SUMMARY
This study aims to describe in detail the temporal dynamics of E. coli O157 shedding and risk factors for shedding in a grass-fed beef herd. During a 9-month period, 23 beef cows were sampled twice a week (58 sampling points) and E. coli O157 was enumerated from faecal samples. Isolates were screened by PCR for presence of rfbE, stx1 and stx2. The prevalence per sampling day ranged from 0% to 57%. This study demonstrates that many members of the herd were concurrently shedding E. coli O157. Occurrence of rainfall (P < 0·01), feeding silage (P < 0·01) and lactating (P < 0·01) were found to be predictors of shedding. Moving cattle to a new paddock had a negative effect on shedding. This approach, based on short-interval sampling, confirms the known variability of shedding within a herd and highlights that high shedding events are rare.
Key words: Cattle, enumeration, Escherichia coli O157, longitudinal, risk factors, shedding
INTRODUCTION
Infection with Escherichia coli O157 in humans is uncommon but potentially fatal [1]. The organism is a common commensal in the gastrointestinal tract of cattle [2]. A great deal of research on E. coli O157 in cattle has been completed in the last 30 years [3–5], although it is uncertain whether this has resulted in lower rates of human illness. No decrease in reported clinical cases with E. coli O157 has been noted in Scotland over the last 10 years [6] and foodborne outbreaks still frequently occur in many countries. Whether humans become infected through foodborne or environmental pathways, cattle faeces are considered the primary source of infection [7]. Therefore understanding the dynamics of shedding of this organism in cattle and the factors that influence shedding are priorities for improving the prevention of human infection.
E. coli O157 is invariably detected in cattle populations that are subjected to sufficient sampling and testing. However, the prevalence of E. coli O157 within herds is variable, with estimates ranging from 0·7% to 28% in the USA [8, 9], 3·4–21·8% in the UK [6] and 1·9–13% in Australia [10, 11]. While some studies have described peaks in prevalence during late spring, summer and early autumn [12, 13], others, all carried out in Great Britain, report a lack of convincing seasonal pattern [14, 15]. Thus, whether or not shedding is a seasonal phenomenon is uncertain. In addition to variation in location, which may drive differences, most studies have not sampled for a sufficient length of time to allow them to conclude whether peaks in prevalence are part of an ongoing cyclical pattern or are simply a manifestation of the particular period of observation.
Describing the epidemiology of E. coli O157 in cattle is challenging because of the cost and inconvenience of measuring the concentration of the pathogen in faeces. Intermittent patterns of shedding have been shown in calves [16, 17], but little is known about the shedding pattern of live adult beef cattle. This is especially so for pasture-based production systems which are not well represented in the literature. While a few longitudinal studies have been performed in adult beef cattle [15, 18–22], data describing the shedding patterns of individual animals that have been repeatedly sampled at short time intervals (<7 days) are missing. Most longitudinal studies used fortnightly or monthly sampling intervals during a 3- to 12-month period [18, 19, 21]. Some studies have assessed the concentration of E. coli O157 in faeces and several others featured repeated observations on the same animals [15, 18, 19, 21]. A study that combines these aspects of repeated sampling and enumeration but with the repetition occurring at short time intervals will provide new and potentially useful information about shedding of E. coli O157 by cattle.
In this study 23 cattle were intensively studied over 9 months while managed in a temperate grazing system. They were repeatedly assessed at 3- to 7-day intervals for the presence and concentration of E. coli O157 in faeces. The overall objective was to describe in detail the temporal dynamics of E. coli O157 shedding in individual cattle managed on pasture and to identify possible risk factors for shedding of this pathogen.
METHODS
Beef herd characteristics
A longitudinal study was performed from 4 October 2012 to 20 June 2013. The study subjects comprised 23 primiparous and multiparous Hereford cattle pastured at Charles Sturt University's beef farm, located at Wagga Wagga, NSW, Australia (latitude 35° S, longitude 147° E). Rainfall is distributed relatively evenly at 48 mm/month. Average daily minimum and average daily maximum temperatures for summer are 16·2 °C and 31·2 °C and for winter 2·7 °C and 12·5 °C, respectively. From October to 28 March the cattle were sampled twice a week, which was reduced to once a week during the months of April, May, and June because of a reduction in the number of cattle shedding (⩽1 animal per day) on three sampling occasions in the last weeks of March.
During the study period the cattle were grazed on different paddocks, varying between 3·5 ha and 7 ha in area, according to the availability of grass, herd nutritional requirements and environmental management considerations. The cattle were fed cereal and ryegrass silage every Monday, Wednesday and Friday when no fresh grass was available, starting from 30 December 2012, and continuing until the last sampling day (Fig. 2). The herd was used biweekly in March and bimonthly in April and May by veterinary students of the university for teaching purposes. Twenty-one cows and heifers calved in July 2012, including two pairs of twins, resulting in 23 calves at foot and nursing until weaning on 24 January 2013. Thus the calves were only present for part of the study period.
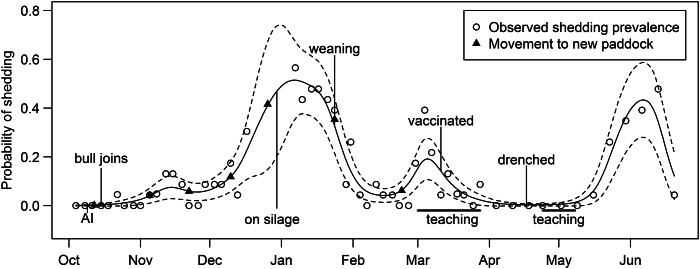
Fig. 2.
Temporal change in probability of animals shedding E. coli O157 (and 95% confidence intervals), the timing of management variables and the timing of movement of animals between paddocks.
Ethical standards
The use of animals in this study was approved by Charles Sturt University Animal Care and Ethics Committee Protocol number 12/060.
Sample collection
Animals
A minimum of 10 g of faeces were collected from each cow by rectal palpation or during defecation while the cows were restrained in a crush in the cattle yards. For each cow a new disposable sleeve glove was used and each faecal sample was individually placed into a Whirl-Pak bag (Nasco, Australia). Transportation occurred on ice within 2 h of the first sample being taken and samples were processed directly upon arrival at the laboratory.
Rectal temperature, faecal score and hide contamination were recorded for each cow at every sampling. Rectal temperature was measured after the faecal grab was taken. Faecal score was categorized from 1 to 4 (runny/loose/soft/dry) [23] and hide contamination from 1 to 5 (clean and dry/slightly dirty/dirty/very dirty/extremely dirty and wet) [24]. Weight (kg) and body condition score (score 1–5) [25] were recorded every 14 days. The first author undertook all sampling and data recording.
Environment
Rainfall (mm in 24 h prior to sampling), ambient temperature (mean °C in 24 h prior to sampling), day length (hours between sunrise and sunset on the day of sampling), humidity (% relative humidity at 09:00 hours) and hours of bright sunshine (bright sunshine in the 24 h to midnight prior to sampling) were obtained for each day throughout the study from the Bureau of Meteorology (BOM). Rainfall data was obtained from the BOM weather station located <2 km from the herd location, while all other data was retrieved from the BOM weather station located <15 km away. The quantity and quality of the pastures were recorded every 14 days. Quantity was measured by assessing the average height, using methods described by Morris & Kenyon [26]. Quality was determined by the stage of pasture growth, e.g. short actively growing vegetative pasture (80%), tall actively growing vegetative pasture (75–80%), early flowering (70–75%) or late flowering (65–70%). Contamination of drinking water with E. coli O157 was analysed coinciding with the pasture estimates. Water samples, from all drinking-water troughs in the paddock used at the time of sampling, were collected into 120 ml sterile containers (Sarstedt, Australia), directly transported to the laboratory and analysed together with the faecal samples.
Management
Management variables that changed during the period of study were recorded. This included feeding of supplements, artificial insemination, joining with a bull, weaning of calves, spread of fertilizer on the pastures, relocation to other paddocks, medical treatments and vaccinations. The dates of veterinary teaching classes (teaching of cow handling, claw trimming and casting) were recorded.
Sample processing
Faecal samples
Ten grams of faeces from each sample were homogenized in 90 ml of sterile buffered peptone water (BPW) (Oxoid, Australia). For each homogenized broth 100 μl was plated directly on sorbitol MacConkey+5-bromo-4-chloro-3-indolyl-b-d-glucuronide agar (Oxoid) containing cefixime (0.05 mg/l) and potassium tellurite (2.5 mg/l) (Oxoid) (CT-SMAC+BCIG) and incubated for 18–24 h at 37 °C for direct culture [27]. Each homogenized broth was incubated at 37 °C for 6 h [28]. The enriched broth was stored at 4 °C until immunomagnetic separation (IMS) was performed the next day.
Confirmation and storage
After incubation of the CT-SMAC+BCIG plates, all plates were screened for non-sorbitol fermenting and β-glucuronidase-negative colonies (straw-coloured colonies). Up to ten suspect colonies were tested using an E. coli O157 latex test (Oxoid) to confirm the colonies as O157 or otherwise [29]. When positive colonies on direct culture plates were found, the number of colony-forming units (c.f.u.) was counted and the c.f.u./g of faeces estimated after accounting for the dilution factor. Pure isolates were stored at −80 °C in BPW with 20% glycerol for later analysis by polymerase chain reaction (PCR).
Water samples
The water samples were filtered with cellulose nitrate membrane filters with pore size 0.2 μm (Bio-Rad, Australia). The entire filters were added to 90 ml sterile BPW, direct plated and enriched as described for the faecal samples.
IMS
For all faecal and water samples that were negative after direct plating, manual IMS was performed. Two ml of each enriched sample was centrifuged for 2 min at 52 g prior to IMS. A 24-well plate was set up with 20 μl of anti E. coli O157 immunomagnetic beads (Invitrogen, Australia). One thousand μl of the supernatant of each enriched centrifuged sample was transferred to a well [11] and thoroughly mixed with the beads on a gyratory mixer (Ratek, Australia) for 1 h. The plate was then placed on a magnet for 2 min to immobilize the beads after which the enrichment solution was removed while the plate remained on the magnet. After removing the plate from the magnet, the beads were washed in 1000 μl of phosphate buffered saline (PBS) (MP Biomedicals, Australia) with 0·05% Tween 20 (Sigma, Australia) (PBST). The solution was mixed for 2 min and placed back on the magnet for 2 min to immobilize the beads. Again the wash solution was removed and the wash step repeated. After the second wash the beads were removed from the magnet and re-suspended in 100 μl PBST. The resulting solution was split and inoculated onto CT-SMAC + BCIG plates and incubated for 18–24 h at 37 °C [11, 28].
PCR
All isolates were screened by PCR for the presence of rfbE, which encodes the E. coli O157 serotype [30], and for the virulence genes stx1 and stx2 [31] (Table 1). Colony PCR was performed on samples, using OneTaq DNA polymerase (New England Biolabs, USA). A multiplex PCR assay was set up for detection of the above-mentioned genes using a multiplex PCR plus kit (Qiagen, Australia). Thermocycling was performed in a Bio-Rad S1000 Thermal Cycler (Bio-Rad) following the cycling conditions from Paton & Paton [31]. PCR products were then visualized on a 2% agarose gel containing SYBR Safe DNA gel stain (Invitrogen).
Table 1.
Primers used for detection of E. coli O157 genes
| Primer | Sequence | Target gene | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| rfbE_F | CTACAGGTGAAGGTGGAATGG | rfbEO157 | 327 | Wang et al. [30] |
| rfbE_R | ATTCCTCTCTTTCCTCTGCGG | |||
| Stx1_F | ATAAATCGCCATTCGTTGACTAC | stx1 | 180 | Paton & Paton [31] |
| Stx1_R | AGAACGCCCACTGAGATCATC | |||
| Stx2_F | GGCACTGTCTGAAACTGCTCC |
stx2 (including stx2 variants) |
255 | |
| Stx2_R | TCGCCAGTTATCTGACATTCTG |
Descriptive analysis
Graphical analysis was performed to assess relationships between the probability of cattle shedding E. coli O157 and a range of animal and environmental variables. Data describing each individual animal's shedding status at each sampling point was summarized for the herd to give the observed proportion of cattle shedding E. coli O157. These proportions were then fitted to a cubic spline model to provide a smoothed curve (and accompanying 95% confidence interval) defining the probability of shedding through time and providing a basis for visually assessing the relationship between various factors (animal, environment, management) and the occurrence of shedding (Figs 3–5). The graph displaying probability of shedding through time was annotated with the timing of key management events such as movement to new paddocks, weaning, vaccination and handling (Fig. 2).
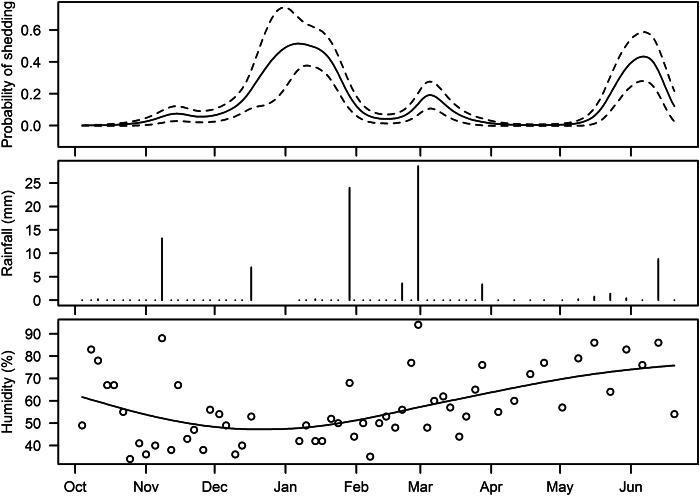
Fig. 3.
Temporal change in probability of animals shedding E. coli O157, rainfall (mm) in the 24 h prior to sampling, and relative humidity (%) at 09:00 hours on the day of sampling.
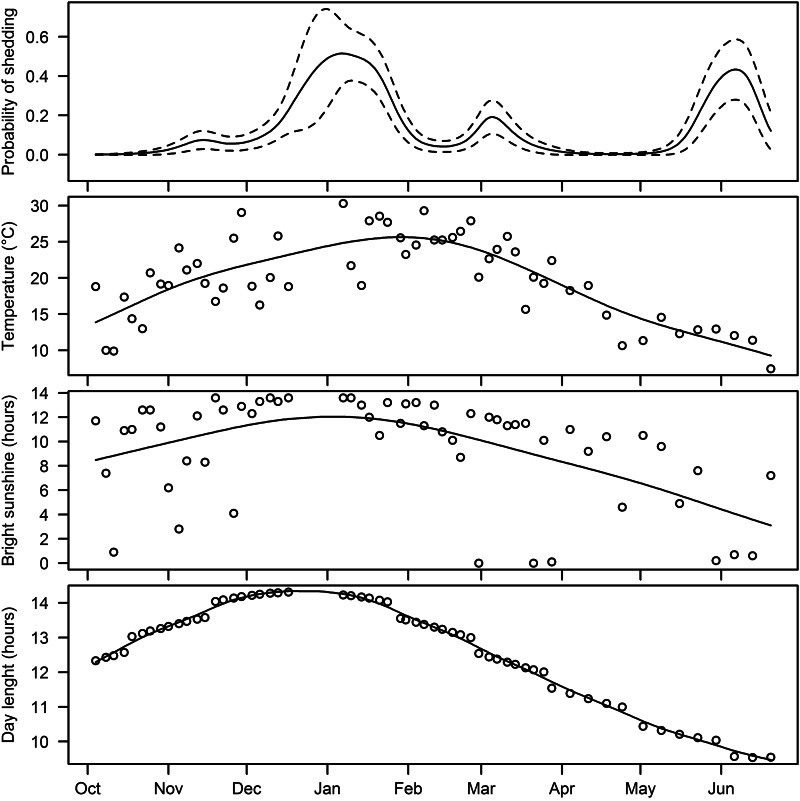
Fig. 4.
Temporal change in probability of animals shedding E. coli O157, mean environmental temperature (°C) in the 24 h to 09:00 hours, bright sunshine (hours) in the 24 h to midnight prior to sampling, and day length on the day of sampling.
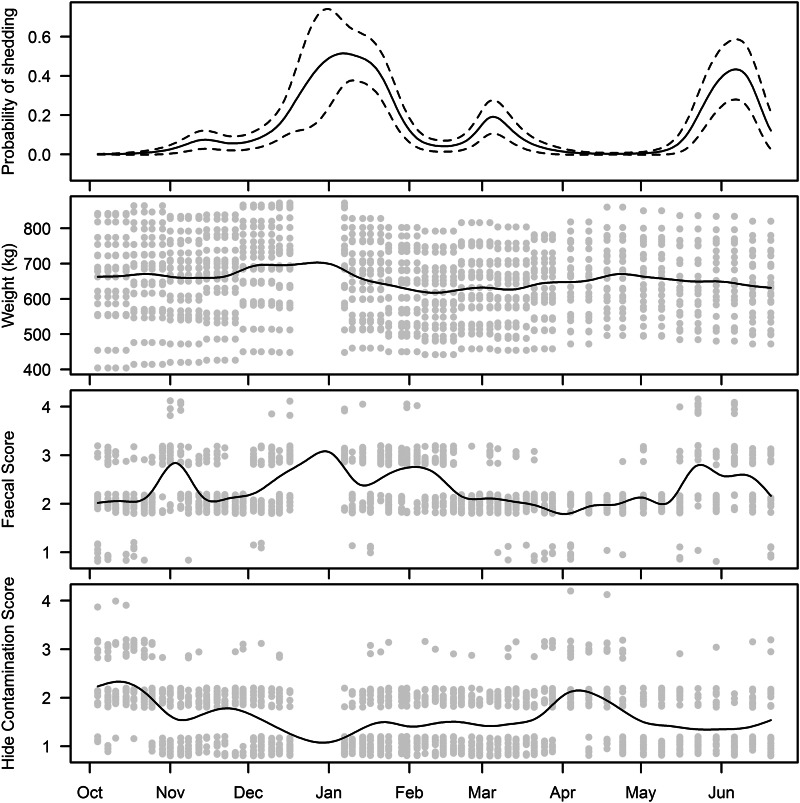
Fig. 5.
Temporal change in probability of animals shedding E. coli O157, mean body weight, mean faecal score, and mean hide contamination score, accompanied by the data for each individual animal.
Statistical analysis
Herd level
Trend in the proportion of animals testing positive to E. coli O157 on each sampling day was estimated by a cubic spline model. Visual assessment of this curve, overlaid with animal, environment and management variables, was performed to explore any putative associations in the first instance. Visual analysis of this curve revealed that one minor and three substantial discrete peaks in shedding occurred over the period under study. The three events with estimated peak proportion of herd shedding at >20% were described as a ‘herd-shedding event’ and defined as the period where the estimated probability of shedding exceeded 0·1. Reassessing the data in light of herd-shedding events allowed all sampling points (days on which sampling occurred) to be classified as occurring within six discrete events comprised of three shedding events alternating with three non-shedding events.
Generalized linear mixed models were used to assess whether variation in each animal, environmental and management variable at each sampling point could be associated with a period of time identified previously as a ‘herd-shedding event’. For animal-level variables [hide contamination score, faecal consistency score, body weight, body condition score, rectal temperature, pregnancy status (Y = 1/N = 0) and lactating (Y = 1/N = 0)], the animal-level variable was specified as the outcome, while herd-shedding event (Y/N) was included as a fixed effect. The event number (1–6) and cow identifier (1–23) within each event were estimated as random effects reflecting the nested sampling structure of observations within cows within events. For management and environmental variables taken at the herd level [movement between paddocks (Y = 1/N = 0), feeding of silage (Y = 1/N = 0), handled by students (Y = 1/N = 0), calves at foot (Y = 1/N = 0), quality and quantity of the pasture, rainfall, humidity, temperature, hours of bright sunshine and day length] herd-shedding event was estimated as a fixed effect and event number as a random effect. These model structures were adopted in order to account for the inclusion of herd-specific random effects and to allow for more biologically meaningful interpretation of coefficients. Null hypothesis significance tests for the fixed effects were conducted by calculation of the analysis of variance table for each model. Least squares estimates and standard errors of average responses under each shedding event class were calculated from each model.
Individual animal level
Logistic regression analysis was used to explore the effect of potential animal-, environmental- and management-level risk factors on individual animal shedding. First, univariable analyses were performed. Variables with P < 0·05 were jointly analysed using a forward selection procedure in multiple regressions. As repeated observations of individual animals may not be independent, a random effect of cow identifier was included.
Based on the estimated counts from faecal samples, animals were categorized as negative, low-level (<103 c.f.u./g) or high-level (⩾103 c.f.u./g) shedders of E. coli O157. These data were analysed by using a generalized linear model with presence of a high shedding as the binomial outcome, low shedding as a fixed effect and cow identifier as a random effect.
RESULTS
Descriptive and statistical analysis
During the 9-month study period, a total of 1323 faecal samples were collected from 23 Hereford cows over 58 sampling days. E. coli O157 was isolated from 168 (12·7%) samples with 21 (1·6%) positive on direct enumeration plates and 147 (11·1%) positive only through IMS. The amount of E. coli O157 shed in faeces ranged from <100 to 20 700 c.f.u./g faeces. All cows within the herd shed E. coli O157 at least once during this study (Fig. 1). A large variation in the frequency of shedding among animals was identified. E. coli O157 was shed intermittently by all animals and the number of occasions on which an individual cow was identified as shedding E. coli O157 ranged between 2 and 15 (out of 58 sampling points in total). The maximum number of consecutive sampling points that an animal was found to be positive (by either direct culture or IMS) was seven (3·5 weeks). The prevalence per sampling day ranged from 0% to 56·5% (Fig. 2). Out of 168 isolates screened by PCR, all were positive for the O157 rfb gene, 166 (98·8%) were positive for stx1 and stx2, and two (1·2%) were positive for stx1 only.
Fig. 1.
Plot of the individual shedding patterns. Samples in which E. coli O157 was detected only by immunomagnetic separation (grey dots) are represented as 50 c.f.u./g for graphical purposes. Sampling points for which enumeration was possible are indicated by black dots and samples in which no E. coli O157 was detected are represented by white dots.
E. coli O157 was isolated from 1 of 62 water samples that were collected during the study. The isolate identified within water tested positive for the O157 rfb, stx1 and stx2 genes by PCR. This positive water sample was found on 11 November 2012 when none of the cows were positive for E. coli O157.
Temporal data on shedding events and management events are shown in Figure 2. This figure shows three distinct peaks in probability of shedding. There was a major peak in mid-summer (mid-December to late January), a minor peak in autumn (late February to mid-March) and a second major peak in early winter (late May to late June). None of the management variables appeared to be associated with any of the peaks in shedding.
Time-based change in the probability of shedding is also shown in Figure 3 accompanied by the temporal patterns of rainfall and humidity. Although all of the peaks in shedding are immediately preceded by rainfall this can also be said for many periods where shedding was not detected. Thus, descriptive evidence alone does not support a relationship between variation in rainfall and the presence or absence of a ‘herd-shedding event’. However, the generalized linear mixed model showed that rainfall is a risk factor (P = 0·03): the mean of rainfall in the 24 h prior to sampling during herd-shedding events (3·4 mm) differed significantly from that for non-shedding events (0·6 mm) (Table 2). Visually, there was no apparent association between humidity and shedding, which was confirmed by statistical analysis.
Table 2.
Variables relating animal, management and environmental factors with the mean and standard error (s.e.) for shedding vs. non-shedding events and the P value
| Variable | Shedding events mean (s.e.) | Non-shedding events mean (s.e.) | P value |
|---|---|---|---|
| Animal | |||
| Hide contamination (1/2/3/4/5) | 1·39 (0·08) | 1·69 (0·08) | 0·04 |
| Faecal score (1/2/3/4) | 2·42 (0·15) | 2·21 (0·14) | 0·22 |
| Temperature | 37·95 (0·06) | 37·97 (0·06) | 0·81 |
| Pregnant (yes/no) | 1·61 (0·06) | 1·60 (0·06) | 0·39 |
| Lactating (yes/no) | 0·25 (0·28) | 0·30 (0·28) | 0·88 |
| Weight | 644·53 (12·28) | 648·05 (12·26) | 0·77 |
| Body condition score (1/2/3/4/5) | 3·87 (0·08) | 3·89 (0·08) | 0·63 |
| Management | |||
| Move paddock (yes/no) | 0·09 (0·07) | 0·11 (0·05) | 0·88 |
| Silage (yes/no) | 0·91 (0·25) | 0·66 (0·25) | 0·43 |
| Handled by students (yes/no) | 0·13 (0·16) | 0·20 (0·15) | 0·69 |
| Calves at foot (yes/no) | 0·27 (0·30) | 0·33 (0·30) | 0·88 |
| Pasture quality (0/65/80) | 11·32 (16·21) | 22·00 (16·01) | 0·50 |
| Pasture quantity | 107·70 (221·93) | 305·73 (219·66) | 0·43 |
| Environment | |||
| Rainfall previous 24 h (mm) | 3·35 (1·12) | 0·58 (0·84) | 0·03 |
| Mean environmental temperature | 19·72 (3·64) | 20·34 (3·59) | 0·73 |
| Day length | 12·09 (1·02) | 12·62 (1·02) | 0·96 |
| Sun | 8·61 (2·12) | 9·48 (2·06) | 0·90 |
| Humidity | 60·38 (6·25) | 57·64 (5·90) | 0·99 |
Temporal change in the probability of shedding in Figure 4 is accompanied by the temporal patterns of temperature, hours of bright sunshine and day length. All three variables showed a similar temporal curve. Based on this descriptive evidence the variation in the environmental variables could not be explained by the presence or absence of a ‘herd-shedding event’. This was supported by statistical analysis in which none of these variables was a significant risk factor (Table 2).
Time-based change in the probability of shedding is shown in Figure 5 together with the temporal patterns of the individual animal variables: body weight, faecal consistency and hide contamination. Visually, there was no apparent association between any of the three variables and the probability of shedding E. coli O157. Statistical analysis of the animal variables in a generalized linear mixed model showed that neither the variation in live weight nor in faecal consistency was associated with the presence or absence of a ‘herd-shedding event’ (Table 2). The average level of hide contamination score during shedding events (1·4) did differ from non-shedding events (1·7) (P = 0·04).
Univariable analysis to explore potential risk factors associated with individual animal shedding resulted in eight variables that were jointly analysed using multiple regression. The final logistic model is shown in Table 3. Four variables remained in the final model. In order of statistical significance the risk factors were: feeding of silage (P < 0·01), lactating (P < 0·01), rainfall in the week prior to sampling (P < 0·01) and movement between paddocks (P = 0·04).
Table 3.
Final model from multiple regression on individual animal level
| Variable | Estimated effect (s.e.) | OR | 95% CI | P value |
|---|---|---|---|---|
| Silage (yes/no) | 2·36 (0·273) | 10·56 | 6·18–18·03 | <0·001 |
| Lactating (yes/no) | 2·07 (0·225) | 7·89 | 5·08–12·26 | <0·001 |
| Rainfall previous week (mm) | 0·67 (0·214) | 1·95 | 1·28–2·97 | 0·002 |
| Move paddock (yes/no) | −1·12 (0·531) | 0·33 | 0·12–0·92 | 0·035 |
OR, Odds ratio; CI, confidence interval; s.e., standard error.
In total, E. coli O157 was isolated from 168 samples on 38 of the 58 sampling days. Analysis showed that the presence of a high-level shedder was significantly (P = 0·04) associated with a higher proportion of low-shedding animals on the same day (odds ratio 1·3, standard error 0·14).
DISCUSSION
The current study focused on the temporal dynamics of E. coli O157 shedding in adult grass-fed beef cattle. Although marked variation of shedding between and within individuals occurred, an obvious pattern in shedding over time was identified. The outstanding feature of the variation in the probability of shedding was the three distinctive peaks indicating the herd experienced discrete shedding events where many animals were concurrently shedding, referred to as synchronization of shedding. Previous studies have reported peaks of shedding in summer and early autumn, which is in agreement with the first two peaks in shedding in the current study; however, the third peak of shedding occurred in winter. Most longitudinal studies performed previously sampled only once a month [18, 20], which only provides crude information on temporal shedding patterns within a herd. Our findings suggest that if more frequent sampling had been used in earlier studies, e.g. twice a week, a different seasonal pattern might have been detected. Moreover, with the use of individual animal data it is evident that the synchronization of shedding in cows is accompanied by a marked elevation in the concentration of the pathogen in faeces. This indicates that shedding at high levels does not occur independently of other animals shedding low levels of the same pathogen within the herd. Previous studies have also observed an association between the presence of a high-level shedder in the herd and a high proportion of low shedders [32, 33]. Whether high-level shedding is the cause of low-level E. coli O157 shedding, or vice versa, is not known.
Intermittent shedding patterns have also been reported previously in calves [16, 17]. However, in this study very few cattle shed at high levels and the duration was very short. Robinson and colleagues [16] observed animals shedding at high levels for extended periods but this study looked at a group of calves purposely selected because they were known to be shedding. The current study is unique because it studies shedding and non-shedding animals in detail over time in a natural setting. There are two other notable explanations for the lower frequency in high-level shedding and the lower concentrations found in this study compared to other studies. One is an effect due to the age of the animals, since younger animals (aged 2–6 months) are associated with a higher prevalence of E. coli O157 than adults [34]. Second, the majority of previous studies involved feedlot cattle, where the high density of housing is likely to increase animal-to-animal transmission [35], compared to animals at pasture. Next to this, E. coli O157 in cattle faeces is not evenly distributed and its density can vary between sites in a faecal pat [36], which might influence the concentration of the organism.
Previously, only Williams et al. [37] identified an association between increasing rainfall and increased shedding of E. coli O157 in calves in Sydney, Australia (a higher rainfall environment compared to the present study). Rainfall events could feasibly promote shedding by making conditions beneficial for multiplication of the bacteria in the environment, changing pasture conditions to increase exposure of cattle to E. coli O157, or by somehow affecting forage composition and nutrition leading to alterations in the gut favouring proliferation of E. coli O157. Previous research in other countries has not found a relationship between rainfall and shedding. However, these studies did not rely on short-interval sampling and were conducted in different climates and production systems.
The negative association between hide score and probability of shedding in this study was unexpected and does not appear to be biologically plausible. It is very likely that this association is a spurious finding arising from either chance or measurement error.
Feeding silage was demonstrated to be associated with E. coli O157 shedding at the individual animal level. Many studies on diet and E. coli O157 shedding have been performed, often presenting contradictory results. Whether the significant association with silage found in the present study is due to a direct effect of silage on the microbiome or due to changes in the environment (e.g. drought) which were the reason for feeding silage, is not known.
Lactating was also positively associated with shedding of the pathogen. This effect has not been reported in literature before. It is possible that stress or hormonal changes influence E. coli O157 shedding cattle.
This study found that when animals were moved to another paddock in the week prior to sampling they were less likely to shed E. coli O157. This might have resulted from the absence of fresh faecal pats in a new paddock and reduced E. coli O157 contamination.
The advantage of using a smoothed curve (rather than raw data on animal shedding) is that it takes account of noise due to sampling variation arising from the small number of individuals, and, at each time point it uses data from proximal time points to support the estimation of probability of shedding [38]. In this way, it was possible to avoid potential false-positive significant correlations due to chance associations that frequently occur when comparing two time series of events [39]. It must be noted that when analysis of variance was applied on score variables, such as hide contamination score, that these scores should not be seen as ‘ranks', but as proxy measurements for an underlying true scale of dirtiness, and averages and standard errors do supply useful information.
Because this study was based on a single herd, care must be taken when extrapolating interpretation to the broader population of cattle. Nevertheless, the major findings are helpful because, for example, they suggest that shedding patterns vary substantially between production systems (e.g. pasture beef, dairy, feedlot beef), and that animals within a herd can be synchronized in their shedding (and non-shedding) which may have practical implications for the risk of human exposure to E. coli O157. Moreover, the extent of individual animal variability revealed in this work and in previous studies [16, 40] casts doubt on findings of risk factor and intervention studies where outcomes are based on single assessment of dichotomous shedding status. Findings here have also highlighted the lack of precision in the present understanding of shedding patterns in beef cattle and indicate the need to now focus on describing the extent of daily variation in shedding within individual animals.
ACKNOWLEDGEMENTS
The authors acknowledge farm manager Jim Mellor for his cooperation throughout study, and the assistance of James Stephens and Tony Hobbs (Charles Sturt University) in sampling of the animals. We are grateful to Franziska Pilger and Saliya Gurusinghe for technical assistance and Robert Barlow for supplying E. coli O157 and Stx-positive isolates.
This work was supported by Meat and Livestock Australia Ltd (grant number A.MFS.0247).
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Pennington H. Escherichia coli O157. Lancet 2010; 376: 1428–1435. [DOI] [PubMed] [Google Scholar]
- 2.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clinical Microbiology Reviews 1998; 11: 142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur TM, et al. Super shedding of Escherichia coli O157:H7 by cattle and the impact on beef carcass contamination. Meat Science 2010; 86: 32–37. [DOI] [PubMed] [Google Scholar]
- 4.Chase-Topping M, et al. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nature Reviews Microbiology 2008; 6: 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng J, et al. Enterohemorrhagic Escherichia coli. In: Doyle M, Buchanan R, eds. Food Microbiology: Fundamentals and Frontiers, 4th edn. Washington: ASM Press, 2013, pp. 287–309. [Google Scholar]
- 6.Pearce MC, et al. Temporal and spatial patterns of bovine Escherichia coli O157 prevalence and comparison of temporal changes in the patterns of phage types associated with bovine shedding and human E. coli O157 cases in Scotland between 1998–2000 and 2002–2004. BMC Microbiology 2009; 9: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman PA, et al. Cattle as a possible source of verocytotoxin-producing Escherichia coli O157 infections in man. Epidemiology and Infection 1993; 111: 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elder RO, et al. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proceedings of the National Academy of Sciences USA 2000; 97: 2999–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeJeune JT, et al. Comparison of E. coli O157 and Shiga toxin-encoding genes (stx) prevalence between Ohio, USA and Norwegian dairy cattle. International Journal of Food Microbiology 2006; 109: 19–24. [DOI] [PubMed] [Google Scholar]
- 10.Cobbold R, Desmarchelier P. A longitudinal study of Shiga-toxigenic Escherichia coli (STEC) prevalence in three Australian dairy herds. Veterinary Microbiology 2000; 71: 125–137. [DOI] [PubMed] [Google Scholar]
- 11.Fegan N, et al. The prevalence and concentration of Escherichia coli O157 in faeces of cattle from different production systems at slaughter. Journal of Applied Microbiology 2004; 97: 362–370. [DOI] [PubMed] [Google Scholar]
- 12.Barkocy-Gallagher GA, et al. Seasonal prevalence of Shiga toxin-producing Escherichia coli, including O157:H7 and non-O157 serotypes, and Salmonella in commercial beef processing plants. Journal of Food Protection 2003; 66: 1978–1986. [DOI] [PubMed] [Google Scholar]
- 13.Heuvelink AE, et al. Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. Journal of Clinical Microbiology 1998; 36: 3480–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis-Iversen J, et al. Temporal patterns and risk factors for Escherichia coli O157 and Campylobacter spp. in young cattle. Journal of Food Protection 2009; 72: 490–496. [DOI] [PubMed] [Google Scholar]
- 15.Smith RP, Paiba GA, Ellis-Iversen J. Longitudinal study to investigate VTEC O157 shedding patterns in young cattle. Research in Veterinary Science 2010; 88: 411–414. [DOI] [PubMed] [Google Scholar]
- 16.Robinson SE, et al. Intermittent and persistent shedding of Escherichia coli O157 in cohorts of naturally infected calves. Journal of Applied Microbiology 2004; 97: 1045–1053. [DOI] [PubMed] [Google Scholar]
- 17.Widiasih DA, et al. Duration and magnitude of faecal shedding of Shiga toxin-producing Escherichia coli from naturally infected cattle. Epidemiology and Infection 2004; 132: 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arthur TM, et al. Longitudinal study of Escherichia coli O157:H7 in a beef cattle feedlot and role of high-level shedders in hide contamination. Applied and Environmental Microbiology 2009; 75: 6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobbold RN, et al. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Applied and Environmental Microbiology 2007; 73: 1563–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo S, et al. Longitudinal prevalence and molecular typing of Escherichia coli O157:H7 by use of multiple-locus variable-number tandem-repeat analysis and pulsed-field gel electrophoresis in fecal samples collected from a range-based herd of beef cattle in California. American Journal of Veterinary Research 2010; 71: 1339–1347. [DOI] [PubMed] [Google Scholar]
- 21.Lahti E, et al. Longitudinal study of Escherichia coli O157 in a cattle finishing unit. Applied and Environmental Microbiology 2003; 69: 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeJeune J, et al. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Applied and Environmental Microbiology 2004; 70: 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alberta Dairy Management (ADM). (http://www.agromedia.ca/ADM_Articles/content/manure.pdf). Accessed 30 November 2013.
- 24.Food Standards Agency (FSA) report. (http://www.food.gov.uk/multimedia/pdfs/publication/redmeatsafety.pdf). Accessed 30 November 2013.
- 25.Department of Agriculture Fisheries and Forestry (DAFF). (http://www.daff.qld.gov.au/__data/assets/pdf_file/0015/53520/Animal-HD-Investigation-Condition-scores.pdf). Accessed 18 June 2014.
- 26.Morris S, Kenyon P. The effect of litter size and sward height on ewe and lamb performance. New Zealand Journal of Agricultural Research 2004; 47: 275–286. [Google Scholar]
- 27.Okrend A, Rose B, Lattuada C. Use of 5-bromo-4-chloro-3-indoxyl-beta-d-glucuronide in MacConkey sorbitol agar to aid in the isolation of Escherichia coli O157:H7 from ground beef. Journal of Food Protection (USA) 1990; 53: 941–943. [DOI] [PubMed] [Google Scholar]
- 28.Dunn JR, Keen JE, Thompson RA. Prevalence of Shiga-toxigenic Escherichia coli O157:H7 in adult dairy cattle. Journal of the American Veterinary Medical Association 2004; 224: 1151–1158. [DOI] [PubMed] [Google Scholar]
- 29.Cernicchiaro N, et al. Assessment of diagnostic tools for identifying cattle shedding and super-shedding Escherichia coli O157:H7 in a longitudinal study of naturally infected feedlot steers in Ohio. Foodborne Pathogens and Disease 2011; 8: 239–248. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, Clifford GC, Frank GR. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. Journal of Clinical Microbiology 2002; 40: 3613–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx 1, stx 2, eaeA, enterohemorrhagic E. coli hlyA, rfb O111, and rfb O157. Journal of Clinical Microbiology 1998; 36: 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chase-Topping ME, et al. Risk factors for the presence of high-level shedders of Escherichia coli O157 on Scottish farms. Journal of Clinical Microbiology 2007; 45: 1594–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Low JC, et al. Rectal carriage of enterohemorrhagic Escherichia coli O157 in slaughtered cattle. Applied and Environmental Microbiology 2005; 71: 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paiba GA, et al. Prevalence of faecal excretion of verocytotoxigenic Escherichia coli O157 in cattle in England and Wales. Veterinary Record 2003; 153: 347–353. [DOI] [PubMed] [Google Scholar]
- 35.Jeon SJ, et al. Evaluation of animal genetic and physiological factors that affect the prevalence of Escherichia coli O157 in cattle. PLoS ONE 2013; 8: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearce M, et al. Temporal shedding patterns and virulence factors of Escherichia coli serogroups O26, O103, O111, O145, and O157 in a cohort of beef calves and their dams. Applied and Environmental Microbiology 2004; 70: 1708–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams KJ, et al. Risk factors for Escherichia coli O157 shedding and super-shedding by dairy heifers at pasture. Epidemiology and Infection 2015; 143: 1004–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verbyla AP, et al. The analysis of designed experiments and longitudinal data by using smoothing splines. Journal of the Royal Statistical Society: Series C (Applied Statistics) 1999; 48: 269–311. [Google Scholar]
- 39.Aldrich J. Correlations genuine and spurious in Pearson and Yule. Statistical Science 1995; 10: 364–376. [Google Scholar]
- 40.Matthews L, et al. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proceedings of the National Academy of Sciences USA 2006; 103: 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]