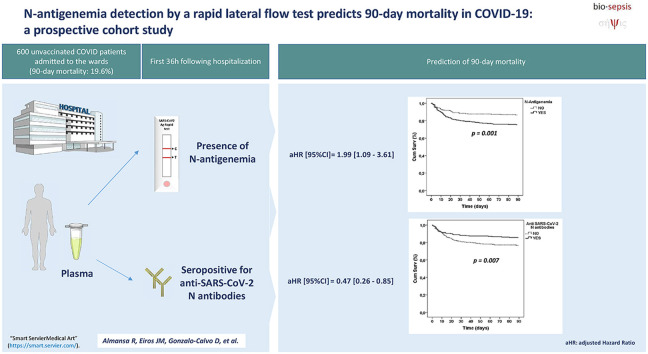
Abstract
Objectives
To evaluate if the detection of N antigen of SARS-CoV-2 in plasma by a rapid lateral flow test predicts 90-day mortality in COVID-19 patients hospitalized at the wards.
Methods
The presence of N-antigenemia was evaluated in the first 36 hours after hospitalization in 600 unvaccinated COVID-19 patients, by using the Panbio COVID-19 Ag Rapid Test Device from Abbott (Abbott Laboratories Inc., Chicago, IL, USA). The impact of N-antigenemia on 90-day mortality was assessed by multivariable Cox regression analysis.
Results
Prevalence of N-antigenemia at hospitalization was higher in nonsurvivors (69% (82/118) vs. 52% (250/482); p < 0.001). The patients with N-antigenemia showed more frequently RNAemia (45.7% (148/324) vs. 19.8% (51/257); p < 0.001), absence of anti-SARS-CoV-2 N antibodies (80.7% (264/327) vs. 26.6% (69/259); p < 0.001) and absence of S1 antibodies (73.4% (240/327) vs. 23.6% (61/259); p < 0.001). The patients with antigenemia showed more frequently acute respiratory distress syndrome (30.1% (100/332) vs. 18.7% (50/268); p = 0.001) and nosocomial infections (13.6% (45/331) vs. 7.9% (21/267); p = 0.026). N-antigenemia was a risk factor for increased 90-day mortality in the multivariable analysis (HR, 1.99 (95% CI,1.09–3.61), whereas the presence of anti-SARS-CoV-2 N-antibodies represented a protective factor (HR, 0.47 (95% CI, 0.26–0.85).
Discussion
The presence of N-antigenemia or the absence of anti-SARS-CoV-2 N-antibodies after hospitalization is associated to increased 90-day mortality in unvaccinated COVID-19 patients. Detection of N-antigenemia by using lateral flow tests is a quick, widely available tool that could contribute to early identify those COVID-19 patients at risk of deterioration.
Keywords: Antigenemia, COVID-19, Hospitalized, Mortality, Rapid test
Graphical abstract
Introduction
The presence of SARS-CoV-2 RNA in plasma (RNAemia) is associated to host-dysregulated responses, critical illness, and death in COVID-19 [[1], [2], [3]]. Dissemination of viral components to the blood could reflect severe alveolitis with damage to the alveolar-vascular barrier [4]. In turn, viral components could contribute to induce extra-pulmonary disease by stimulating innate immunity responses and/or mediating endothelial and tissue damage [2,5]. Although current evidence linking SARS-CoV-2 RNAemia with severe disease and poor outcome is solid, the potential influence of antigenemia (the presence of viral antigens in blood) on the prognosis of COVID-19 patients has been poorly explored yet [6,7]. Herein, we evaluated if the detection of N antigen of SARS-CoV-2 in plasma by a rapid lateral flow test predicted 90-day mortality in COVID-19 patients hospitalized at the wards.
Methods
The inclusion criteria was the following: consecutive adult patients with a positive nasopharyngeal swab PCR for SARS-CoV-2 admitted to the wards from 2 July 2020 to 10 March 2021 for whom an informed consent to participate in the study was feasible to obtain from the patient or his/her legal representative in the first 36 hours after admission. The plasma from EDTA blood was obtained in these first 36 hours and stored at –80ºC. The exclusion criteria was the following: patients showing concomitant infections at admission, those who had received any dose of a SARS-CoV-2 vaccine, and those for whom informed consent could not be requested/obtained. The study finally involved 600 patients out of the 1333 COVID-19 patients admitted to the participant wards during this period. This was a sub-study of the CIBERES-UCI-COVID project (Clinicaltrials.gov NCT04457505). Approval of the study protocol was obtained from the ethics committees of the participant hospitals. This work has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. Samples were processed by the BioSepsis laboratory and by the IRB-Lleida Biobank (B.0000682)/“Plataforma Biobancos PT17/0015/0027". N-antigenemia was defined as a positive result for the presence of N antigen of SARS-CoV-2 in plasma by using the Panbio COVID-19 Ag Rapid Test (Abbott Laboratories Inc., Chicago, IL, USA). Anti-SARS-CoV-2 S1 and N-antibodies were profiled using the SARS-CoV-2 IgG II Quant/SARS-CoV-2 IgG assays on an Alinity platform (Abbott Laboratories Inc.) Viral RNA load in plasma was profiled using droplet digital PCR as previously described [2]. Statistical analysis was performed using IBM SPSS Statistics Version 25.0 (IBM Corp., Armonk, NY, USA). The level of significance was set at p = 0.05. The factors associated to 90-day mortality were identified by multivariable Cox regression analysis. Those variables of the Table 1 yielding p < 0.100 in the univariable analysis were used as adjusting variables.
Table 1.
Clinical characteristics of the patients
| Clinical characteristics and outcomes | All cohort | 90-Day mortality |
||
|---|---|---|---|---|
| Survivors | Nonsurvivors | p | ||
| N | 600 | 482 | 118 | — |
| Age, median years (IQR) | 72.0 (24.0) | 67.5 (23.0) | 85.0 (10.0) | <0.001 |
| Male, n (%) | 335 (55.8) | 273 (56.6) | 62 (52.5) | 0.422 |
| Smoking, n (%) | 23 (3.8) | 20 (4.1) | 3 (2.5) | 0.415 |
| Comorbidities | ||||
| Hypertension, n (%) | 321 (53.5) | 243 (50.4) | 78 (66.1) | 0.002 |
| Dyslipidemia, n (%) | 214 (35.7) | 180 (37.3) | 34 (28.8) | 0.083 |
| Diabetes, n (%) | 132 (22.0) | 103 (21.4) | 29 (24.6) | 0.451 |
| Obesity, n (%) | 118 (19.7) | 102 (21.2) | 16 (13.6) | 0.063 |
| Chronic cardiovascular disease, n (%) | 100 (16.7) | 64 (13.3) | 36 (30.5) | <0.001 |
| Chronic cerebrovascular disease, n (%) | 36 (6.0) | 23 (4.8) | 13 (11.0) | 0.010 |
| Chronic atrial fibrillation, n (%) | 73 (12.2) | 46 (9.5) | 27 (22.9) | <0.001 |
| Chronic renal disease, n (%) | 70 (11.7) | 44 (9.1) | 26 (22.0) | <0.001 |
| Chronic respiratory disease, n (%) | 86 (14.3) | 64 (13.3) | 22 (18.6) | 0.136 |
| Cancer, n (%) | 63 (10.5) | 50 (10.4) | 13 (11.0) | 0.838 |
| Status at hospital admission | ||||
| Days since symptoms onset to hospital admission, median years (IQR) a596 | 5.0 (6.0) | 6.0 (5.0) | 3.0 (5.0) | <0.001 |
| SOFA score, median (IQR) | 2.0 (2.0) | 2.0 (1.0) | 2.5 (3.0) | <0.001 |
| Sepsis, n (%) | 340 (56.7) | 256 (53.1) | 84 (71.2) | <0.001 |
| Bilateral pneumonia in the chest x-ray, n (%) | 369 (61.6) | 289 (60.00) | 80 (68.4) | 0.100 |
| PaO2/FIO2 (<400), n (%) | 192 (32.0) | 161 (33.4) | 31 (26.3) | 0.137 |
| MAP (<70 mmHg), n (%) a599 | 568 (94.8) | 455 (94.6) | 113 (95.8) | 0.608 |
| Glasgow (<15), n (%) | 40 (6.7) | 19 (3.9) | 21 (17.8) | <0.001 |
| Laboratory parameters at hospital admission | ||||
| Hyperglycemia (glucose >126 mg/dL), n (%) | 254 (42.3) | 194 (40.2) | 60 (50.8) | 0.037 |
| Creatinine ≥1.2 mg/dL, n (%) | 141 (23.5) | 88 (18.3) | 53 (44.9) | <0.001 |
| Bilirrubin ≥1.2 mg/dL, n (%) a599 | 30 (5.0) | 23 (4.8) | 7 (6.0) | 0.590 |
| Hypertransaminasemia (ALT >40 UI/L), n (%) a596 | 157 (26.3) | 137 (28.7) | 20 (16.9) | 0.010 |
| Hypernatremia (Na >145 mmol/L), n (%) | 43 (7.20) | 13 (2.70) | 30 (25.40) | <0.001 |
| LDH >250 UI/L, n (%) a588 | 397 (67.50) | 314 (66.50) | 83 (71.60) | 0.300 |
| Lactate >2 mmol/L, n (%) | 116 (19.30) | 83 (17.20) | 33 (28.00) | 0.008 |
| Hemoglobin <13 g/dL, n (%) | 463 (77.20) | 384 (79.70) | 79 (66.90) | 0.003 |
| D-Dimers >500 ng/mL, n (%) a592 | 319 (53.90) | 237 (49.80) | 82 (70.70) | <0.001 |
| Thrombocytopenia (platelets <150 cell × 103/μL), n (%) | 185 (30.08) | 141 (29.30) | 44 (37.30) | 0.090 |
| C-reactive protein >150 mg/L, n (%) | 101 (16.80) | 69 (14.30) | 32 (27.10) | 0.001 |
| Lymphopenia <1000 cells/mm3, n (%) | 322 (53.70) | 245 (50.80) | 77 (65.30) | 0.005 |
| Neutrophilia >7500 cells/mm3, n (%) a599 | 119 (19.90) | 80 (16.60) | 39 (33.10) | <0.001 |
| Monocytopenia <200 cells/mm3, n (%) a599 | 41 (6.80) | 31 (6.40) | 10 (8.50) | 0.434 |
| Positive N-antigenemia (Abbott), n (%) | 332 (55.30) | 250 (51.90) | 82 (69.50) | <0.001 |
| RNAemia (YES), n (%) a581 | 199 (34.30) | 140 (29.90) | 59 (52.70) | <0.001 |
| Viral RNA load in plasma (copies N1/mL) a581 | 0.00 (209.92) | 0.00 (142.03) | 158.24 (932.45) | <0.001 |
| Viral RNA load in plasma (copies N2/mL) a581 | 0.00 (252.34) | 0.00 (187.50) | 134.52 (1206.57) | <0.001 |
| Seropositive for anti-SARS-CoV-2 N antibodies ≥1.4 AU/mL, n (%) a586 | 253 (43.20) | 217 (46.00) | 36 (31.60) | 0.005 |
| anti-SARS-CoV-2 N antibodies, AU/mL a586 | 0.68 (4.25) | 0.96 (4.49) | 0.19 (2.81) | 0.006 |
| Seropositive for anti-SARS-CoV-2 S1 antibodies ≥50 AU/mL, n (%) a586 | 285 (48.60) | 243 (51.50) | 42 (36.80) | 0.005 |
| anti-SARS-CoV-2 S1 antibodies, AU/mL a586 | 36.45 (362.17) | 61.10 (455.13) | 6.35 (178.25) | 0.001 |
| Treatments | ||||
| Remdesivir, n (%) | 58 (9.70) | 51 (10.60) | 7 (5.90) | 0.126 |
| Heparin, n (%) a599 | 440 (73.50) | 361 (74.90) | 79 (67.50) | 0.105 |
| Corticoids, n (%) a599 | 443 (74.00) | 351 (72.80) | 92 (78.60) | 0.199 |
| Tocilizumab, n (%) | 97 (16.20) | 80 (16.60) | 17 (14.40) | 0.562 |
| Azithromycin, n (%) | 270 (45.00) | 206 (42.70) | 64 (54.20) | 0.024 |
| Complications | ||||
| ARDS, n (%) | 150 (25.00) | 118 (24.50) | 32 (27.10) | 0.553 |
| Acute cardiac failure, n (%) ∗598 | 42 (7.00) | 26 (5.40) | 16 (13.80) | 0.001 |
| Acute renal failure, n (%) ∗598 | 35 (5.90) | 25 (5.20) | 10 (8.60) | 0.157 |
| Acute arrhythmia, n (%) | 43 (7.20) | 24 (5.00) | 19 (16.10) | <0.001 |
| Nosocomial infection, n (%) ∗598 | 66 (11.00) | 46 (9.50) | 20 (17.20) | 0.018 |
| ICU admission, n (%) | 57 (9.50) | 43 (8.90) | 14 (11.90) | 0.328 |
| Length of hospital stay, median days (IQR) | 8.00 (9.00) | 8.00 (7.00) | 10.50 (11.00) | 0.008 |
The continuous variables are represented as median (IQR) and the categorical variables as absolute count (%). The differences between groups were assessed using the chi-squared or Fisher's Exact Tests for the categorical variables and the Mann-Whitney U test for the continuous variables.
Abbreviations: ARDS, acute respiratory distress syndrome; ALT, alanine aminotransferase; AU: arbitrary units; ICU, intensive care unit; LDH, lactic acid dehydrogenase; MAP, mean arterial pressure; SOFA, Sequential Organ Failure Assessment.
aFor those variables with missing values, the sample size is detailed following the superscipt letter. Significant p values are highlighted in bold letter.
Results
Patients dying in the first 90 days after hospitalization (19.6%, 118/600) were older than the survivors, presented more frequently hypertension, cardiovascular disease, cerebrovascular disease, atrial fibrillation and renal disease (Table 1). Nonsurvivors arrived to the hospital earlier since the onset of the symptoms and presented with more severe disease, showing slightly higher Sequential Organ Failure Assessment (SOFA) scores. Of the patients, 9.5% (57/600) were transferred to the intensive care unit (ICU) over the course of hospitalization to the wards (Table 1).
The prevalence of N-antigenemia in the first 36 hours after hospitalization was higher in nonsurvivors (69% (82/118) vs. 52% (250/482); p < 0.001) who showed also higher viral RNA levels in plasma and lower concentrations of SARS-CoV-2 anti-N and anti-S1 antibodies (Table 1). Interestingly, the patients with N-antigenemia presented earlier at the hospital since disease onset (5 days vs. 6 days in median, p = 0.003), showed with more frequency viral sepsis at hospitalization (63.6% (211/332) vs. 48.1% (129/268); p < 0.001) (as defined by the SEPSIS-3 consensus) [8], along with higher levels of C-reactive protein (CRP) (81 [91] vs. 68 [108] mg/L; p = 0.050). Lactic acid dehydrogenase (LDH) (343 [273] vs. 297 [258] UI/L; p = 0.012), and lower concentrations of lymphocytes (0.8 [0·7] vs. 1.0 [0.7] × 1000 cells/mm3; p < 0.001), monocytes (0.4 [0.4] vs. 0·5 [0.4] cells/mm3; p < 0.001) and platelets (159 [73] vs. 223 [132] 1000 cells × 103/μL; p < 0.001) (values are provided as median [IQR]). Patients with N-antigenemia showed more frequently RNAemia, but were less frequently seropositive for anti-SARS-CoV-2 N and S1 antibodies (see Supplementary material, File 1). Developing ARDS was more common in patients with N-antigenemia (30.1% [100/332] vs. 18.7% [50/268]; p = 0.001). They also suffered more often from nosocomial infections (13.6% [45/331] vs. 7.9% [21/267]; p = 0.026).
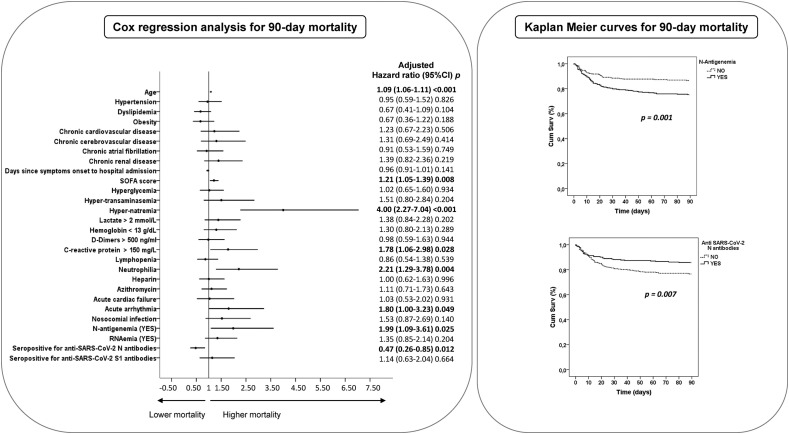
The multivariable analysis showed higher odds of 90-day mortality associated with the presence of N-antigenemia, whereas anti-SARS-CoV-2 N antibodies represented a protective factor (Fig. 1 and Supplementary materisl, File 2). N-antigenemia, or the absence of anti-N antibodies, translated into a significant reduction in survival time (Fig. 1). Other factors independently associated with mortality were age, Sequential Organ Failure Assessment score, hyper-natremia, high CRP or neutrophil levels, and developing an acute arrythmia (Fig. 1 and Supplementary material, File 2).
Fig. 1.
Left: Forest plot showing the adjusted HR from the Cox multivariate analysis to predict 90-day mortality (see Supplementary material, File 2). Right: Kaplan-Meier curves for 90-day mortality.
Discussion
The presence of SARS-CoV-2 N-antigenemia at admission to the hospital wards is a stand-alone predictor of 90-day mortality in COVID-19. Using either single molecule array, ELISA or CLEIA based tests, other authors had already evidenced the link between antigenemia and COVID-19 severity. Ogata et al. reported that high concentrations of S1 in plasma upon presentation to the hospital correlate with cases of COVID-19 requiring immediate intubation [6]. Perna et al. observed that the serum levels of SARS-CoV-2 N antigen were higher in COVID-19 patients admitted to ICU [7]. Wang et al. found that plasma antigen concentration at COVID-19 diagnosis was associated with ICU admission [9]. As far as we know, our study was the first in demonstrating higher odds of 90-day mortality associated to N-antigenemia. Antigenemia was accompanied by a number of signatures indicating severity—shorter course of the disease before hospitalization, higher frequency of viral sepsis at admission [10], and ARDS and nosocomial infections over the course of hospitalization, lower platelet, lymphocyte and monocyte counts, along with the activation of the inflammatory response paralleling tissue destruction, denoted by the presence of higher levels of CRP and LDH. Perna et al. had already reported that the concentration of N antigen in serum correlated with CRP levels in COVID-19 patients [7]. Olea et al. found significantly higher serum levels of ferritin, LDH, CRP, and D-dimers in ICU patients with positive SARS-CoV-2 N antigen in plasma [11]. Our results evidenced that patients with N-antigenemia admitted to the wards presented frequently with RNAemia and the absence of anti-SARS-CoV-2 antibodies, as reported also in critically ill COVID-19 patients [12]. This suggested that patients with N-antigenemia have impaired immune responses leading to uncontrolled viral replication. Interestingly, the presence of anti-N antibodies represented a protective factor against mortality.
We did not evaluate whether N-antigenemia responded to the presence of live virus in blood, although mounting evidence supports the infection of distant tissues by SARS-CoV-2 in some patients [[13], [14], [15]]. The results have to be validated also in the current scenario of predominant circulation of Omicron.
In summary, the presence of N-antigenemia or the absence of anti-SARS-CoV-2 N antibodies after hospitalization is associated to increased 90-day mortality in COVID-19. Detection of N-antigenemia by using lateral flow tests is a widely available tool that could contribute to early identify those patients at risk of deterioration. N-antigenemia could represent an important factor to understand the effect of antivirals in this disease.
Transparency declaration
AT, JME, FB, MDG, APT, RA and JFBM have a patent application on SARS-CoV-2 antigenemia. The remainder authors declare no conflicts of interest regarding this work.
Conflict of interest
The authors declare that they have no conflicts of interest.
Funding
This work was possible thanks to the financial support from Instituto de Salud Carlos III (Subvenciones de concesión directa para proyectos y programas de investigación del virus SARS-CoV-2, causante del COVID-19, FONDO - COVID19, code COV20/00110, Instituto de Salud Carlos III, CIBERES, 06/06/0028) (AT) co-funded by the European Social Fund (ESF) /“(A Way to Make Europe). The work was also supported by Fundació La Marató de TV3 (ajudes Econòmiques a Projectes de Recerca sobre Covid-19 - La Marató 2020, code 202108-30-31) (DdGC, JFBM), in addition by an ESCMID Research Grant 2020 (APT) and finally by Institut Català de la Salut and Gestió de Serveis Sanitaris (project COVIDPONENT) (FB). DdGC, AdF, and APT have received financial support from Instituto de Salud Carlos III (Miguel Servet 2020: CP20/00041/PFIS: FI20/00278 /Sara Borrell: CD18/00123), co-funded by the European Social Fund (ESF) /“A way to make Europe” /“Investing in your future”.
Author's contributions
RA, JFBM, JME, and DdGC designed the study. AT coordinated the study implementation. RLI, GT, TRA, JFA, AAD, JA, JGB, LI, FdC, and FB recruited the patients. LGF, ONGP, MJV, SC, AY, FRJ, and JG collected the samples. LGG, TLG, AMM, and CGP collected the clinical data. AdF, NJ, TP, AO, WT, MDG, and RA developed the laboratory works. NGM and APT analyzed the viral load in plasma. RA and JFBM performed the statistical analysis and wrote the manuscript. JFBM and LGG verified the data. All the authors critically revised the manuscript. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Acknowledgements
The authors want to thank Mr. Albert Gabarrus (statistician from Hospital Clinic/IDIBAPS, Barcelona, Spain) for his technical advice with the statistical analysis of this study.
Editor: Andre Kalil
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2022.05.023.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Li H., Gu X., Li H., Gong F., Xu J., Wang Y., et al. Risk factors of viral RNAaemia and its association with clinical prognosis among patients with severe COVID-19. Chest. 2021;159:1382–1386. doi: 10.1016/j.chest.2020.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bermejo-Martin J.F., González-Rivera M., Almansa R., Micheloud D., Tedim A.P., Domínguez-Gil M., et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care. 2020;24:691. doi: 10.1186/s13054-020-03398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang K., Wu L., Luo Y., Gong B. Quantitative assessment of SARS-CoV-2 RNAemia and outcome in patients with Coronavirus Disease 2019. J Med Virol. 2021;93:3165–3175. doi: 10.1002/jmv.26876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGonagle D., Kearney M.F., O’Regan A., O’Donnell J.S., Quartuccio L., Watad A., et al. Therapeutic implications of ongoing alveolar viral replication in COVID-19. Lancet Rheumatol. 2022;4:e135–e144. doi: 10.1016/S2665-9913(21)00322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birra D., Benucci M., Landolfi L., Merchionda A., Loi G., Amato P., et al. COVID 19: a clue from innate immunity. Immunol Res. 2020;68:161–168. doi: 10.1007/s12026-020-09137-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogata A.F., Maley A.M., Wu C., Gilboa T., Norman M., Lazarovits R., et al. Ultra-sensitive serial profiling of SARS-CoV-2 antigens and antibodies in plasma to understand disease progression in COVID-19 patients with severe disease. Clin Chem. 2020;66:1562–1572. doi: 10.1093/clinchem/hvaa213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perna F., Bruzzaniti S., Piemonte E., Maddaloni V., Atripaldi L., Sale S., et al. Serum levels of SARS-CoV-2 nucleocapsid antigen associate with inflammatory status and disease severity in COVID-19 patients. Clin Immunol. 2021;226 doi: 10.1016/j.clim.2021.108720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H., Hogan C.A., Verghese M., Solis D., Sibai M., Huang C., et al. SARS-CoV-2 nucleocapsid plasma antigen for diagnosis and monitoring of COVID-19. Clin Chem. 2021;68:204–213. doi: 10.1093/clinchem/hvab216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karakike E., Giamarellos-Bourboulis E.J., Kyprianou M., Fleischmann-Struzek C., Pletz M.W., Netea M.G., et al. Coronavirus Disease 2019 as cause of viral sepsis: a systematic review and meta-analysis. Crit Care Med. 2021;49:2042–2057. doi: 10.1097/CCM.0000000000005195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olea B., Albert E., Torres I., Gozalbo-Rovira R., Carbonell N., Ferreres J., et al. SARS-CoV-2 N-antigenemia in critically ill adult COVID-19 patients: frequency and association with inflammatory and tissue-damage biomarkers. J Med Virol. 2022;94:222–228. doi: 10.1002/jmv.27300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin-Vicente M., Almansa R., Martínez I., Tedim A.P., Bustamante E., Tamayo L., et al. Low anti-SARS-CoV-2 S antibody levels predict increased mortality and dissemination of viral components in the blood of critical COVID-19 patients. J Intern Med. 2022;291:232–240. doi: 10.1111/joim.13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schurink B., Roos E., Radonic T., Barbe E., Bouman C.S.C., de Boer H.H., et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe. 2020;1:e290–e299. doi: 10.1016/S2666-5247(20)30144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorward D.A., Russell C.D., Um I.H., Elshani M., Armstrong S.D., Penrice-Randal R., et al. Tissue-specific immunopathology in fatal COVID-19. Am J Respir Crit Care Med. 2021;203:192–201. doi: 10.1164/rccm.202008-3265OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Recalde-Zamacona B., García-Tobar L., Argueta A., Álvarez L., Andrea C.E.D., Alonso M.F., et al. Histopathological findings in fatal COVID-19 severe acute respiratory syndrome: preliminary experience from a series of 10 Spanish patients. Thorax. 2020;75:1116–1118. doi: 10.1136/thoraxjnl-2020-215577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.