Abstract
The development and progression of solid tumors is dependent on cancer cell autonomous drivers and the tumor microenvironment (TME). Cancer-associated fibroblasts (CAFs) in the TME possess both tumor-promoting and tumor-restraining functions. In the current study, we interrogated the role of αSMA+ CAFs in a genetic mouse model of metastatic colorectal cancer (CRC). Selective depletion of αSMA+ CAFs resulted in increased tumor invasiveness, lymph node metastasis, and reduced overall survival. Depletion of αSMA+ CAFs reduced BMP4 and increased TGFβ1 secretion from stromal cells, and was associated with increased Lgr5+ cancer stem-like cells (CSCs) and the generation of an immunosuppressive TME with increased frequency of Foxp3+ regulatory T cells and suppression of CD8+ T cells. This study demonstrates that αSMA+ CAFs in CRC exert tumor-restraining functions via BMP4/TGFβ1 paracrine signaling that serves to suppress Lgr5+ CSCs and promote anti-tumor immunity, ultimately limiting CRC progression.
Introduction
The development and progression of solid tumors is dependent on oncogenic events in cancer cells and signaling in an evolving tumor microenvironment (TME). Activated fibroblasts, hereafter referred to as cancer-associated fibroblasts (CAFs), are an abundant cell type in the TME with a complex secretome, including extracellular matrix proteins (e.g., collagens), matrix metalloproteinases, cytokines, and chemokines [1, 2]. Although heterogeneous, CAFs can be identified by their expression of several cardinal gene products, including α-smooth muscle actin (αSMA), fibroblast activation protein, and platelet-derived growth factor receptor-β, among others [3, 4]. αSMA is commonly used to identify myofibroblasts [5] and this marker captures a significant fraction of CAFs in various solid tumor types including pancreatic, breast, and colorectal cancers (CRCs) [1, 3, 5, 6]. Although our previous work in breast and pancreatic cancer demonstrated αSMA+ CAFs as tumor-restraining cells [7, 8], the diversity of CAFs across distinct tumor types has underscored their distinct biological mechanisms [1-3, 9]. The biological complexity αSMA+ CAFs prompted us to explore their roles in CRC.
CRC is one of the most common malignancies in the United States, in both men and women [10]. CRC results from the progression of adenoma to adenocarcinoma and is associated with the accumulation of oncogenic events. In familial adenomatous polyposis and most sporadic cases of CRC, deactivation of the tumor suppressor adenomatous polyposis coli (APC) initiates transformation, followed by mutation in KRAS and loss of the tumor suppressor TP53 [11]. CRC has been classified into different subtypes based on gene expression profiles and response to treatments [12-17]. These studies have identified a correlation between the most aggressive tumor subtype, consensus molecular subtype 4 (CMS4), and strong stromal presence, suggesting that high stromal content confers a worse prognosis for CRC patients. Other studies have specifically correlated a higher presence of CAFs with worse outcomes [18, 19]. In an effort to characterize the CRC microenvironment, singlecell RNA sequencing of the mouse intestinal mesenchyme identified four distinct fibroblast populations, only one of which expressed αSMA [20]. Collectively, these studies highlight CAF heterogeneity in CRC, albeit with very limited insight on its functions in CRC progression. Here we employed well-characterized genetic tools [7, 8, 21] to define the critical role of αSMA+ CAFs in a genetic mouse model of metastatic CRC, which recapitulates the evolution of human disease [22].
Materials and methods
Laboratory mice
All mice used in these studies were of C57BL/6 background. The Villin-Cre-ERT2; Apcflox/flox; Trp53flox/flox; tetO-LSL-KrasG12D; Rosa26-LSL-Luc; Rosa26-LSL-rtTA-GFP or iKAP mice were previously published [22] and were a gift from Dr. Ronald DePinho (MD Anderson Cancer Center). iKAP mice were crossed with αSMA-TK [21] (Jackson Laboratory Stock 029921) and αSMA-RFP [23] (Jackson Laboratory Stock 031159). For tumor induction, mice were given an intrarectal 4-OH tamoxifen injection (50 mg/kg) and were placed on doxycycline diet (1.5 g/kg). Tumors were determined by endoscopy at week 4 post-tumor induction. Briefly, anesthesia was administered through a nose cone, with regulated flow of isoflurane vapor (1–2%). The colon was then flushed with a warm phosphate buffered saline (PBS) enema (10 ml), proceeded by insufflation with air from a standard 10-gallon air pump to allow for endoscopy. Images were acquired with a Storz Veterinary endoscope. After confirmation of the presence of tumors, mice were given daily ganciclovir (GCV) intraperitoneal injections (50 mg/kg), to deplete αSMA+ myofibroblasts, until moribundancy for survival studies. Continuous GCV administration ensures a progressive depletion of proliferating αSMA+ cells by preventing their accumulation; this approach does not target non-proliferating αSMA+ cells. At endpoint, tissues were collected for further analysis. The number of tumors was determined by macroscopic evaluation. Colons were processed for paraffin or optimal cutting temperature compound embedding for subsequent histological staining. In some cases, tumors were dissected and cells dissociated for flow cytometry analysis as described below. Both male and female mice were used for analysis. Sample size was estimated based on an effect size of 1, α-error probability of 0.05, and power of 0.95. Randomization and blinding was not performed and no animals were excluded from analysis. All experiments were approved by the Institutional Animal Care and Use Committee at MD Anderson Cancer Center.
Immunohistochemistry and immunofluorescence
For histopathological and immunohistochemical (IHC) analyses, mouse colon and liver tissues were fixed in 10% neutral buffered formalin, dehydrated, processed in paraffin, and 5 μm sections were prepared by deparaffinization and rehydration. Antigen retrieval was performed in 10 mM sodium citrate buffer (pH 6.0) or TE buffer (pH 9.0, for BMP4 immunofluorescence and transforming growth factor-β1 (TGFβ1) staining) in a molecular pathology microwave. Following blocking, tissue sections were incubated with primary antibodies against αSMA (Dako #M0851, 1:200), Ki67 (Thermo #RM-9106, 1:400), collagen I (Southern Biotech #1310-01, 1:200), desmin (Abcam #ab15200, 1:100), β-catenin (Abcam #ab32572, 1:200), lymphatic vessel endothelial receptor 1 (LYVE-1) (Abcam #ab14917, 1:200), Zeb1 (Novus Biologicals #NBP1-05987, 1:100), E-cadherin (Cell Signaling #3195, 1:400), CD44 (Invitrogen #14-0441-81, 1:50), TGFβ1 (R&D Systems #MAB240, 1:200), BMP4 (Allele Biotech #ABP-PAB-10444, 1:50 for IHC), and phospho-Akt (Ser473, Abcam #ab81283, 1:100) overnight at 4 °C, followed by incubation with biotin-conjugated secondary antibodies and ABC reagent (Vector #PK-6100). The sections were then developed using DAB staining (Thermo #750118 and Thermo #34065 for desmin-stained sections) according to the manufacturer’s instructions. For αSMA and BMP4 costaining, slides were incubated with BMP4 antibody (GeneTex #GTX100874, 1:500), followed by incubation with rabbit-on-mouse horseradish peroxidase (HRP) polymer (BioCare Medical) and tyramide signal amplification (TSA) reagent (Opal 520, Akoya Biosciences). Antigen retrieval was repeated and slides incubated with αSMA antibody, mouse-on-mouse HRP polymer (BioCare Medical), and TSA reagent (Opal 570, Akoya Biosciences). A similar protocol was followed for CD45 and TGFβ1 costaining, where slides were incubated with CD45 antibody (Cell Signaling #70257, 1:100), followed by incubation with rabbit-on-mouse HRP polymer (BioCare Medical) and TSA reagent (Opal 520, Akoya Biosciences). Antigen retrieval was repeated and slides incubated with TGFβ1 antibody (R&D Systems #MAB240, 1:200), mouse-on-mouse HRP polymer (BioCare Medical), and TSA reagent (Opal 570, Akoya Biosciences). DNA was stained with Hoechst 33342 (Invitrogen #H3570) and slides mounted with Vectashield mounting media (Vector H-1000). Slides were imaged with a Zeiss LSM800 confocal microscope. Sirius red staining was done following the manufacturer’s protocol (Sigma-Aldrich #365548). Immunostaining for DCLK1 was performed on frozen sections. Fixation was performed using acetone for 10 min at 4 °C, proceeded by blocking with 5% donkey serum in PBS with 0.1% Tween-20 at room temperature (RT) for 1 h. Sections were then incubated with anti-DCLK1 antibody (Cell Signaling #62257, 1:200) overnight at 4 °C, followed by incubation with fluorescently labeled secondary antibodies for 1 h at RT. Slides were mounted in medium containing DAPI (Vector #H-1200). Quantification of staining was done with the NIH Image J Analysis Software, using the same threshold for all images being compared. For each staining, three to five visual fields per slide were used in quantification. The sample number is indicated in the figure legend. For staining of cultured cells, primary CAFs were seeded on collagen-coated glass coverslips in 24-well plates. After overnight culture, the cells were fixed with −20 °C methanol, blocked with 5% goat serum, and incubated with primary antibodies against αSMA (Dako #M0851, 1:100), vimentin (Cell Signaling #5741, 1:100), and FSP-1/S100A4 (Dako #A5114, 1:100) for 2 h at RT. Following incubation with fluorescently labeled secondary antibodies (ThermoFisher #A11008 (Alexa Fluor 488) and #A11004 (Alexa Fluor 568), 1:500) for 1 h at RT, the DNA was stained with Hoechst 33342 (Invitrogen #H3570) following the manufacturer’s instructions. The coverslips were mounted on slides with Vectashield mounting medium (Vector H-1000) and staining was visualized on a Zeiss Axio Observer Z1 fluorescent microscope.
RNA-sequencing analysis
Total RNA was isolated from control (n = 3) and depleted (n = 3) endpoint tumors, as well as from the colon epithelium of wild-type (WT) mice (n = 3), and submitted to the Advanced Genomics Core Facility at MD Anderson Cancer Center. For RNA-sequencing analyses, Cutadapt (Version 2.8) [24, 25] was used for aligning raw reads to mouse genome (build mm10) and RSEM (version 1.3.1) (DOI: 10.1186/1471-2105-12-323) was used to quantify the transcript counts for each gene. R Package, DESeq2 (version 1.28.1) [26] was used to perform differential expression analyses for the gene count data (p < 0.05, fold-change > 1.5). Pathway analyses of differentially expressed genes (adjusted p < 0.05, fold-change > 2) were conducted by WebGestalt 2019 (DOI: 10.1093/nar/gkz401). Significantly enriched pathways (false discovery rate (FDR) < 0.05) were selected. Gene set enrichment analysis (GSEA) was performed based on differentially expressed genes (FDR q-value < 0.1) in GSEA 4.1.0 using the Hallmarks gene sets database. Additional raw and processed data have been uploaded to the Gene Expression Omnibus database (accession number GSE164300).
Co-culture of iKAP cancer cells and CAFs
iKAP cancer cells were isolated as previously described [22] and were a gift from Dr. Ronald DePinho. iKAP cancer cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) (Gemini) and 1% penicillin–streptomycin (Corning). For the isolation of iKAP CAFs, iKAP tumors were collected in PBS and briefly rinsed in 1% bleach. Tumors were cut into small pieces (~1 mm2) and digested in 400 U/ml Collagenase IV (Thermo #17104019) in DMEM at 37 °C for 24 h. Following digestion, the media was replaced with DMEM containing 20% FBS and 1% penicillin–streptomycin, and the cells were grown until 85% confluent. Cells were not short tandem repeat marker validated or tested for mycoplasma. Fibroblasts were maintained and used for experiments until passage 6. For evaluation of iKras recombination, DNA was isolated from cells using DNeasy Blood & Tissue kit according to manufacturer’s instructions. Genotyping PCR for Villin-Cre-ERT2, iKras, and iKras recombination was performed using previously described primers [22] listed in Table 1 and PCR products were visualized using QIAxcel capillary gel electrophoresis. For co-culture experiments, cancer cells were seeded on 24-well plates at 5 × 104 cells per well. After 24 h, 2 × 105 CAFs were seeded on collagen-coated Transwell inserts (Corning #353057) on top of the cancer cells and cultured for 48 h. Following co-culture, cancer cells were collected for RNA isolation and quantitative PCR (qPCR) analysis.
Table 1.
Genotyping PCR primer sequences.
| Target | Sequence | Expected product size (bp) |
|---|---|---|
| Villin-Cre-ERT2 | TG Forward: 5′-CCTGGAAAATGCTTCTGTCCG-3′, TG Reverse: 5′-CAGGGTGTTATAAGCAATCCC-3′; WT Forward: 5′-CAATGGTAGGCTCACTCTGGGAGATGATA-3′, WT Reverse: 5′-AACACACACTGGCAGGACTGGCTAGG-3′ | TG: 400 bp, WT: 300 bp |
| iKras | Forward: 5′-ATGACTGAGTATAAACTTGTGGTGG-3′, Reverse: 5′-TCACATAACTGTACACCTTGTCCTTGAC-3′ | 600 bp |
| iKras recombination | Forward: 5′-CGGTGGGAGGTCTATATAAGCA-3′, Reverse: 5′-CCACCACAAGTTTATACTCAGTCAT-3′ | Unrecombined: 1000 bp, Recombined: 180 bp |
Quantitative real-time PCR
RNA was isolated from cells using TRIzol reagent (ThermoFisher #15596026). cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Thermo-Fisher #4368814). SYBR Green PCR Master Mix was used for qPCR reactions in a Quantstudio 7 real-time PCR system (Applied Biosystems). Actb was used as the house-keeping gene. Fold-change in gene expression was determined using the ΔΔCT method. Technical triplicates were used for each measurement and statistical analyses were done on the ΔCT values. The primer sequences for the genes analyzed are listed in Table 2.
Table 2.
qPCR primer sequences.
| Gene (mouse) | Forward sequence | Reverse sequence |
|---|---|---|
| Vimentin | 5′-CTTGAACGGAAAGTGGAATCCT-3′ | 5′-GTCAGGCTTGGAAACGTCC-3′ |
| Zeb1 | 5′-GCTGGCAAGACAACGTGAAAG-3′ | 5′-GCCTCAGGATAAATGACGGC-3′ |
| Twist1 | 5′-CTGCCCTCGGACAAGCTGAG-3′ | 5′-CTAGTGGGACGCGGACATGG-3′ |
| Lgr5 | 5′-CTTCCGAATCGTCGATCTTC-3′ | 5′-AACGATCGCTCTCAGGCTAA-3′ |
| Dclk1 | 5′-TCAATGAGGACCAGCTCCAG-3′ | 5′-TCCGAGAGAGTTCGGGTCA-3′ |
| Cd44 | 5′-AGCAGCGGCTCCACCATCGAGA-3′ | 5′-TCGGATCCATGAGTCACAGTG-3′ |
| Prom1 | 5′-TTGGTGCAAATGTGGAAAAG-3′ | 5′-ATTGCCATTGTTCCTTGAGC-3′ |
| Bmp2 | 5′-GGGACCCGCTGTCTTCTAGT-3′ | 5′-TCAACTCAAATTCGCTGAGGAC-3′ |
| Bmp4 | 5′-CTTCAACCTCAGCAGCATCC-3′ | 5′-GATGAGGTGTCCAGGAACCA-3′ |
| Bmp7 | 5′-ACGGACAGGGCTTCTCCTAC-3′ | 5′-ATGGTGGTATCGAGGGTGGAA-3′ |
| Actb | 5′-GGACCTGACGGACTACCTCATG-3′ | 5′-TCTTTGATGTCACGCACGATTT-3′ |
Flow cytometry
Colon tumors were cut into 1–2 mm3 pieces and digested by incubation at 37 °C for 1 h in RPMI-1640 medium containing 0.15 μg/ml of DNase I (Invitrogen #8174), 40 U of Dispase I (Life Technologies #17105041), and 100 U/ml of Collagenase IV (Thermo #17104019). Tumor pieces were next vortexed extensively followed by incubation at 37 °C for 5 min, until tissue was digested completely. The resulting solution was filtered with a 100 μm cell strainer and washed with RPMI-1640 medium. The cells were resuspended with Fc blocking reagent and stained with fluorochrome-conjugated antibodies against CD45 (eBioscience #15-0451), CD4 (BD Pharmingen #563331), CD8 (BioLegend #100742), and CD3e (BD Pharmingen #563123). For Foxp3 staining, cells were fixed and permeabilized followed by incubation with anti-mouse Foxp3 antibody (eBioscience #45-5773) and were analyzed using BD LSR Fortessa X-20 Cell Analyzer (BD Biosciences). All acquired data were analyzed using FlowJo V10 (BD Biosciences). Following live/dead staining (ThermoFisher #L34957), singlets were gated on CD45 positivity, followed by CD3e+, then CD4+ and CD8+ cells. After gating for CD4+ T cells, the percentage of FoxP3+ Tregs was determined.
Detection of circulating tumor cells
Blood was collected retro-orbitally (100 μl) from control and depleted mice, and incubated in 1 ml of ACK lysing buffer (ThermoFisher #A1049201) at RT for red blood cell lysis. The cells were then washed twice with cold PBS and resuspended in PBS containing 2% FBS. Flow cytometry analysis for GFP+ circulating tumor cells (CTCs) was performed using BD LSR Fortessa X-20 Cell Analyzer (BD Biosciences). The data were recorded as the percentage of GFP+ cells per 105 total cells.
Statistical analysis
Nonparametric (Mann–Whitney test comparing two samples) and parametric (unpaired t-test) tests were performed for comparison between two groups, and unpaired t-test with Welch’s correction performed for comparison of two groups with unequal variances. One-way analysis of variance with Sidak’s multiple comparisons test (parametric) or nonparametric Kruskall–Wallis test with Dunn’s multiple comparison test (nonparametric) were used for comparison of more than two groups. For pericolonic inflammation (PI), Fisher’s exact test was used. For survival analysis, the Kaplan–Meier method was employed and statistical significance was determined with log-rank Mantel–Cox test. The performed statistical tests are indicated in the figure legends. P < 0.05 was used to define statistical significance.
Results
αSMA+ CAFs exert tumor-restraining functions in CRC
To determine the function of αSMA+ CAFs in CRC progression, we used the iKAP mouse model of CRC, which is driven by inducible expression of KrasG12D and conditional deletion of Apc and Trp53 [22]. iKAP mice were crossed to αSMA-RFP transgenic mice for visualization of αSMA+ cells and to the αSMA-TK transgenic mice, where expression of viral thymidine kinase (TK) is driven by the αSMA promoter (a list of mouse genotypes is included in Supplementary Table 1). With this model, administration of GCV results in the depletion of proliferating αSMA+ cells [3, 7, 8].
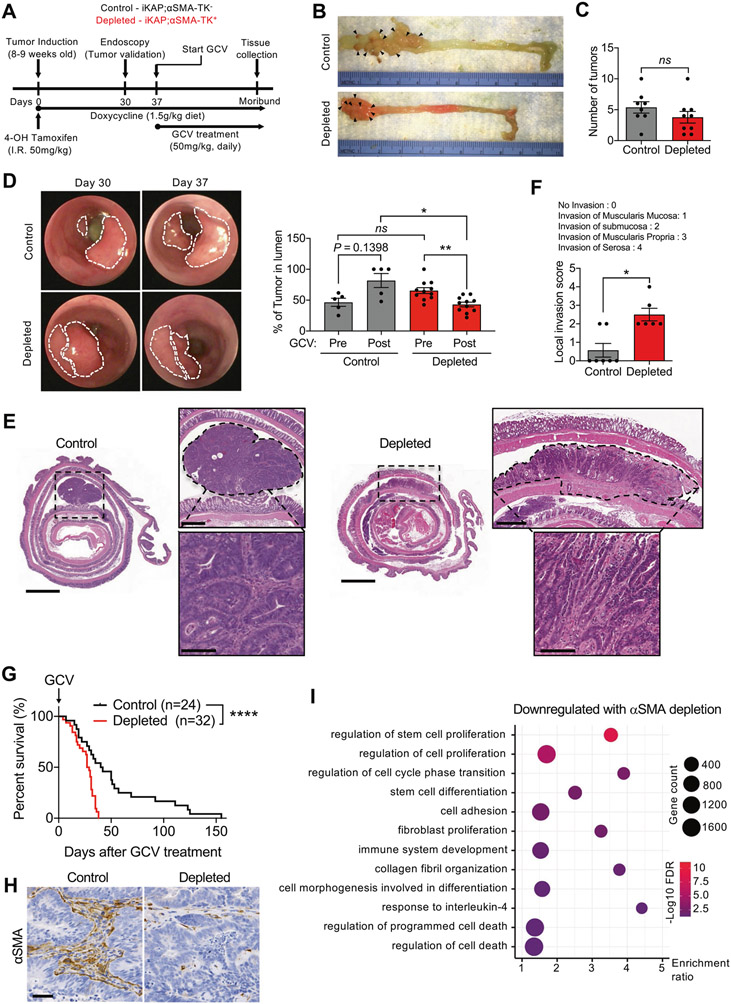
CRC induction in iKAP;αSMA-TK− (control) and iKAP; αSMA-TK+ (depleted) mice was elicited with a single intrarectal injection of 4-OH tamoxifen at 8–10 weeks of age, and the mice were fed a doxycycline-supplemented diet (Fig. 1A). Tumor formation was verified by endoscopy at 4 weeks post-tumor induction. When colon tumors were detected, GCV treatment was initiated and continued until the endpoint of the study. As previously reported, tumors arose primarily in the distal colon (Fig. 1B). Mice with depletion of αSMA+ CAFs presented with a similar number of tumor nodules when compared to control mice (Fig. 1C); however, endoscopic analysis of tumors prior to and following GCV administration showed that tumor growth, as defined by luminal protrusion, was greater in control tumors compared to αSMA+ CAF-depleted tumors (Fig. 1D). We suspected these findings were indicative of a switch to inward growth and invasion through the muscularis mucosa into the submucosa when αSMA+ CAFs are depleted. Histopathological analysis supported that αSMA+ CAF-depleted tumors were more undifferentiated and invasive compared to control tumors (Fig. 1E, F). The local invasiveness of CRC tumors is generally determined and classified based on how deep into the tissue layers the tumor cells invade [27]. Scoring for local invasion, defined by the depth of invasion of tumor cells in the colon parenchyma, indicated that αSMA+ CAF-depleted tumors presented with significantly higher invasion score compared to control (Fig. 1F). The increased invasion noted with depletion of αSMA+ CAFs was also associated with significantly decreased survival in tumor-bearing mice (Fig. 1G). Loss of body weight was observed in both groups as mice developed tumors and body weight at endpoint was higher in αSMA+ CAF-depleted mice compared to control mice (Supplementary Fig. 1A), possibly reflecting pericolonic inflammation (PI, vide infra).
Fig. 1. Depletion of αSMA+ CAFs leads to increased tumor invasion and decreased survival in CRC model.
A Schematic diagram for tumor induction and GCV treatment in the iKAP CRC mouse model. B Images of distal colon tumors from representative αSMA+ CAF-depleted and control mice. Arrow heads point to tumor foci. C Quantification of the number of tumors in control (n = 8) and depleted (n = 9) mice at endpoint. D Representative endoscopy images of CRC tumors before and after GCV treatment, and quantification of tumor in lumen. Control, n = 5; depleted, n = 11. E Representative images of H&E-stained colon sections with tumors outlined. Scale bar for colon section: 2000 μm; scale bar for top right zoomed-in image: 500 μm; scale bar for bottom right zoomed-in image: 100 μm. F Local invasion score of tumors in control (n = 7) and depleted (n = 6) tumors. G Survival analysis of αSMA+ CAF-depleted (n = 32) and control (n = 24) mice. H Representative immunohistochemistry for αSMA in CAF-depleted (n = 5) and control (n = 4) tumors. Scale bar: 50 μm. I Dot plot of the pathways downregulated in depleted compared control tumors. The data are presented as mean ± SEM. FDR: false discovery rate; GCV: ganciclovir. C Unpaired t-test performed; D Kruskall–Wallis test with Dunn’s multiple comparison test performed; F Mann–Whitney test performed; G Log-rank Mantel–Cox test performed. *P < 0.01, **P < 0.01, ****P < 0.0001, ns: not significant.
Depletion of αSMA+ CAFs was confirmed by IHC and revealed 70% reduction in αSMA compared to control mice (Fig. 1H and Supplementary Fig. 1B). Moreover, expression of αSMA was maintained in the muscularis mucosa of untransformed areas of the colon (Supplementary Fig. 1C). Perivascular (desmin+) immunolabeling revealed no significant changes in pericyte coverage in tumors depleted of αSMA+ cells compared to control (Supplementary Fig. 1D), supporting that the depletion strategy predominantly targeted αSMA+ CAFs. Surprisingly, type I collagen content was increased in depleted tumors (Supplementary Fig. 1E), suggesting a potential compensation, either by other stromal cells or cancer cells, upon depletion of αSMA+ CAFs. Approximately halfway between induction of GCV and lethal disease, IHC analysis for Ki67 revealed the proliferative index was significantly greater in tumors with αSMA+ CAF depletion, compared to their control counterparts (Supplementary Fig. 1F). When compared at endpoint, the proliferative index between control and αSMA+ CAF-depleted tumors did not differ significantly (Supplementary Fig. 1F).
Global gene expression profiling of endpoint tumors (Supplementary Fig. 2A) revealed a downregulation of genes associated with the regulation of stem cell proliferation, fibroblast proliferation, and collagen fibril organization in αSMA+ CAF-depleted tumors compared to control (Fig. 1I). Compared to control tumors, depleted tumors contained enrichment of genes associated with cell migration and tumor invasion, such as Mpp3 [28] (Supplementary Fig. 2B). GSEA identified pathways associated with malignancy (hypoxia and Kras signaling) enriched in depleted tumors compared to control tumors (Supplementary Fig. 2C and Supplementary Table 2). Collectively, our results indicate that the depletion of αSMA+ CAFs in CRC generates more invasive and lethal disease, and support that αSMA+ CAFs serve a tumor-restraining function in CRC.
αSMA+ CAF depletion promotes CRC lymphatic invasion
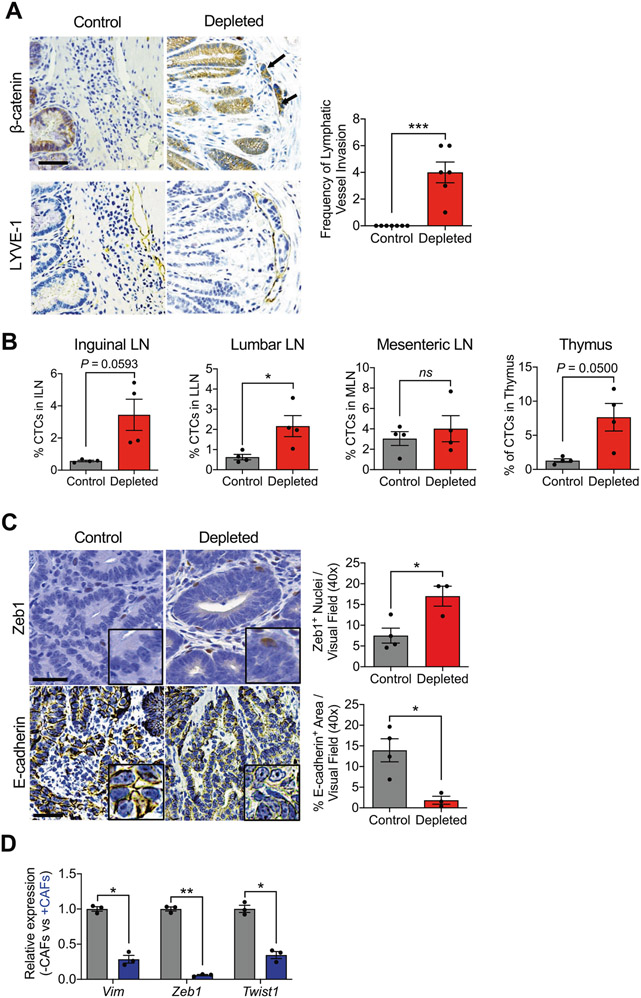
To investigate whether the depletion of αSMA+ CAFs had an impact on cancer cell dissemination, the number of GFP+ CTCs from control and depleted mice was determined by fluorescence-activated cell sorting (FACS) analysis. We found that depleted mice had a significantly increased number of CTCs compared to control mice (Supplementary Fig. 3A). The first site of metastasis for CRC cells is the lymph nodes; we next determined whether depletion of αSMA+ CAFs had an impact on lymphatic metastasis. Immunostaining of serial slides for LYVE-1 and β-catenin identified cancer cells present in the lymphatic system, indicative of lymph node metastasis, strictly in αSMA+ CAF-depleted mice (Fig. 2A). Next, the presence of GFP+ tumor cells was determined in lymphoid organs including the inguinal, luminal, and mesenteric lymph nodes, as well as the thymus (Fig. 2B and Supplementary Fig. 3B). The lumbar lymph node showed a significant increase of GFP+ tumor cells (Fig. 2B and Supplementary Fig. 3B). Immunostaining for markers of epithelial to mesenchymal transition (EMT), including Zeb1 and E-cadherin, showed a significant induction of EMT (increased Zeb1, decreased E-cadherin) in the depleted tumors (Fig. 2C). These effects of CAFs on cancer cells were confirmed in vitro. CAFs were isolated from the tumors of iKAP mice and verified by lack of endogenous green fluorescent protein (GFP) expression (expressed by cancer cells), expression of the fibroblast markers αSMA, vimentin, and FSP-1 (Supplementary Fig. 3C), and lack of cancer cell-specific gene recombination via genomic DNA PCR analysis (Supplementary Fig. 3D). iKAP CAFs were co-cultured with cancer cells (from iKAP tumors), which resulted in lower expression of Vim, Zeb1, and Twist1 when compared to cancer cells alone (Fig. 2D), suggesting that αSMA+ CAFs limit cancer cells’ expression mesenchymal gene transcripts. Metastatic disease, identified by CD44 immunostaining and histopathological analyses, had increased prevalence in the livers of depleted compared to control mice (Supplementary Fig. 3E).
Fig. 2. Loss of αSMA+ CAFs increases rates of local invasion.
A Representative immunohistochemistry for β-catenin and LYVE-1, and quantification of the frequency of lymphatic vessel invasion in control (n = 7) and depleted (n = 6) mice. Arrows point to β-catenin+ cancer cells within LYVE-1+ lymphatic vessels. B Percentage of GFP+ tumor cells in the thymus, inguinal (ILN), lumbar (LLN), and mesenteric (MLN) lymph nodes of control (n = 4) and depleted (n = 4) mice. C Representative immunohistochemistry for Zeb1 and E-cadherin in control (n = 4) and depleted (n = 3) tumors, and quantification of staining. Zeb1 scale bar: 20 μm, E-Cadherin scale bar: 50 μm, inset: digital zoom. D Transcript levels for Vim, Zeb1, and Twist1 in cancer cells in monoculture (−CAFs) or co-culture with iKAP CAFs (+CAFs). n = 3 biological replicates. Statistics were performed based on ΔCT values. The data are presented as mean ± SEM. A Mann–Whitney test performed; B (first and fourth panels), D unpaired t-test with Welch’s correction performed; B (second and third panels), C unpaired t-test performed. *P < 0.05, **P < 0.01, ns: not significant.
αSMA+ CAFs secretome controls CSCs frequency
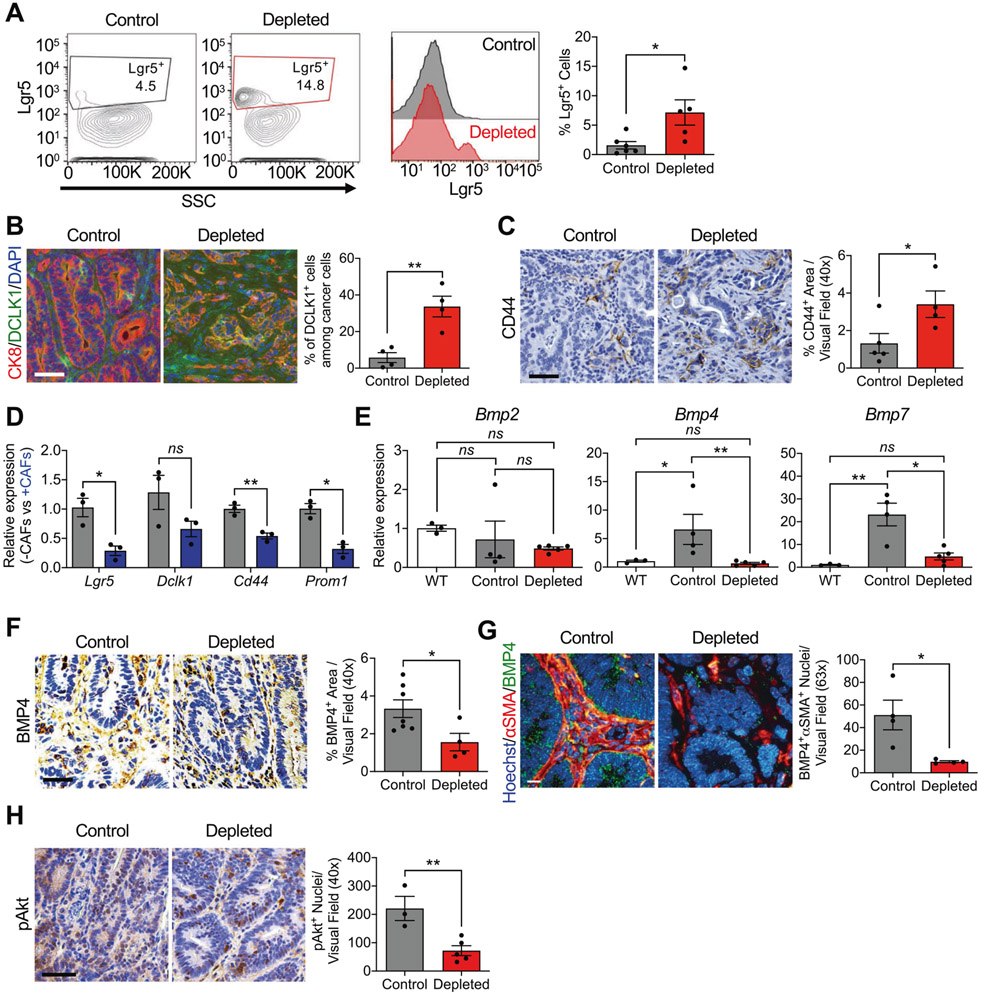
The increased frequency of CSCs in CRC is associated with cancer invasiveness and resistance to therapy [29-31]. FACS analysis in tumors from depleted mice compared to their counterpart controls revealed increased Lgr5 expression on tumor cells (Fig. 3A). Expression of DCLK1, a CSC-specific marker previously shown to play an important role in CRC tumorigenesis and not present in normal intestinal stem cells (ISCs) [32], was also found to be increased in depleted mice (Fig. 3B). The CSC marker CD44 also revealed an increase in the expression in αSMA+ CAF-depleted tumors compared to control (Fig. 3C). When co-cultured with CAFs, cancer cells showed reduced expression of CSC markers Lgr5, Cd44, and Prom1 (Fig. 3D). These results support that αSMA+ CAFs limit the frequency of CSCs in CRC and depletion of αSMA+ CAFs allows CSCs to increase in number. CSC transcriptomic profile in cancer cells is, at least in part, modulated by the secretome of CAFs. Given that αSMA+ myofibroblasts generate bone morphogenic protein (BMP) ligands in the intestine, which are identified as promoters of the undifferentiated phenotype of CSCs, suppressing their proliferation [33-35], we analyzed the expression of BMP ligands Bmp2, Bmp4, and Bmp7 in control and depleted tumors, as well as WT colonic epithelium (Fig. 3E). The expression of Bmp4 and Bmp7 was decreased in depleted tumors compared to control tumors (Fig. 3E). Given the established role of BMP4 in the negative regulation of Lgr5+ stem cells [34], we examined its expression in control and depleted tumors by IHC. The depleted tumors exhibit decreased BMP4, compared to control tumors (Fig. 3F). Costaining demonstrated that αSMA+ CAFs express BMP4 and this colocalization is reduced upon depletion of αSMA+ CAFs (Fig. 3G). BMP4 signaling has been implicated in promoting the differentiation and apoptosis of CRC stem cells by engaging non-canonical BMP signaling pathways [36]. We observed suppression of the downstream non-canonical BMP target pAKT in epithelial cells concurrent with depletion of αSMA+ CAFs and suppression of BMP4 signaling (Fig. 3H). These findings suggest that αSMA+ CAFs likely play a functional role in promoting the differentiation of CSCs in CRC at least partially through BMP4-mediated signaling.
Fig. 3. αSMA+ CAF-depleted CRC tumors display an enhanced CSC phenotype.
A Representative contour plots, histograms, and quantification of Lgr5+ cells from control (black, n = 6) and depleted (red, n = 5) tumors. Number in gate indicate the % Lgr5+ cells. B Representative immunostaining of colon tumor sections for CK8 (red) and DCLK1 (green) in control (n = 4) and depleted (n = 4) tumors, and quantification of staining. DAPI (blue) was used as nuclear stain. Scale bar: 50 μm. C Representative immunohistochemistry of colon tumor sections for CD44 in control (n = 5) and depleted (n = 4) tumors, and quantification of staining. Scale bar: 50 μm. D Transcript levels of CSC markers Lgr5, Dclk1, Cd44, and Prom1 in cancer cells following co-culture with iKAP CAFs. n = 3 biological replicates. Statistics were performed based on ΔCT values. E Transcript levels of Bmp2, Bmp4, and Bmp7 in WT colon (n = 3), control tumors (n = 4), and depleted tumors (n = 5). F Representative immunohistochemistry of colon tumor sections for BMP4 and quantification of staining in control (n = 7) and depleted (n = 4) tumors. Scale bar: 50 μm. G Representative immunohistochemistry of colon tumor sections for BMP4 and αSMA, and quantification of staining of control (n = 4) and depleted (n = 4) tumors. Scale bar: 10 μm. H Representative immunohistochemistry of colon tumor sections for phosphorylated AKT (pAKT) and quantification of staining of control (n = 3) and depleted (n = 5) tumors. Scale bar: 50 μm. SSC: side scatter. The data are presented as mean ± SEM. A Mann–Whitney test performed; B, C, F–H unpaired t-test performed; D unpaired t-test with Welch’s correction performed; E one-way ANOVA with Sidak’s multiple comparison test performed based on ΔCT values. *P < 0.05, **P < 0.01, ns: not significant.
αSMA+ CAF depletion generates an immunosuppressive TME
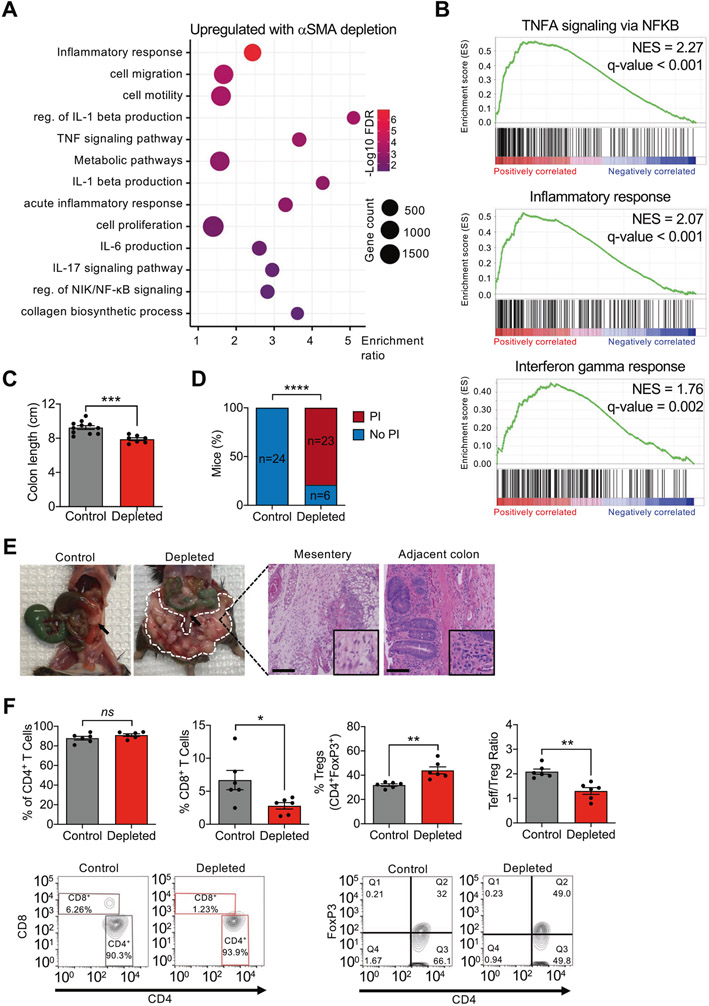
RNA-sequencing analyses revealed upregulation of pathways associated with inflammation, including tumor necrosis factor signaling, interleukin (IL)-6 and IL-1β production, and IL-17 signaling (Fig. 4A, B). GSEA also identified enrichment of inflammatory response and interferon-γ response pathways in the depleted tumors, indicative of an immunosuppressive phenotype (Fig. 4B). Gross analysis of the colons of control and depleted mice at endpoint revealed a significant reduction in colon length in the depleted mice, indicative of potential increase in inflammation in the context of αSMA+ CAF depletion (Fig. 4C). Microscopic analysis of the mesenteric adipose tissue surrounding the tumors demonstrated the presence of PI in the majority of mice with depletion of αSMA+ CAFs, compared to control mice (Fig. 4D, E). Inflammation was also observed in adjacent untransformed colon (Fig. 4E).
Fig. 4. Depletion of αSMA+ myofibroblasts induces an immunosuppressive phenotype.
A Dot plot of the pathways upregulated in depleted compared to control tumors. B Selected GSEA plots of gene sets associated with immune pathways. C Measurement of colon length at endpoint in control (n = 11) and depleted (n = 7) mice. D Quantification of incidence of pericolonic inflammation in depleted mice (n = 29) compared to control (n = 24). E Representative images of the gross appearance and H&E images of mesentery and adjacent colon. Arrows point to the tumor; encircled in dashed white line is pericolonic inflammation. Mesentery scale bar: 100 μm; adjacent colon scale bar: 50 μm. F Percentage of CD4+ T cells, CD8+ cytotoxic T cells, CD4+FoxP3+ Treg cells, and Teff/Treg ratio in tumors from control (n = 6) and depleted (n = 6) mice, and representative FACS contour plots. The values in quadrant Q2 indicate the percentage of FoxP3+CD4+ Tregs. FDR: false discovery rate; NES: normalized enrichment score; PI: pericolonic inflammation; reg. of: regulation of. The data are presented as mean ± SEM. C, F (first and fourth panels) unpaired t-test performed; D Fisher’s exact test performed; F (second and third panels) unpaired t-test with Welch’s correction performed. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns: not significant.
To further investigate whether depletion of αSMA+ CAFs leads to an immunosuppressive TME, we analyzed the tumors for T-cell populations. The percentage of CD4+ T cells did not change significantly; however, the population of cytotoxic (CD8+) T cells was significantly decreased in the tumors from depleted mice (Fig. 4F). We also observed an increased percentage of regulatory T cells (Tregs) in the depleted tumors (Fig. 4F) and an overall reduction in the ratio of Teff to Treg (Teff/Treg) in the αSMA+ CAF-depleted tumors (Fig. 4F). This immunosuppressive phenotype was consistent with that previously observed following depletion of αSMA+ CAFs in a pancreatic cancer model [7]. Overall, these results show that depletion of αSMA+ CAFs leads to an immunosuppressive TME with a decrease in cytotoxic CD8+ tumor-infiltrating lymphocytes.
TGFβ1 is known to play a role in inflammation and excessive accumulation of TGFβ1 is associated with chronic inflammation and fibrosis [37]; thus, we hypothesized that depletion of αSMA+ CAFs may result in increased TGFβ1. IHC analysis revealed an increase in TGFβ1 protein expression in the tumor stroma following αSMA+ CAF depletion (Supplementary Fig. 4A). Moreover, costaining revealed increased colocalization of TGFβ1 with CD45 in depleted mice (Supplementary Fig. 4B). Consequently, the increased TGFβ1 may act to alter the immune microenvironment in depleted mice to hinder T-cell function, as TGFβ1 has previously been described to suppress the proliferation and effector function of T cells [38].
Discussion
Previous studies indicated a correlation between the increased incidence of αSMA+ CAFs and tumor aggressiveness in different solid malignancies [18, 39, 40]; however, a functional genetic study of their role in CRC had not been performed. Employing the iKAP CRC model engineered with a somatic lineage depletion system, we demonstrate that αSMA+ CAF depletion leads to increased tumor aggressiveness and a decrease in overall survival. A recent study identified four main fibroblast populations in the intestinal mesenchyme, only one of which expresses αSMA [20]. Other studies identified Grem1 and Islr as marking distinct CRC fibroblast subsets, with αSMA overlap with Grem1 occurring more frequently than with Islr [41]. How these populations change over the course of CRC progression and the precise functional roles of CAF subsets in CRC remain open questions. This study demonstrates that αSMA+ CAFs function to restrain tumor progression as part of the protective host response against neoplasia and malignancy.
Our observation of increased collagen deposition in tumors with depleted myofibroblasts is consistent with a recent study by Wu et al. [42], showing that elevated type I collagen expression positively correlated with increased stemness and incidence of metastases in CRC patients. Type 1 collagen was found to promote mesenchymal gene expression and cancer cell invasion [43], and was associated with the tumor invasive margin [44]. Another study identified COL1A1 as a potential target and diagnostic marker for metastatic CRC, due to its high expression in patients [45]. We observe increased collagen deposition in the αSMA+ CAF-depleted tumors, suggesting that other cell types, including cancer cells, are likely responsible for collagen production and/or compensate for the loss of the potential collagen production by αSMA+ CAFs [46]. We observe evidence for increased local invasiveness, increased CTCs, and increased lymph node metastasis. Moreover, the metastatic disease rate was increased in αSMA+ CAF-depleted compared to control mice, as previously reported in breast tumor models [8]. Although our studies in distinct tumor models do not support significant off-target effects in the αSMA-TK transgenic mice, it is possible that αSMA+ stromal cells display functions that are concurrently suppressed with our genetic strategy.
Collectively, our study suggests that αSMA+ CAFs likely use diverse mechanisms to control the rate of oncogenic Kras and loss of Apc- and Trp53-driven CRC. We show that αSMA+ CAFs produce BMP4 and suppress the production of TGFβ1 by other stromal cells, including immune cells, in the TME. BMPs belong to the TGFβ superfamily of ligands and play an important role in limiting the hyperproliferation of the intestinal epithelium [33, 47]. Altogether, the studies reported here offer an insight into the functional role of the αSMA+ CAF in the progression of CRC.
It was recently reported that Smad1/Smad4 mediated repression of the ISC gene signature, including Lgr5 expression [34]. BMP4 secreted by αSMA+ myofibroblasts in the intestinal mesenchyme promotes the differentiation of the ISCs as ISCs move up the crypt and away from Wnt signals promoting stem cell maintenance, and where BMP inhibitors Gremlin and Noggin are expressed [34, 47-49]. The reduction in BMP4 levels in αSMA+ CAF-depleted tumors suggests a pathway where CSCs remain undifferentiated, leading to invasive CRC.
The iKAP tumors are associated with a TGFβ1 signaling phenotype, which correlates with the CRC CMS4 subtype [22]. TGFβ1 has a well-established role in tumor invasion and immune suppression [50-52]. Increased TGFβ1 in αSMA+ CAF-depleted tumors likely contributes to the increased invasiveness of the tumors and increased Tregs, associated with decreased CD8+ T cells. Associated with this observation, we also document increased inflammation, with increased IL-6, IL-1β, and IL-17-mediated signaling pathways. Altogether, our studies demonstrate that αSMA+ CAFs restrain the progression rate of CRC, exemplifying the potential functional heterogeneity of fibroblasts and their secretome [3, 9, 53], and offering an appreciation of potential protective host response against malignancy.
Supplementary Material
Acknowledgements
This study was supported by research funds provided by the MD Anderson Cancer Center. KV-A is supported by Ruth L. Kirschstein National Research Service Award 5T32CA186892. We thank Dr. Ronald DePinho for providing the iKAP mouse model and for insightful comments on the manuscript. We thank Dr. Adam Boutin for his generous help with the project, Dr. Krishnan Mahadevan for advice on proliferative index determination, and Patience Kelly for assistance with slide scanning and staining. The Advanced Technology Genomics Core is supported by the core grant CA016672(ATCG).
Footnotes
Conflict of interest The authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41388-021-01866-7.
Data availability
The source data for all the figures in the manuscript can be downloaded at: https://doi.org/10.17632/w8kmnbpyfw.1. Raw and processed RNA-sequencing data are deposited in the Gene Expression Omnibus database (accession number GSE164300).
References
- 1.Kalluri R The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–98. [DOI] [PubMed] [Google Scholar]
- 2.LeBleu VS, Kalluri R. A peek into cancer-associated fibroblasts: origins, functions and translational impact. Dis Models Mech. 2018;11:dmm029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeBleu VS, Neilson EG. Origin and functional heterogeneity of fibroblasts. FASEB J. 2020;34:3519–36. [DOI] [PubMed] [Google Scholar]
- 4.Nurmik M, Ullmann P, Rodriguez F, Haan S, Letellier E. In search of definitions: cancer-associated fibroblasts and their markers. Int J Cancer. 2020;146:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otranto M, Sarrazy V, Bonte F, Hinz B, Gabbiani G, Desmouliere A. The role of the myofibroblast in tumor stroma remodeling. Cell Adh Migr. 2012;6:203–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gascard P, Tlsty TD. Carcinoma-associated fibroblasts: orchestrating the composition of malignancy. Genes Dev. 2016;30:1002–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Özdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker LM, O’Connell JT, Vo AP, Cain MP, Tampe D, Bizarro L, et al. Epigenetic reprogramming of cancer-associated fibroblasts deregulates glucose metabolism and facilitates progression of breast cancer. Cell Rep. 2020;31:107701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20:174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Center for Disease Control and Prevention. Colorectal cancer statistics. Center for Disease Control and Prevention; 2019. [Google Scholar]
- 11.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. [DOI] [PubMed] [Google Scholar]
- 12.De Sousa EMF, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LP, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19:614–8. [DOI] [PubMed] [Google Scholar]
- 13.Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadanandam A, Wang X, de Sousa EMF, Gray JW, Vermeulen L, Hanahan D, et al. Reconciliation of classification systems defining molecular subtypes of colorectal cancer: interrelationships and clinical implications. Cell Cycle. 2014;13:353–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woolston A, Khan K, Spain G, Barber LJ, Griffiths B, Gonzalez-Exposito R, et al. Genomic and transcriptomic determinants of therapy resistance and immune landscape evolution during anti-EGFR treatment in colorectal cancer. Cancer Cell. 2019;36:35–50. e39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320–9. [DOI] [PubMed] [Google Scholar]
- 19.Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47:312–9. [DOI] [PubMed] [Google Scholar]
- 20.Roulis M, Kaklamanos A, Schernthanner M, Bielecki P, Zhao J, Kaffe E, et al. Paracrine orchestration of intestinal tumorigenesis by a mesenchymal niche. Nature 2020;580:524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeBleu VS, Taduri G, O’Connell J, Teng Y, Cooke VG, Woda C, et al. Origin and function of myofibroblasts in kidney fibrosis. Nat Med. 2013;19:1047–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boutin AT, Liao WT, Wang M, Hwang SS, Karpinets TV, Cheung H, et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev. 2017;31:370–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeBleu VS, Teng Y, O’Connell JT, Charytan D, Muller GA, Muller CA, et al. Identification of human epididymis protein-4 as a fibroblast-derived mediator of fibrosis. Nat Med. 2013;19:227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10–12. [Google Scholar]
- 26.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haggitt RC, Glotzbach RE, Soffer EE, Wruble LD. Prognostic factors in colorectal carcinomas arising in adenomas: implications for lesions removed by endoscopic polypectomy. Gastroenterology. 1985;89:328–36. [DOI] [PubMed] [Google Scholar]
- 28.Ma H, Cai H, Zhang Y, Wu J, Liu X, Zuo J, et al. Membrane palmitoylated protein 3 promotes hepatocellular carcinoma cell migration and invasion via up-regulating matrix metalloproteinase 1. Cancer Lett. 2014;344:74–81. [DOI] [PubMed] [Google Scholar]
- 29.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren G 2nd, et al. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69:1951–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. [DOI] [PubMed] [Google Scholar]
- 32.Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet. 2013;45:98–103. [DOI] [PubMed] [Google Scholar]
- 33.He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet. 2004;36:1117–21. [DOI] [PubMed] [Google Scholar]
- 34.Qi Z, Li Y, Zhao B, Xu C, Liu Y, Li H, et al. BMP restricts stemness of intestinal Lgr5(+) stem cells by directly suppressing their signature genes. Nat Commun. 2017;8:13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veschi V, Mangiapane LR, Nicotra A, Di Franco S, Scavo E, Apuzzo T, et al. Targeting chemoresistant colorectal cancer via systemic administration of a BMP7 variant. Oncogene. 2020;39:987–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lombardo Y, Scopelliti A, Cammareri P, Todaro M, Iovino F, Ricci-Vitiani L, et al. Bone morphogenetic protein 4 induces differentiation of colorectal cancer stem cells and increases their response to chemotherapy in mice. Gastroenterology. 2011;140:297–309. [DOI] [PubMed] [Google Scholar]
- 37.Wahl SM. Transforming growth factor beta: the good, the bad, and the ugly. J Exp Med. 1994;180:1587–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan YY, Flavell RA. ‘Yin-Yang’ functions of transforming growth factor-beta and T regulatory cells in immune regulation. Immunol Rev. 2007;220:199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L, Liu L, Yao HH, Zhu ZQ, Ning ZL, Huang Q. Stromal myofibroblasts are associated with poor prognosis in solid cancers: a meta-analysis of published studies. PLoS ONE. 2016;11:e0159947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiori ME, Di Franco S, Villanova L, Bianca P, Stassi G, De Maria R. Cancer-associated fibroblasts as abettors of tumor progression at the crossroads of EMT and therapy resistance. Mol Cancer. 2019;18:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi H, Gieniec KA, Wright JA, Wang T, Asai N, Mizutani Y, et al. The balance of stromal BMP signaling mediated by GREM1 and ISLR drives colorectal carcinogenesis. Gastro-enterology. 2021;160:1224–39. e1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X, Cai J, Zuo Z, Li J. Collagen facilitates the colorectal cancer stemness and metastasis through an integrin/PI3K/AKT/Snail signaling pathway. Biomed Pharmacother. 2019;114:108708. [DOI] [PubMed] [Google Scholar]
- 43.Vellinga TT, den Uil S, Rinkes IH, Marvin D, Ponsioen B, Alvarez-Varela A, et al. Collagen-rich stroma in aggressive colon tumors induces mesenchymal gene expression and tumor cell invasion. Oncogene. 2016;35:5263–71. [DOI] [PubMed] [Google Scholar]
- 44.Blockhuys S, Agarwal NR, Hildesjo C, Jarlsfelt I, Wittung-Stafshede P, Sun XF. Second harmonic generation for collagen I characterization in rectal cancer patients with and without preoperative radiotherapy. J Biomed Opt. 2017;22:1–6. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z, Fang C, Wang Y, Zhang J, Yu J, Zhang Y, et al. COL1A1: a potential therapeutic target for colorectal cancer expressing wild-type or mutant KRAS. Int J Oncol. 2018;53:1869–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Damaghi M, Mori H, Byrne S, Xu L, Chen T, Johnson J, et al. Collagen production and niche engineering: a novel strategy for cancer cells to survive acidosis in DCIS and evolve. Evol Appl. 2020;13:2689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Que J. BMP signaling in development, stem cells, and diseases of the gastrointestinal tract. Annu Rev Physiol. 2020;82:251–73. [DOI] [PubMed] [Google Scholar]
- 48.Powell DW, Pinchuk IV, Saada JI, Chen X, Mifflin RC. Mesenchymal cells of the intestinal lamina propria. Annu Rev Physiol. 2011;73:213–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stzepourginski I, Nigro G, Jacob JM, Dulauroy S, Sansonetti PJ, Eberl G, et al. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc Natl Acad Sci USA. 2017;114:E506–e513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Céspedes MV, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu S, Zhang N, Yopp AC, Chen D, Mao M, Chen D, et al. TGF-beta induces Foxp3 + T-regulatory cells from CD4 + CD25 − precursors. Am J Transplant. 2004;4:1614–27. [DOI] [PubMed] [Google Scholar]
- 52.Moo-Young TA, Larson JW, Belt BA, Tan MC, Hawkins WG, Eberlein TJ, et al. Tumor-derived TGF-beta mediates conversion of CD4+Foxp3+ regulatory T cells in a murine model of pancreas cancer. J Immunother. 2009;32:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karagiannis GS, Poutahidis T, Erdman SE, Kirsch R, Riddell RH, Diamandis EP. Cancer-associated fibroblasts drive the progression of metastasis through both paracrine and mechanical pressure on cancer tissue. Mol Cancer Res. 2012;10:1403–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source data for all the figures in the manuscript can be downloaded at: https://doi.org/10.17632/w8kmnbpyfw.1. Raw and processed RNA-sequencing data are deposited in the Gene Expression Omnibus database (accession number GSE164300).