Ethics Committee of the School of Public Health of Fudan University approved this study. All participants provided written informed consent to allow their information to be stored and used for research.
SNP typing
Sequence types (STs) were determined based on the 10 synonymous SNPs, which were sufficient to classify Beijing strains obtained globally [15]. The chromosomal positions in the whole-genome sequence of H37Rv [28] were 797736, 909166, 1477596, 1548149, 1692069, 1892017, 2376135, 2532616, 2825581, and 4137829. STs were designated as described previously [15, 29].
15-locus MIRU-VNTR
The 15 classical MIRU-VNTR loci were selected and individually amplified in all 163 Beijing isolates as described previously [14, 30]. The resulting typing pattern was used to create a 15-digit allelic profile for each isolate. The discrimination ability of the locus combinations was calculated using the Hunter–Gaston discriminatory index (HGDI) [31].
Data retrieved from previous reports
We searched the studies in which the strains were collected from venues in East Asia between 1 January 2000 and 30 December 2010; literature with the absence of ST or 15-MIRU-VNTR data were excluded from the analysis. ST data were retrieved for isolates obtained from patients in Taiwan [32] and Japan [18] to compare population structures. In addition, 15-MIRU-VNTR data and subfamily classifications were retrieved for isolates obtained from patients in Shanghai [33], Taiwan [32], and Japan [18]. Their origins and the years during which the isolates were obtained are as follows: 191 isolates from Shanghai, collected between 2007 and 2010 in the Chongming District [33]; 338 isolates from Taiwan, collected between 2003 and 2007 from four geographical regions of Taiwan (north, east, central, south) [32]; 355 isolates from Japan, collected between 2001 and 2004 in Osaka and Kobe [18]. The MIRU-VNTR data from Funing and these three other East Asian regions were compared by constructing a MST using Bionumerics software v. 7.1 (Applied Maths, Belgium).
RESULTS
Classification of Beijing strains by SNP typing
The 163 Beijing strains could be classified into 13 STs (Fig. 1), with most of the strains classified into four major Beijing sublineages: ST10 (53·4%), ST19 (11·7%), ST22 (10·4%) and ST11 (10·4%). Based on 10-loci SNP typing, the sequence of ST11 was consistent with the standard reference strain H37Rv, suggesting a close genetic distance. By contrast, ST22 and ST10 had many mutations relative to H37Rv, indicating a greater genetic distance. These findings suggest that the order of appearance of the STs in phylogenetic evolution was ST11, ST26, ST3, STK, ST19, ST10 and ST22.
Fig. 1.
SNP genotyping results of 163 Mtb strains of Beijing genotype in Funing County, China. Alleles indicate SNP positions of H37Rv: 909166, 797736, 2825581, 1892017, 4137829, 1477596, 2532616, 2376135, 1692069, and 1548149.
Comparison of ST profiles from Funing, Taiwan, and Japan
To better understand the extent of geographical variations in STs of Mtb isolates, the ST profiles of Beijing strains from the present study were compared with profiles from strains isolated in Taiwan and Japan. The 856 strains isolated in these three studies included 403 ancient and 453 modern Beijing isolates (Table 1). Most of the Beijing genotypes from Funing (78·5%) and Taiwan (73·4%) belonged to the modern subfamily, whereas most of the strains from Japan (78·9%) were of the ancient subfamily. The proportion of strains classified as ST10 was significantly higher in Funing (53·4%) and Taiwan (53·3%) than in Japan (17·2%). Of the strains isolated from Funing and Taiwan, 75·5% and 82·6%, respectively, belonged to three major Beijing sublineages, ST10, ST19 and ST22. By contrast, 68·4% of strains from Japan belonged to the sublineages ST3, ST19 and STK.
Table 1.
Sequence types of Mycobacterium tuberculosis isolates from Funing, Taiwan and Japan
| Beijing sublineage | No. (%) of strains | ||
|---|---|---|---|
| Funing | Taiwan | Japan | |
| ST11 | 17 (10·4) | 4 (1·2) | 4 (1·1) |
| ST3 | 4 (2·5) | 13 (3·9) | 84 (22·7) |
| ST26 | 9 (5·5) | 27 (8·0) | 28 (7·9) |
| ST19 | 19 (11·7) | 50 (14·8) | 111 (31·3) |
| ST25 | 0 (0·0) | 7 (2·1) | 2 (0·6) |
| ST10 | 87 (53·4) | 180 (53·3) | 61 (17·2) |
| ST22 | 17 (10·4) | 49 (14·5) | 14 (3·94) |
| STK | 1 (0·6) | 4 (1·2) | 51 (14·4) |
| NEW | 9 (5·5) | 0 (0·0) | 0 (0·0) |
| STN | 0 (0·0) | 4 (1·2) | 0 (0·0) |
| Ancient* | 35 (21·6) | 88 (26·0) | 280 (78·9) |
| Modern | 128 (78·5) | 250 (73·4) | 75 (21·1) |
| Total | 163 | 338 | 355 |
* The proportions of ancient and modern Beijing strains were calculated by determining the presence of the RD181 deletion in strains from Funing and an IS6110 insertion in the NTF chromosomal region in strains from Taiwan and Japan.
Comparison of MIRU-VNTR from Funing, Shanghai, Taiwan and Japan
To determine whether certain MIRU-VNTR loci could be used to discriminate between Beijing strains, we analysed published MIRU-VNTR profiles and calculated the HGDI of Beijing strains from three other East Asian regions – Shanghai, Taiwan and Japan (Table 2). We found that the HGDI of Qub11b was >0·6 across the four areas, while the HGDIs of MIRU26, Mtub21, Qub26, and Mtub04 were each >0·3 in all four regions.
Table 2.
HGDI values of Mycobacterium tuberculosis isolates from Funing, Shanghai, Taiwan and Japan
| MIRU-VNTR locus | Funing | Shanghai | Taiwan | Japan |
|---|---|---|---|---|
| ETR C | 0·061 | 0·142 | 0·097 | 0·028 |
| MIRU4 | 0·130 | 0·072 | 0·047 | 0·05 |
| ETR A | 0·204 | 0·178 | 0·220 | 0·145 |
| Mtub30 | 0·266 | 0·086 | 0·107 | 0·317 |
| Mtub39 | 0·325 | 0·180 | 0·207 | 0·185 |
| Qub4156 | 0·326 | 0·550 | 0·554 | 0·622 |
| MIRU16 | 0·340 | 0·153 | 0·086 | 0·307 |
| MIRU40 | 0·390 | 0·244 | 0·224 | 0·269 |
| Mtub04 | 0·483 | 0·333 | 0·651 | 0·474 |
| Qub26 | 0·590 | 0·633 | 0·566 | 0·675 |
| Mtub21 | 0·592 | 0·582 | 0·565 | 0·459 |
| MIRU31 | 0·650 | 0·168 | 0·587 | 0·296 |
| Qub11b | 0·663 | 0·662 | 0·710 | 0·767 |
| MIRU10 | 0·684 | 0·152 | 0·325 | 0·436 |
| MIRU26 | 0·744 | 0·539 | 0·456 | 0·341 |
HGDI, Hunter–Gaston discriminatory index.
MST based on 15-MIRU-VNTR genotyping and ST profiles of Beijing family isolates from Funing
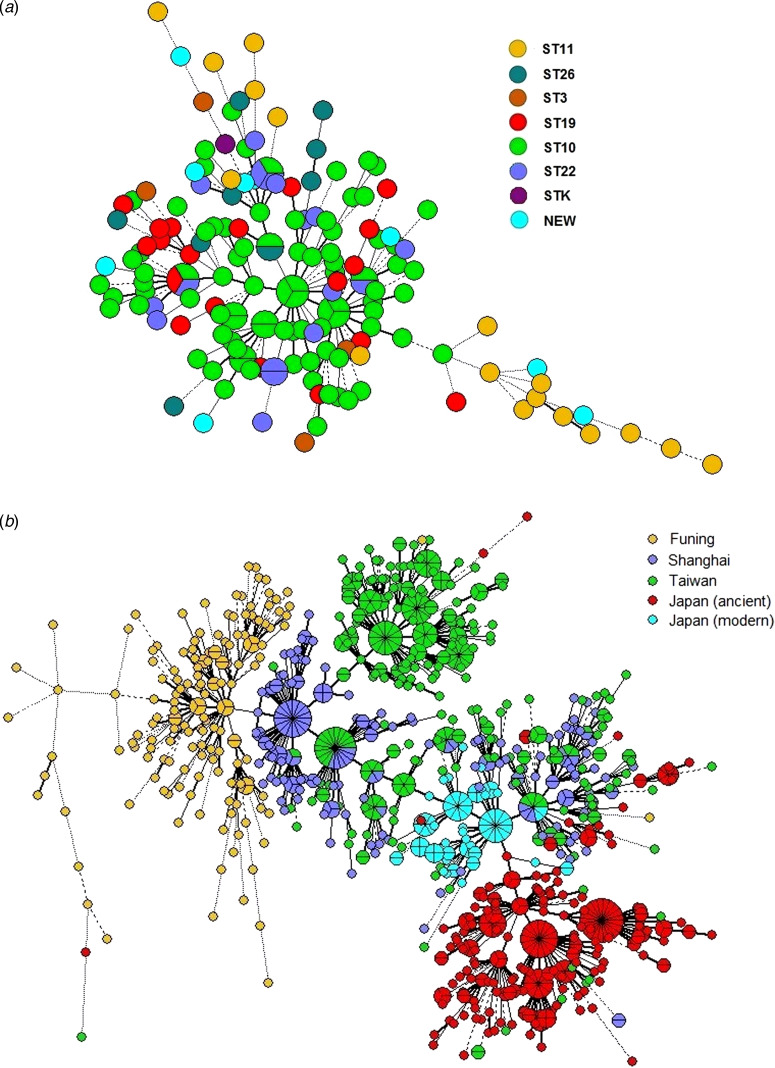
To determine the consistency between MIRU-VNTR genotypes and STs classified by SNP, we constructed a MST based on 15-MIRU-VNTR genotyping and identified the SNP information by colour coding (Fig. 2a). The isolates ST10 and ST22 almost overlapped in the tree, indicating their close genetic relationship. By contrast, ST11 and ST26 were positioned away from the major MIRU-VNTR genotypes and had greater genetic distances to ST10. The entire structure of the MST was consistent with the sublineages defined by SNP typing. However, some of the MIRU-VNTR clusters contained multiple SNP subtypes, suggesting that there was homoplasy within the Beijing strains examined with 15-MIRU-VNTR.
Fig. 2.
Minimum spanning trees based on 15-MIRU-VNTR genotyping of (a) 163 Mtb strains of Beijing genotype in Funing, and (b) Mtb isolates from Funing, Shanghai, Taiwan and Japan. Each circle indicates a different MIRU-VNTR genotype, with the size of the circle indicating the quantity of the isolate. Heavy lines connecting two MIRU-VNTR types denote single-locus variants; thin lines denote double-locus variants; dashed lines indicate triple-locus variants; and dotted lines indicate variants at more than three loci. The coloured symbols in (a) denote different sequence types of Mtb isolates, whereas the coloured symbols in (b) denote Mtb isolates from Funing, Shanghai, Taiwan and Japan.
Phylogenetic analysis of Beijing family isolates from Funing, Shanghai, Taiwan and Japan
Figure 2b shows the genotype distributions in Funing, Shanghai, Taiwan and Japan. The genetic distance of strains from Funing and Shanghai was relatively close, whereas the distances of strains from Funing compared to strains from Taiwan and Japan were longer. However, there were overlaps for genotypes from Shanghai, Taiwan and Japan. Specifically, strains from the modern subfamily in Japan overlapped with strains from Taiwan and Shanghai, whereas strains from the ancient subfamily were crowded together and located at some distance from the strains isolated in other regions.
DISCUSSION
To our knowledge, this study is the first to assess the genetic diversity of Mtb from different regions in East Asia. This study showed that the modern subfamily in Shanghai overlapped with strains from other countries, whereas the ancient subfamily was genetically differentiated across several countries. In addition, strains of the modern subfamily, especially ST10, showed especially wide distribution in populations in these East Asian countries. Qub11b, with an HGDI >0·6, and four other loci, MIRU 26, Mtub21, Qub26, and Mtub04, each with an HGDI >0·3, were able to be distinguished among Beijing strains in a given geographical region.
The Beijing strain is one of the predominant Mtb lineages in East Asia as well as in other countries throughout the world [34, 35]. Beijing strains can be divided into several sublineages that may differ in their ability to spread and cause disease. Methods to subdivide Beijing strains are important for exploration of their phylogenetic evolution and propagation. Although Mtb phylogenetic evolution was first assessed by 15-MIRU-VNTR and 24-MIRU-VNTR in China [36], these methods yielded uncertain results with relatively large errors [33]. Mtb gene sequencing showed the advantages and accuracy of SNP typing in phylogenetic analyses [37]. Although the discriminatory index of SNPs is not very high [38], SNP typing is regarded as an optimal method for phylogenetic research [39], allowing Beijing strains to be subdivided into different STs. ST11, ST26, ST3, STK, ST19 and ST25, each of which possesses intact RD181 and NTF regions, correspond to ancient sublineages; whereas ST10 and ST22 correspond to modern sublineages, with all possessing an RD181 region deletion and some having an IS6110 insertion on the right side of the NTF region [15, 17, 25, 40]. The order of appearance of the STs in phylogenetic evolution was consistent from previous studies, implying the direction of genetic mutations in Mtb [15].
This study used 15-MIRU-VNTR genotyping to construct an MST of Beijing strain, with this MST validated by profiling of SNPs. Each MIRU-VNTR genotype was roughly concordant with an ST classification. However, some strains belonging to different ST sublineages were found to be mixed and not to be classified correctly in the same MIRU-VNTR clusters. Other highly discriminatory loci are required to make MIRU-VNTR results agree better with SNP information. Due to the high homoplasy of Beijing strains, the MIRU-VNTR method is less than optimal in genotyping these strains. The MST suggested that combining SNP and MIRU-VNTR typing may be superior to either method alone in assessing Mtb epidemiology in areas dominated by Beijing strains.
This study found that strains of the modern subfamily, especially ST10, were highly prevalent. Interestingly, ST10 was also dominant in Taiwan (53·3%) [18] and Thailand (57·7%) [41], suggesting that ST10 may be epidemiologically successful, with Asian populations being especially susceptible to infection by, and transmission of, these strains. The genotypes of Beijing strains from Shanghai, Taiwan and Japan showed overlaps and relatively short genetic distances. The reason for the apparent global success of the Beijing strains is not yet fully understood, but may be related to a variety of host-related factors, including movements of human populations [22]. The modern Beijing sublineages are more predominant than the ancient sublineages throughout Asian countries, including South Korea, Vietnam, Malaysia, Thailand and China [1, 32, 41], whereas most of the Beijing strains in Japan belong to the ancient subfamily [42]. We found that strains of the modern subfamily in Japan overlapped with strains from Taiwan and Shanghai, while ancient subfamily strains were genetically unequal across countries. These findings suggest that the modern genotype has a greater ability to expand throughout East Asia, even around the world, compared to the ancient genotype. However, caution should be exercised when drawing conclusions because the sampling period and study population were different in the four studies. Interestingly, the phylogenetic analysis according to SNPs supported the monophyletic clade structure and suggests different Beijing STs across the different Asian sites. However, the order in which all of the above evolutionary events occurred remains to be determined, in particular we did not include an analysis of temporal evolution of the strains across the study sites within such a short period.
CONCLUSION
In conclusion, we found that Beijing strains of the modern subfamily, especially ST10, were highly prevalent. Qub11b, with an HGDI >0·6, and four other loci, MIRU 26, Mtub21, Qub26, and Mtub04, each with an HGDI >0·3, were able to discriminate among Beijing strains within a given geographical region. The combination of SNP and MIRU-VNTR typing was better than either alone for genotyping in areas dominated by Beijing strains.
ACKNOWLEDGEMENTS
We thank department of Tuberculosis Control, Yinzhou Center for Disease Control and Prevention, Ningbo, China for providing the samples. This study was funded by the Chinese National Science Foundation (30800937), the China-UK Global public health research programme (GHSP-CS-OP 302) and the programme of Key Discipline Development of Public Health of Shanghai (grant no. 12GWZX0101).
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Bifani PJ, et al. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends in Microbiology 2002; 10: 45–52. [DOI] [PubMed] [Google Scholar]
- 2.Glynn JR, et al. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerging Infectious Diseases 2002; 8: 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mokrousov I. Genetic geography of Mycobacterium tuberculosis Beijing genotype: a multifacet mirror of human history? Infection, Genetics and Evolution 2008; 8: 777–785. [DOI] [PubMed] [Google Scholar]
- 4.Singh J, et al. Genetic diversity and drug susceptibility profile of Mycobacterium tuberculosis isolated from different regions of India. Journal of Infection 2015; 71: 207–219. [DOI] [PubMed] [Google Scholar]
- 5.Brudey K, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiology 2006; 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filliol I, et al. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. Journal of Clinical Microbiology 2003; 41: 1963–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parwati I, van Crevel R, van Soolingen D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infectious Diseases 2010; 10: 103–111. [DOI] [PubMed] [Google Scholar]
- 8.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infectious Diseases 2007; 7: 328–337. [DOI] [PubMed] [Google Scholar]
- 9.Anh DD, et al. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerging Infectious Diseases 2000; 6: 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang JR, et al. Genotypic analysis of genes associated with transmission and drug resistance in the Beijing lineage of Mycobacterium tuberculosis. Clinical Microbiology and Infection 2011; 17: 1391–1396. [DOI] [PubMed] [Google Scholar]
- 11.Schouls LM, et al. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. Journal of Bacteriology 2004; 186: 5496–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zozio T, et al. Genotyping of Mycobacterium tuberculosis clinical isolates in two cities of Turkey: description of a new family of genotypes that is phylogeographically specific for Asia Minor. BMC Microbiology 2005; 5: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boxrud D, et al. Comparison of multiple-locus variable-number tandem repeat analysis, pulsed-field gel electrophoresis, and phage typing for subtype analysis of Salmonella enterica serotype Enteritidis. Journal of Clinical Microbiology 2007; 45: 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Supply P, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. Journal of Clinical Microbiology 2006; 44: 4498–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filliol I, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. Journal of Bacteriology 2006; 188: 759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutacker MM, et al. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. Journal of Infectious Diseases 2006; 193: 121–128. [DOI] [PubMed] [Google Scholar]
- 17.Iwamoto T, et al. Population structure analysis of the Mycobacterium tuberculosis Beijing family indicates an association between certain sublineages and multidrug resistance. Antimicrobial Agents and Chemotherapy 2008; 52: 3805–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wada T, Iwamoto T, Maeda S. Genetic diversity of the Mycobacterium tuberculosis Beijing family in East Asia revealed through refined population structure analysis. FEMS Microbiology Letters 2009; 291: 35–43. [DOI] [PubMed] [Google Scholar]
- 19.Beggs ML, Eisenach KD, Cave MD. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. Journal of Clinical Microbiology 2000; 38: 2923–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Soolingen D, et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. Journal of Clinical Microbiology 1995; 33: 3234–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, et al. Genotypes and characteristics of clustering and drug susceptibility of Mycobacterium tuberculosis isolates collected in Heilongjiang Province, China. Journal of Clinical Microbiology 2011; 49: 1354–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mokrousov I, et al. Origin and primary dispersal of the Mycobacterium tuberculosis Beijing genotype: clues from human phylogeography. Genome Research 2005; 15: 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warren RM, et al. Patients with active tuberculosis often have different strains in the same sputum specimen. American Journal of Respiratory and Critical Care Medicine 169: 610–614. [DOI] [PubMed] [Google Scholar]
- 24.Comas I, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nature genetics 2013; 45: 1176–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dou HY, et al. Associations of Mycobacterium tuberculosis genotypes with different ethnic and migratory populations in Taiwan. Infection, Genetics and Evolution 2008; 8: 323–330. [DOI] [PubMed] [Google Scholar]
- 26.Gagneux S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences USA 2006; 103: 2869–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dou HY, Huang SC, Su IJ. Prevalence of Mycobacterium tuberculosis in Taiwan: a model for strain evolution linked to population migration. International Journal of Evolutionary Biology 2011; 2011: 937434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole ST, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998; 393: 537–544. [DOI] [PubMed] [Google Scholar]
- 29.Iwamoto T, et al. Population structure dynamics of Mycobacterium tuberculosis Beijing strains during past decades in Japan. Journal of Clinical Microbiology 2009; 47: 3340–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Supply P, et al. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Molecular Microbiology 2000; 36: 762–771. [DOI] [PubMed] [Google Scholar]
- 31.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. Journal of Clinical Microbiology 1988; 26: 2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen YY, et al. Genetic diversity of the Mycobacterium tuberculosis Beijing family based on SNP and VNTR typing profiles in Asian countries. PLoS ONE 2012; 7: e39792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo T, et al. Combination of single nucleotide polymorphism and variable-number tandem repeats for genotyping a homogenous population of Mycobacterium tuberculosis Beijing strains in China. Journal of Clinical Microbiology 2012; 50: 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanekom M, et al. Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis 2011; 91: 510–523. [DOI] [PubMed] [Google Scholar]
- 35.Schurch AC, et al. SNP/RD typing of Mycobacterium tuberculosis Beijing strains reveals local and worldwide disseminated clonal complexes. PLoS ONE 2011; 6: e28365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allix-Beguec C, et al. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. Journal of Clinical Microbiology 2008; 46: 2692–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dan-Dan D, Qian G. Mycobacterium tuberculosis genetic diversity and differential virulence. Journal of Microbes and Infections 2010; 5: 106–110. [Google Scholar]
- 38.Cardoso Oelemann M, et al. The forest behind the tree: phylogenetic exploration of a dominant Mycobacterium tuberculosis strain lineage from a high tuberculosis burden country. PLoS ONE 2011; 6: e18256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Comas I, et al. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS ONE 2009; 4: e7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dou HY, et al. Molecular epidemiology and evolutionary genetics of Mycobacterium tuberculosis in Taipei. BMC Infectious Diseases 2008; 8: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faksri K, et al. Genetic diversity of the Mycobacterium tuberculosis Beijing family based on IS6110, SNP, LSP and VNTR profiles from Thailand. Infection, Genetics and Evolution 2011; 11: 1142–1149. [DOI] [PubMed] [Google Scholar]
- 42.Wada T, Iwamoto T. Allelic diversity of variable number of tandem repeats provides phylogenetic clues regarding the Mycobacterium tuberculosis Beijing family. Infection, Genetics and Evolution 2009; 9: 921–926. [DOI] [PubMed] [Google Scholar]