SUMMARY
Presence of Vibrio cholerae serogroups O1 and O139 in the waters of the rural area of Matlab, Bangladesh, was investigated with quantitative measurements performed with a portable flow cytometer. The relevance of this work relates to the testing of a field-adapted measurement protocol that might prove useful for cholera epidemic surveillance and for validation of mathematical models. Water samples were collected from different water bodies that constitute the hydrological system of the region, a well-known endemic area for cholera. Water was retrieved from ponds, river waters, and irrigation canals during an inter-epidemic time period. Each sample was filtered and analysed with a flow cytometer for a fast determination of V. cholerae cells contained in those environments. More specifically, samples were treated with O1- and O139-specific antibodies, which allowed precise flow-cytometry-based concentration measurements. Both serogroups were present in the environmental waters with a consistent dominance of V. cholerae O1. These results extend earlier studies where V. cholerae O1 and O139 were mostly detected during times of cholera epidemics using standard culturing techniques. Furthermore, our results confirm that an important fraction of the ponds’ host populations of V. cholerae are able to self-sustain even when cholera cases are scarce. Those contaminated ponds may constitute a natural reservoir for cholera endemicity in the Matlab region. Correlations of V. cholerae concentrations with environmental factors and the spatial distribution of V. cholerae populations are also discussed.
Key words: Cholera, infectious disease epidemiology, investigation, Vibrio cholera
INTRODUCTION
In light of recent cholera epidemics that have struck Haiti [1, 2], South American and African countries [3], tools for providing quick and reliable monitoring of surface waters for pathogens can be of vital importance. These tools could help understand local epidemiological dynamics, and may play an important role in the monitoring and control of epidemics. The endeavour of tracking local concentrations of possibly toxigenic Vibrio cholerae O1 and O139 populations is also important to pinpoint specific areas of aquatic contamination in order to either restrict access to them, or to eradicate the bacteria from those sites by improving sanitation conditions.
Standard methods, in particular culturing techniques, have often proved ineffective in detecting the presence of V. cholerae in environmental waters during both epidemic [4, 5] and inter-epidemic [6] periods. Such inability in detecting V. cholerae has been attributed to the fact that V. cholerae can survive in the aquatic environment in a viable but non-culturable state [7]. More complex analyses, for example based on fluorescent staining of cell surfaces or DNA are capable of detecting V. cholerae even in inter-epidemic periods – as first shown by Alam et al. [8], who made use of a microscopy-based approach that, although necessary for qualitative analysis (e.g. detection of clustering), relies on manual counting.
In this respect, portable flow cytometry could be an ideal method for monitoring V. cholerae concentrations in the environment due to the rapidity of sample analysis and the absolute volumetric counting that it reliably provides [9]. Flow cytometry is an automated approach which allows the processing of large sample sizes over a relatively short time period. Regarding accuracy, Hammes et al. [10] showed that flow-cytometric total bacterial cell counts were within 5% of the true target values in drinking-water samples and could be considered reliable also at concentrations as low as 100–200 cells/ml [9]. By contrast, it should be noted that recent assessments of other techniques, such as culturing, polymerase chain reaction (PCR) and immunofluorescence microscopy proved to be sensitive only to counts of around 103 colony-forming units/ml [11]. Flow cytometry of V. cholerae has already been applied to in vitro growth studies [12, 13], which confirmed the growth and persistence of V. cholerae in fresh water even in the presence of pre-existing, autochthonous bacterial communities and while subject to variations of environmental conditions.
In this work we aimed to demonstrate that flow cytometry can be used in field-work conditions. To this end, we tested the presence and determined the concentration of V. cholerae at several sites in Matlab (46 ponds, 10 canals, and 10 sites on the Dhonagada river), a rural area of Bangladesh that is a well-known endemic region for cholera. The field campaign was conducted in a limited time window (September–October 2012), falling between the two seasonal cholera peaks described for this region (April–May and November–December). For each sampling site we measured the total concentration of cells concomitantly with basic physico-chemical parameters. The flow-cytometry-based assay we present here allows measurement of both O1 and O139 serogroups of V. cholerae, i.e. the two serogroups associated with cholera epidemics and pandemics. The O139 serogroup was the cause of major outbreaks when it emerged around the Bay of Bengal in 1992 [14], yet reports of cholera cases associated with it are scarce since 1996 (except for sporadic reports [15]).
Our interest lay, in particular, in a procedure that may aid in the field validation of mathematical models of cholera spread, which usually take bacterial concentrations in environmental water reservoirs into account to determine local infection probabilities [16]. Recently, spatially explicit models of cholera have emerged as useful tools to capture the dynamics of propagation of the disease [17–20], but they still lack a thorough description of the local dynamics of V. cholerae concentrations. In particular, it is still uncertain whether coastal environments in general contain a viable, resident population of V. cholerae as in the Bay of Bengal [21–23], or whether pathogens present in those coastal regions are simply a result of the discharge from the hydrological network. This uncertainty holds especially for epidemic and newly invaded areas. Nonetheless, recent studies seem to indicate that resident populations of V. cholerae are indeed present in both endemic (as in the case of rural Bangladesh [24]) and epidemic [25] contexts, although detection of V. cholerae may prove difficult in the latter case [26]. Thus, additional methods to detect the presence and estimate the abundance of V. cholerae, such as the one presented here, may provide an invaluable tool to improve disease surveillance and refine mathematical models. Such models would be important for monitoring and predicting ongoing epidemics in time and space.
MATERIALS AND METHODS
Description of the study area
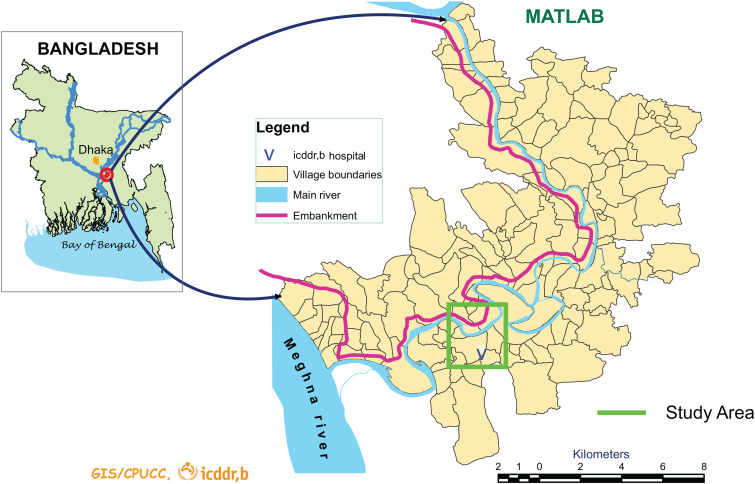
Matlab is located about 55 km southeast of Dhaka, the capital of Bangladesh. In Matlab, the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) has been conducting demographic surveillance for more than 40 years encompassing a population of 200 000 individuals. Seasonal occurrence of cholera is characteristic of this region, with two peaks in incidence every year. The first peak usually occurs in spring around April–May while the second is mostly linked to late autumn (November–December [27]). The surface water system in this area is composed of ponds and irrigation canals that are diverted from the Dhonagada river, a branch of the Meghna river (Fig. 1). Pond waters in rural areas are frequently used by villagers for domestic purposes. However, people living in this area usually treat the environmental water prior to drinking [28]. In close proximity to the villages, ponds are surrounded by elevated ground, which protects the villages from flooding even during the monsoon season and which may impede mixing of river and pond waters. The volume of those ponds can vary greatly between each other and over time so that some dry up completely during times of drought whereas others do not. Survival of V. cholerae in ponds has been associated with the presence of other organisms, such as blue-green algae and other members of the phytoplankton and zooplankton communities [29], as ponds are generally rich in organic matter. The high concentration of organic matter is due to the drainage of latrines and to rainfall run-off causing transport of fertilizers and cattle faeces into the ponds [30].
Fig. 1.
Location of the study area (green square) with respect to Bangladesh and the Matlab area. The location of the icddr,b hospital is also highlighted.
Sample collection and processing
Water samples were collected in the Matlab area (Fig. 1) between September and October 2012. Samples were taken from 46 ponds, 10 locations in irrigation canals and 10 sampling sites along the Dhonagada river. All samples were collected aseptically in sterile dark bottles (Nalgene Nunc International, USA) in the morning and placed in an insulated plastic box with ice packs maintaining the temperature at 4–8 °C. The samples were then transported to the Environmental Microbiology Laboratory of the icddr,b in Dhaka for further processing on the same day. The analysis of the samples was done the following day. From each bottle, nine samples of 1 ml each (preserved in Sarstedt vials) were filtered through tilted nylon gauze filters (CellTrics® Partec, Germany). The pore-size equalled 10 μm in six cases (three samples were used for the estimation of V. cholerae O1 concentration, the remaining three for O139) and 30 μm for the other residual three samples for counting total number of cells. Thus, three parallel measurements per biological sample could be performed.
Total cell count
Enumeration of total cell concentrations was performed according to the method described in Vital et al. [12]. Briefly, the samples processed through the 30 μm filters were mixed with the DNA dye SYBR Green (Molecular Probes Inc., USA; 1x final concentration). The flow cytometer used for these experiments was a portable Cyflow SL Blue (Partec) equipped with a solid-state laser emitter. The green fluorescent signal of the DNA-bound SYBR Green dye was exited at 488 nm and the emission signal was detected at 532 nm.
Specific detection of V. cholerae O1 and O139 via flow cytometry
Specific detection of V. cholerae was performed according to the method described by Vital et al. [12]. On the collection day, all samples (1 ml each) devoted to pathogen count were subjected to UV irradiation inside a laminar air flow cabinet (Clyde-Apac HWS 120/75, Australia) equipped with a UV sterilizing chamber (Philips TUV 30W/G30T8, The Netherlands), in order to arrest growth. Next, 10 μl of the V. cholerae O1 or O139 direct fluorescent antibody (DFA) reagents (New Horizon Diagnostics Corporation, USA) were added to each sample and incubated as described in Vital et al. [12]. To verify the ability of the flow cytometer to correctly measure the concentration of V. cholerae O1 or O139 in water samples containing different populations of microorganisms – aside from standard positive and negative controls – we spiked sample environmental water with known concentrations of V. cholerae spanning between 102 and 105 cells/ml, analysing five replicates for each concentration level. These V. cholerae cells were obtained by overnight culturing on agar plates and initial concentration was estimated by spectrophotometry. The instrumental counts confirmed the order of magnitude of V. cholerae cells expected in each sample in all cases except for negative control samples, in which we observed counts up to 265 false positives/ml, while the average count was 116·5 false positives/ml. This is in agreement with a previous study [9] where the accuracy of the method cannot be tested below a minimum concentration of 100–200 cells/ml. The detection limit threshold was thus conservatively set at 265 cells/ml. Presence of V. cholerae O1 was confirmed by DFA microscopy for the samples showing higher counts. No cross-reaction of the antibodies between V. cholerae O1 and O139 serogroups was observed.
Analysis of environmental samples
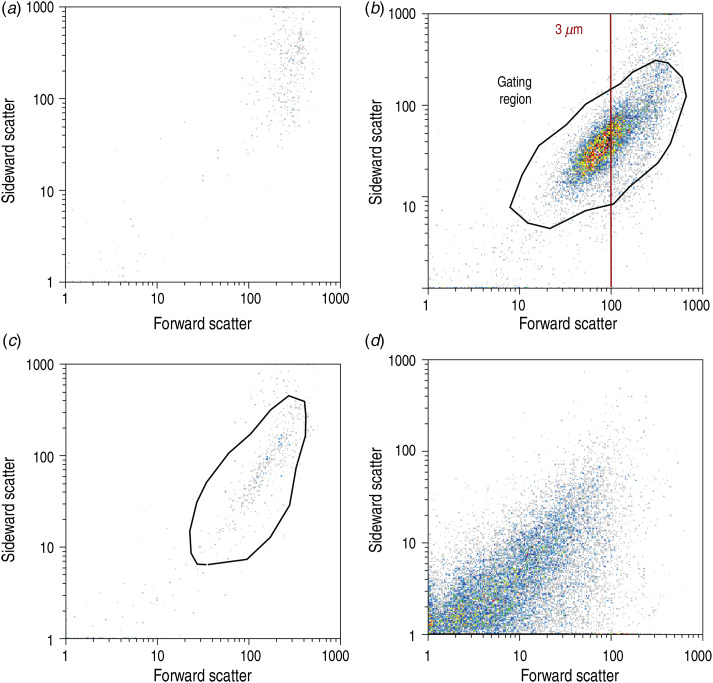
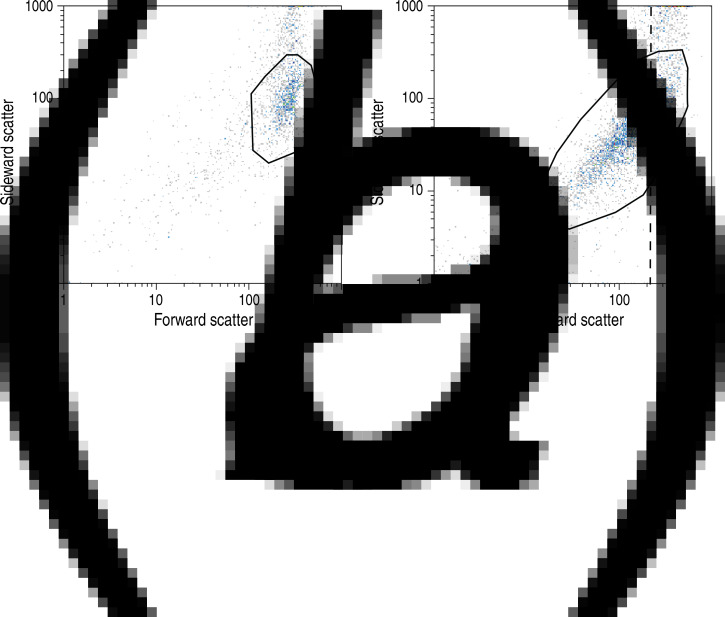
To correctly enumerate the number of V. cholerae cells present in each environmental sample, a procedure that can be applied to safely discriminate between stained cells, other organisms and background noise in the instrumental output had to be devised. Previous studies indicate that the post-processing steps contain many of the possible sources of arbitrariness and error, given that true positives need to be separated from background noise and false positives [9]. Figure 2 shows the typical output signals obtained by flow cytometry using the differently treated samples, which were derived from the same environmental water source. Forward scatter (FS) provides a measure of the dimension of particles/cells and sideward scatter (SS) is related to the irregularity of particles/cells. In order to limit possible errors in the post-processing phase, the enumeration of pathogens was based on the following post-processing steps. The processing software was set to show only particles that emitted a green fluorescent signal. In this respect, non-stained water samples were analysed to identify naturally fluorescing mineral particles or microorganisms, which might constitute false positives in our analysis (Fig. 2a). Next, stained samples were analysed. Figure 2(b, c) shows the ‘cloud’ that is observed in O1- and O139-positive samples, generally centred around a FS value corresponding to the value characteristic of the beads used for calibration. The calibration beads had a fixed diameter of 3 μm (red line in Fig. 2b) and are therefore comparable in size to single cells of V. cholerae. At this point, O1-/O139-positive signals were counted. Replicates with counts lower than the detection threshold of 265 cells/ml were excluded from further analysis. Next, a gating region centred around the typical FS/SS values was conservatively defined to exclude background noise and residual stain, appearing at low values of FS and SS. The enumeration of O1-/O139-positive cells within the gating region was deemed a reliable measure of O1/O139 concentrations. It should be noted that, after gating, the population concentration may obviously be lower than the threshold set for the ungated cell count. Figure 2d, instead, shows positive signals when staining all living cells. Gating was not applied to these samples, as cell count was used here as a mere proxy for the local size of the microorganism community. Table 1 shows an example of our post-processing procedure, while Table 2 summarizes the total excluded replicates/sampling sites for both O1 and O139 serogroups. Some samples showed higher FS values (see Fig. 3) than those shown in Fig. 2(b, c), that are typical of isolated V. cholerae cells. This may indicate the presence of small clusters of V. cholerae cells, a feature that has been observed previously especially in inter-epidemic periods [8]. Alternatively, such an increase in FS could be caused by elongated cells. Note that larger clusters, such as those shown in Alam et al. [8], would be filtered out in this assay due to the small pore-size of the filters used for sample processing. This is a specific requirement of the instrument as the diameter of the orifice measures only 100 μm; therefore filtering must be at lower pore-size to avoid the possible presence of very elongated particles.
Fig. 2.
Representative data obtained by flow-cytometry-based cell counting. Water samples taken from pond 36, analysed for concentrations of V. cholerae O1, O139, and total cells. Colours indicate the number of counts in each pixel in relation to the total count. (a) Negative control to measure background fluorescence in the water sample without antibody or DNA staining. (b) Ten-μm-filtered water sample with O1-specific staining. The red line indicates the calibration value for spherical beads of 3 μm diameter. The black solid line depicts the gated region used for final counting. (c) Ten-μm-filtered water sample stained with O139-specific antibodies. (d) Thirty-μm-filtered water sample stained with the non-specific DNA dye SYBR Green. The counting was done with the green fluorescence trigger on in order to detect the labelled antibody attached to V. cholerae or the SYBR Green-stained DNA within cells.
Table 1.
Example of post-processing, including the application of the detection limit (265 cells/ml). Counts are expressed in cells/ml
| Sample, measurements | Replicas | Positive count | Gated count | Mean cell count |
|---|---|---|---|---|
| Pond 11 | ||||
| O1 cells (DFA stain) | 1 | 10435 | 6900 | |
| 2 | 2755 | 1975 | 3236·7 | |
| 3 | 2690 | 835 | ||
| O139 cells (DFA stain) | 1 | 535 | 140 | |
| 2 | 260 | − | 137·5 | |
| 3 | 480 | 135 | ||
| Total cells (SYBR Green stain) | 1 | 49204050 | n.p. | |
| 2 | 47849750 | n.p. | 49896175 | |
| 3 | 52634725 | n.p. |
n.p., Not performed (see Materials and methods section).
Table 2.
Number of replicates and sampling sites for each serogroup excluded from the analysis because of being below the detection limit
| Site type | Total sites (replicates per serogroup) | nexcl,r,O1 | nexcl,r,O139 | nexcl,s,O1 | nexcl,s,O139 |
|---|---|---|---|---|---|
| Canal | 10 (30) | 18 | 19 | 3 | 4 |
| River | 10 (30) | 6 | 11 | 2 | 2 |
| Pond | 46 (138) | 20 | 31 | 1 | 2 |
nexcl,r,O1/O139, Number of excluded replicates of O1/O139 concentration measures; nexcl,s,O1/O139, number of resulting excluded sampling sites of O1/O139 concentration measures.
Fig. 3.
Flow-cytometry-based V. cholerae O1 analysis of environmental water sampled from two different ponds. (a) The presence of counts in the upper side scatter and forward scatter range (pond 18) is possibly indicative of cluster formation by V. cholerae cells. (b) Water analysis of samples derived from pond 34 suggests co-existence between single and clustered V. cholerae cells, graphically separated by the dashed black line.
Physico-chemical parameters
Air, water temperature and pH were measured with a portable meter (HANNA model HI991001, USA), while conductivity, salinity and total dissolved solid content were measured with a portable Sension 5 (HACH, USA).
Statistical analyses
Wilcoxon rank-sum test (WRST) was used to test the hypothesis that two variables are sampled from distributions with different medians, using a signed test in case such variables were paired. Specifically, WRST was used to detect differences in the median of V. cholerae O1 and O139 concentration estimations, and in the values of population concentrations and physico-chemical parameters in flood-protected and non-protected ponds. Correlation analysis was used to analyse the relationship between V. cholerae O1/O139 concentrations and environmental parameters, in which we set significance at P = 0·01. Linear regression was used for O1/O139 concentrations vs. total cell counts or conductivity measurements. Jarque–Bera and Anderson–Darling tests were used to check for the normality of residuals. Confidence intervals and R2 values of linear fit parameters are reported in the figure legends when applicable. WRST tests, correlation analyses and linear regressions were performed considering the average of the replicates in each sampling location, after the exclusion of replicates below the detection limit for O1/O139 concentrations. We used analysis of variance (ANOVA) to test the independence of samples from each other.
RESULTS AND DISCUSSION
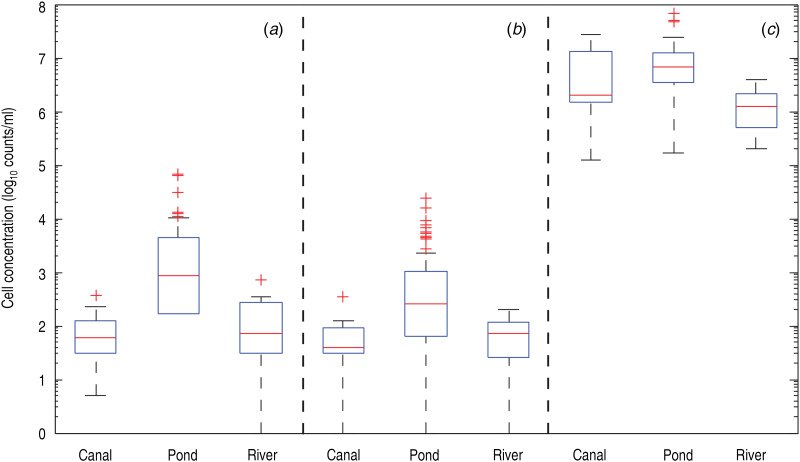
We analysed the distribution of V. cholerae in different water bodies. In particular we tested whether there is any significant difference regarding the presence of V. cholerae O1, O139, and total cell counts in ponds, irrigation canals, and rivers. As depicted in Figure 4, the concentration of V. cholerae O1 in ponds was consistently higher than the concentration of the O139 serogroup (paired WRST – rejecting the null hypothesis that the two distributions have the same median with P < 0·001) with a maximum concentration reaching ~70 000 V. cholerae cells/ml. The concentration range measured here complies with laboratory-based data derived from in vitro and microcosm experiments [12, 13, 23]. Note that the possible presence of small clusters of bacteria – which in this analysis would count as single particles/cells – might lead to a slight underestimation of the actual concentration. The concentrations of O139 serogroup in ponds were in most (91%) cases inferior to those observed for O1 serogroup, possibly indicating co-existence between the two serogroups which favours the latter in environmental conditions. However, differences in the seasonal concentration patterns of the two serogroups could provide an alternative explanation. As further evidence of co-existence between the two serogroups, their concentrations in each pond were found to be strongly correlated (Pearson's ρ = 0·74, P < 0·001). The ratio of O139 to O1 was on average 0·33, but with high deviation (σ = 0·4). However, it should be noted that non-specific attachment of the antibodies might still occur to a certain extent, even though no cross-reactions between the O1- and O139-specific antibodies towards the opposite serogroup have been previously observed, and no cross-reaction against other natural aquatic bacteria were detected in laboratory microcosms [12].
Fig. 4.
Occurrence of V. cholerae O1 and O139 in ponds, irrigation canals, and rivers. Boxplot of measured concentrations of (a) V. cholerae O1 and (b) V. cholerae O139 (10 μm-filtered samples) and of total living cells (c, 30-μm-filtered samples) in water samples of the different water bodies of the study area. The number of sampling sites was 10 for river and canals, and 46 for ponds. The red line indicates the median value, while the edges of the box correspond to the 25th–75th percentile values and the whiskers extend to the extreme datapoints not considered outliers, which are shown with red crosses.
Interestingly, water samples taken from canals and rivers consistently showed negligible V. cholerae counts, with a median concentration of serogroups O1 and O139 below 100 cells/ml (Fig. 4a, b). By contrast, the total cell count measured in parallel from each water sample did not show large differences between the three water sources (ponds, canals, rivers). Indeed, the average concentration was within a range of 106–107 cells/ml (Fig. 4c). Although these figures may indicate that our V. cholera-specific detection did not reach consistently the 1% target-to-background ratio (in our case, the ratio between V. cholerae and total cell concentrations) recommended by Hammes & Egli [9], our total cell count estimation procedure did not include a gating step, as the identification of true positives in this case would have been time-consuming to an extent that this particular study did not allow. The true target-to-background ratio is thus expected to be higher than what is shown in Figure 4.
Our findings confirm that resident populations of V. cholerae O1 and O139 persist in freshwater environments, particularly in ponds. This represents a new result with respect to other studies concerning the survival of these pathogens in the Bay of Bengal region. Those studies have mainly focused on saline aquatic environments as major natural habitats of V. cholerae [22, 23], suggesting that these might act as permanent reservoirs from which the disease can spread inland. Indeed a clear spatial and temporal pattern of cholera spread in Bangladesh has been reported [31, 32]. According to such studies, cholera outbreaks usually start around the coastal area in spring, reaching the inland regions in autumn. It should be noted that the low values of V. cholerae concentration found in river waters do not contradict such observations, as our sampling campaign was conducted during a period of supposedly high flow. However, further insight is needed to understand whether transport of pathogens – as proposed by Bertuzzo et al. [33] – may determine different patterns in different regions of Bangladesh or not, even in presence of resident local populations. Alternatively, spatio-temporal patterns of environmental drivers, such as rainfall (as in the case of Haiti [34–36]) or spatial heterogeneity of social and environmental conditions [2, 18, 37, 38] could contribute to the observed cholera seasonality in Bangladesh.
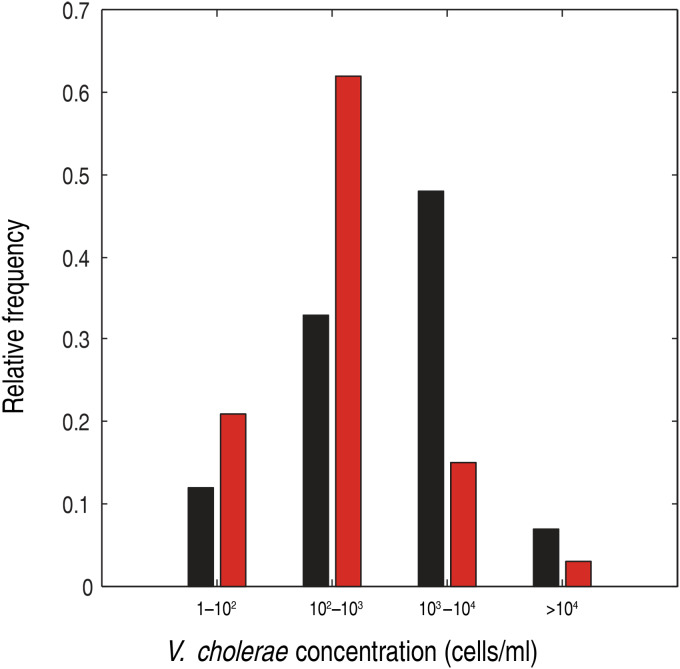
Next we determined the fraction of pond-derived water samples falling into a given concentration range for V. cholerae O1 and O139. As shown in Figure 5 the concentration of V. cholerae O1 was >103 cells/ml in more than half of the sites, and in few cases even >104 cells/ml. By contrast, average concentration of the O139 serogroup exceeded 103 cells/ml in only six of the 46 ponds analysed (Fig. 5). An ANOVA was performed on all three concentration replicates sampled from independent sites in order to discover if the mean concentrations of V. cholerae were significantly different from each other (P < 0·001 for both O1 and O139).
Fig. 5.
Distribution of the observed concentrations for the pond samples. The distribution refers to the averages of the three replicate measurements performed for each location. Black/red bars represent V. cholerae O1/O139 concentrations.
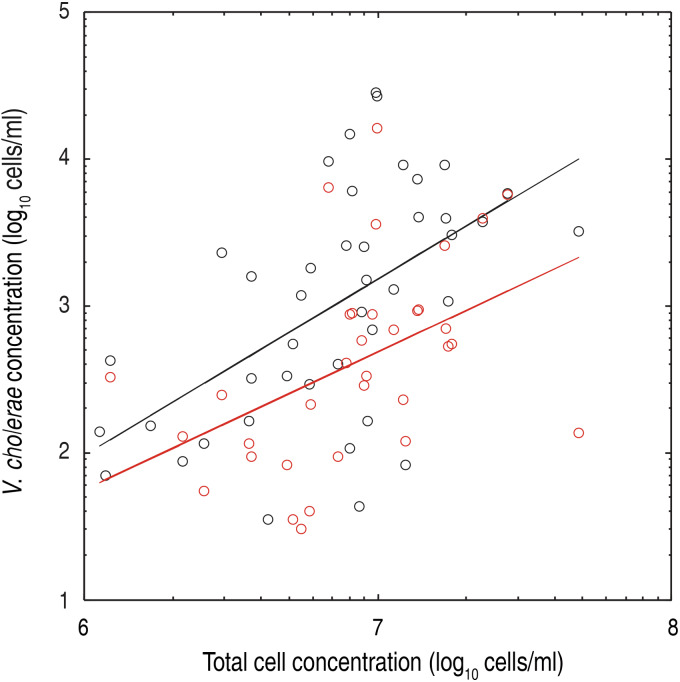
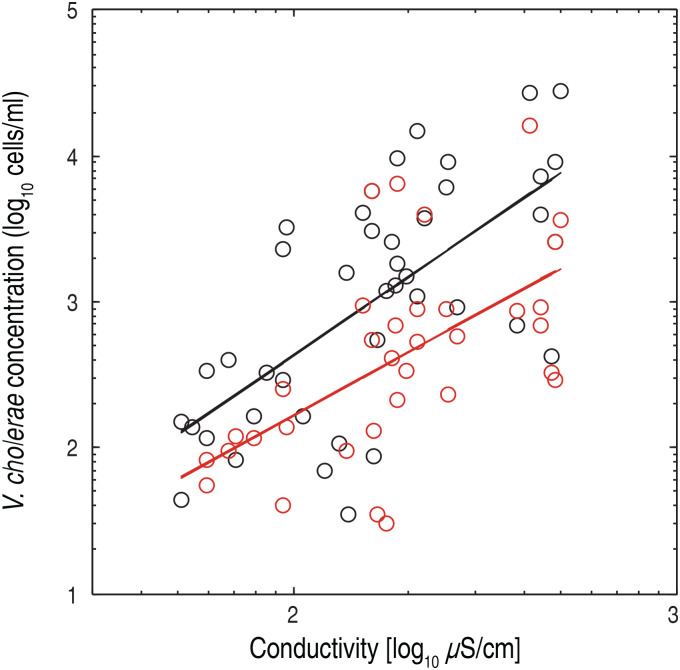
The association of V. cholerae O1 and O139 with other organisms has been a topic of great importance in the study of the ecology of these bacteria, especially with respect to survival strategies in inter-epidemic periods [30, 39–42]. It should be noted that the necessary – for the reasons mentioned above – filtering steps of sample treatment may well have substantially removed planktonic cells. Therefore we cannot assess the association between V. cholerae and other organisms directly, something which standard microscopy techniques are more apt to do. We assume here that total cell concentration – after 30 μm filtering – is a proxy of the concentration of other species of larger size. Figure 6 shows an analysis of the relationship between the concentration of V. cholerae O1 and O139 and the total cell count in the ponds. The relationship between the logarithms of these quantities turned out to be significant for the O1 serogroup in particular (O1: R2 = 0·31, P < 0·001; O139: R2 = 0·21, P < 0·007), showing that at least the orders of magnitude of V. cholerae and total cell concentrations are indeed related. Of interest, correlations between physic-chemical parameters and V. cholerae concentration were significant for most of the tested parameters, i.e. water temperature, total dissolved solid, salinity and conductivity (Table 3). The only parameter which resulted as uncorrelated, i.e. pH, should probably be tested over a longer time span [43]. Figure 7 shows the relationship between V. cholerae O1 and O139 and conductivity, which is a measure of the amount of dissolved solids in the water and of its salinity. Both factors have been shown to influence V. cholerae growth [12]. These findings suggest that environmental factors – those describing water quality in particular – may well provide an indicator of the risk of cholera presence in an endemic area. However, the limited time span of this study does not allow further speculation, and future long-term investigations should shed further light on the association of V. cholerae with abiotic factors.
Fig. 6.
Correlation between V. cholerae O1/O139 and total cell counts. Log-log plot of V. cholerae serogroups O1 (black dots) and O139 (red dots) concentrations compared to total cell counts. Each dot represents the average concentration of the three replicative flow-cytometry measurements from different pond-derived water samples. Regression lines (y = b·xa) show the following parameters, with their respective 5–95% confidence intervals (CI): b = 6·06 × 10−6, CIb = 7·31 × 10−10–0·05; a = 1·20, CIa = 0·63–1·77, R2 = 0·31 (for O1 concentrations, black solid line); b = 1·28 × 10−4, CIb = 3·19 × 10−9–5·12; a = 0·94, CIa = 0·28–1·60, R2 = 0·21 (for O139 concentration, red solid line).
Table 3.
Observed physic-chemical parameters in ponds, correlation coefficients to V. cholerae O1 and O139 concentrations (Pearson's ρ) and their statistical significance
| Parameter | Median value | Range | ρO1 | PO1 | ρO139 | PO139 |
|---|---|---|---|---|---|---|
| Water temperature (°C) | 29·3 | 27·1−31·8 | −0·2893 | 0·0016 | −0·3425 | 0·0035 |
| Salinity (g/l) | 0·1 | 0−0·2 | 0·4837 | <0·001 | 0·3616 | 0·013 |
| Total dissolved solid (mg/l) | 74·35 | 23·60−240 | 0·5704 | <0·001 | 0·3748 | 0·01 |
| Conductivity (μS/cm) | 159·90 | 50·70−498·00 | 0·5627 | <0·001 | 0·3691 | 0·012 |
| pH | 6·88 | 6·60−7·08 | −0·0360 | 0·81 | −0·1104 | 0·4653 |
Fig. 7.
Correlation between V. cholerae O1/O139 and conductivity. Log-log plot of V. cholerae serogroups O1 (black dots) and O139 (red dots) concentrations compared to conductivity measurements (in μS/cm). Each dot represents the average concentration of the three replicative flow cytometry measurements from different pond-derived water samples. Regression lines (y = b·xa) show the following parameters, with their respective 5–95% confidence intervals (CI): b = 0·11, CIb = 4·30 × 10−3–2·88, a = 1·79, CIa = 1·16–2·43, R2 = 0·45 (for O1 concentrations, black solid line); b = 0·21, CIb = 5 × 10−3–8·99, a = 1·45, CIa = 0·74–2·16, R2 = 0·35 (for O139 concentration, red solid line).
Spatial distribution of V. cholerae O1 and O139 concentrations also showed a definite pattern (Fig. 8). The Matlab area is crossed by a branch of the Meghna river, which is embanked on one side to protect settlements close to the river. Our results indicate that ponds located in flood-protected areas host larger populations (with median concentrations of both O1 and O139 serogroups being an order of magnitude higher than in non-protected areas; WRST, P < 0·001 for both serogroups). A correlation between the risk of being infected with cholera and living in flood-controlled areas has previously been described for the same study area, but no further explanations have been given so far [44]. Our results suggest that specific and more favorable environmental/ecological conditions may support survival and growth of V. cholerae in such areas. Table 4 indicates that ponds located in these regions show different values of physico-chemical parameters. In particular, the total dissolved solid content is higher in flood-protected ponds. Flood protection might also prevent the washing out of the system by extreme events of flooding and thus promote the establishment of a thriving resident community of V. cholerae. In fact, dilution of water reservoirs is observed to be a cause of the waning of cholera outbreaks in the Bengal region [31, 45].
Fig. 8.
Spatial distribution of V. cholerae concentration in the study area. Map indicating the location of the sampled ponds and the relative concentration of V. cholerae serogroups (a) O1 and (b) O139. The icddr,b hospital, the regional market place and the outline of the river enbankment are highlighted in white. The heat map in the lower part indicates the concentration range. The satellite image is gathered from the LandSat NASA mission; the dataset is available at http://landsatlook.usgs.gov/.
Table 4.
Comparison of the abundance of V. cholerae O1/O139 and physico-chemical parameters in ponds located in flood-protected and non-protected areas.
| Flood-protected | Non-flood-protected | WRST P value | |
|---|---|---|---|
| O1 conc. (cells/ml) | 5022 (0−28420) | 309·17 (0−5837) | <0·001 |
| O139 conc. (cells/ml) | 870 (215−16330) | 95 (0−5765) | <0·001 |
| Water temperature (°C) | 28·55 (27·1−30·7) | 29·4 (28·4−31·8) | 0·83 |
| Salinity (g/l) | 0·134 (0−0·2) | 0·063 (0−0·2) | 0·36 |
| TDS (mg/l) | 111·2 (25·8−240) | 62·9 (0−225) | 0·05 |
| pH | 6·85 (6·69−7·01) | 6·98 (6·60−7·08) | 0·63 |
WRST, Wilcoxon rank-sum test; TDS, total dissolved solid.
Values given are median values with the minimum and the maximum observed values for each variable within parentheses. P values refer to the significance of a WRST that compares variables recorded in flood-protected and non-protected areas.
CONCLUSIONS
Based on the data presented here we can summarize our main findings with the following statements:
Resident populations of V. cholerae O1 and O139 persist in freshwater environments (especially ponds) in the rural area around Matlab, Bangladesh.
The concentration of V. cholerae O1 in ponds is consistently higher than the concentration of O139 serogroup.
Water samples taken from canals and rivers consistently show negligible counts of V. cholerae concentrations.
The orders of magnitude of V. cholerae O1/O139 and total cell concentrations are positively and significantly correlated.
V. cholerae concentrations are significantly correlated to most of the tested physico-chemical parameters.
Flood-protected ponds consistently show higher concentrations of V. cholerae O1/O139 than non-protected ponds.
We have tested, under field conditions, a procedure for the rapid assessment of V. cholerae O1 and O139 concentrations in diverse water bodies. This flow-cytometry-based approach allowed a rapid test of several water samples within a short time period. The procedure proved effective in detecting the presence of V. cholerae O1 and O139 during an inter-epidemic period, conditions in which culture-based techniques have previously failed. The method allowed measuring of V. cholerae O1 and O139 concentrations, rather than simply detecting the presence/absence of the pathogen, and is thus particularly suitable for epidemic surveillance and control, and in the long-term assessment of eradication efforts.
Further insight from lengthier campaigns is needed for some of our findings, which appear to shed some light on key issues of V. cholerae ecology and epidemiology. The disappearance of epidemics caused by the O139 serogroup from recent years' records seems appears to be reflected by the consistently larger O1 populations in our samples. At the same time, the O139 serogroup is still present in environmental waters and could again originate outbreaks. The presence of resident populations of potentially pathogenic V. cholerae serogroups in pond waters needs to be linked to the spatio-temporal pattern of cholera incidence observed in the Bangladesh region. The relationship between river-based pathogen transport and local cholera outbreaks appears elusive, especially in light of the higher V. cholerae concentrations observed in flood-protected areas. The high number of significantly correlated environmental parameters, including total cell concentration, with respect to V. cholerae concentrations, is also encouraging as environmental proxies could provide an important monitoring tool for the individuation of pathogen reservoirs, especially during the course of the outbreaks.
However, the applicability of this method remains limited, especially in large-scale studies and emergencies, as more cost-effective methods for V. cholerae detection do exist. The efficacy of these methods has been tested especially for diagnostic purposes [46]. However, the method presented here in a field-work situation allows measuring of the concentration, rather than simply detecting the presence of the pathogen, and may thus be more accurate to study the efficacy of eradication efforts. Moreover, the portability of some flow cytometer devices, such as that used in the present study, reduces greatly the costs associated with this kind of analysis, as a single instrument can be carried elsewhere and used for other field campaigns.
Further, the filtering requirement of the instrument stands out as a possible limitation to our method, as cluster and biofilm formation have been observed recurrently in colonies of non-culturable V. cholerae cells [8, 47]. We thus recommend, when possible, to couple flow cytometry with standard techniques (DFA microscopy and PCR) as additional controls. However, this is of limited relevance for the main application of the method in epidemic situations, when single V. cholerae cells are expected to be dominant in the environment [8]. Further efforts should also be devoted especially to the establishment of reliable and fast procedures for the evaluation of the fraction of V. cholerae O1 and O139 harbouring toxigenic genes, which are ultimately responsible for cholera outbreaks – and which this particular procedure does not allow the identification of.
Despite all these caveats, we argue that the procedure tested here could be a useful tool for the field validation of spatially explicit mathematical models of epidemic cholera. Data on pathogen abundance in water environments, including reservoirs such as the ponds sampled in this study, may serve as a much-needed field validation of spatially explicit mathematical models of the spread of infections. Most of these models [16, 19, 20, 34, 48] take into account the abundance of bacteria as a state variable of their system of equations. The possibility of tracking such a variable in space and time would in principle allow improvement of the calibration of still uncertain parameters, such as the contamination rate of infected individuals and the mortality of V. cholerae in the environment. A larger calibration dataset would grant, for instance, an increased robustness of these models when used for prediction and prevention of epidemics. Thus, we wish to emphasize that modern epidemiology of water-borne diseases should be based on a balanced use of field work and mathematics.
ACKNOWLEDGEMENTS
L.R., E.B., L.M., and A.R. acknowledge the support provided by the European Research Council (ERC) advanced grant programme through the project ‘River networks as ecological corridors for species, populations and waterborne disease’ (RINEC 227612). E.B., L.M., and A.R. acknowledge support from the Swiss National Science Foundation (SNF/FNS) project ‘Dynamics and controls of large-scale cholera outbreaks' (DYCHO CR23I2_138104).
The authors thank Professor Thomas Egli, Head of the Environmental Microbiology Laboratory in EAWAG, Zurich, Switzerland, and all the members of his laboratory for their invaluable help in the design of this work. The authors also thank Professor Melanie Blokesch for her thorough observations on the experimental design and all parts of this work.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Piarroux R, et al. Understanding the cholera epidemic, Haiti. Emerging Infectious Diseases 2011; 17: 1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaudart J, et al. Spatio-temporal dynamics of cholera during the first year of the epidemic in Haiti. PLoS Neglected Tropical Diseases 2013; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Annual report on cholera (http://www.who.int/cholera/statistics/en/). Technical report, World Health Organization, 2011.
- 4.Khan M, et al. Presence of vibrios in surface water and their relation with cholera in a community. Tropical and Geographical Medicine 1984; 36: 335–340. [PubMed] [Google Scholar]
- 5.Islam M, Drasar B, Bradley D. Long-term persistence of toxigenic Vibrio cholerae O1 in the mucilaginous sheath of a blue-green alga, Anabaena variabilis. Journal of Tropical Medical Hygiene 1990; 93: 133–139. [PubMed] [Google Scholar]
- 6.Huq A, et al. Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent-monoclonal antibody and culture methods. Applied Environmental Microbiology 1990; 56: 2370–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colwell R, et al. Viable but non-culturable Vibrio cholerae and related pathogens in the environment – implications for release of genetically engineered microorganisms. Nature Biotechnology 1985; 3: 817–820. [Google Scholar]
- 8.Alam M, et al. Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Applied Environmental Microbiology 2006; 72: 2849–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammes F, Egli T. Cytometric methods for measuring bacteria in water: advantages, pitfalls and applications. Analytical and Bioanalytical Chemistry 2010; 397: 1083–1095. [DOI] [PubMed] [Google Scholar]
- 10.Hammes F, et al. Flow-cytometric total bacterial cell counts as a descriptive microbiological parameter for drinking water treatment processes. Water Research 2008; 42: 269–277. [DOI] [PubMed] [Google Scholar]
- 11.Wang D, et al. Detection of Vibrio cholerae O1 and O139 in environmental water samples by an immunofluorescent-aggregation assay. Applied and Environmental Microbiology 2010; 76: 5520–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vital M, et al. Growth of Vibrio cholerae O1 Ogawa Eltor in freshwater. Microbiology 2007; 153: 1993–2001. [DOI] [PubMed] [Google Scholar]
- 13.Vital M, et al. Evaluating the growth potential of pathogenic bacteria in water. Applied Environmental Microbiology 2010; 76: 6477–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albert M, et al. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet 1993; 342: 387–390. [PubMed] [Google Scholar]
- 15.Faruque S, et al. Reemergence of epidemic Vibrio cholerae O139, Bangladesh. Emerging Infectious Diseases 2003; 9: 1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Codeco C. Endemic and epidemic dynamics of cholera: the role of the aquatic reservoir. BMC Infectious Diseases 2001; 1: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertuzzo E, et al. On the space-time evolution of a cholera epidemic. Water Resources Research 2008; 44: W01424. [Google Scholar]
- 18.Mari L, et al. Modelling cholera epidemics: the role of waterways, human mobility and sanitation. Journal of the Royal Society Interface 2012; 9: 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuite A, et al. Cholera epidemic in Haiti, 2010: using a transmission model to explain spatial spread of disease and identify optimal control interventions. Annals of Internal Medicine 2011; 154: 593–601. [DOI] [PubMed] [Google Scholar]
- 20.Chao DL, Halloran ME, Longini IM Jr.. Vaccination strategies for epidemic cholera in Haiti with implications for the developing world. Proceedings of the National Academy of Sciences USA 2011; 108: 7081–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colwell R. Global climate and infectious disease: the cholera paradigm. Science 1996; 274: 2025–2031. [DOI] [PubMed] [Google Scholar]
- 22.Collins A. Vulnerability to coastal cholera ecology. Social Science and Medicine 2003; 57: 1397–1407. [DOI] [PubMed] [Google Scholar]
- 23.Worden A, et al. Trophic regulation of Vibrio cholerae in coastal marine waters. Environmental Microbiology 2006; 8: 21–29. [DOI] [PubMed] [Google Scholar]
- 24.Stine OC, et al. Seasonal cholera from multiple small outbreak, rural Bangladesh. Emerging Infectious Diseases 2008; 14: 831–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohamed AA, et al. Molecular epidemiology of geographically dispersed Vibrio cholerae, Kenya, January 2009–May 2010. Emerging Infectious Diseases 2012; 18: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baron S, et al. No evidence of significant levels of toxigenic V. cholerae O1 in the Haitian aquatic environment during the 2012 rainy season. PLoS Current Outbreaks 2013; 2013; 13: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glass R, et al. Endemic cholera in rural Bangladesh, 1966–1980. American Journal of Epidemiology 1982; 116: 959–970. [DOI] [PubMed] [Google Scholar]
- 28.Huq A, et al. Simple sari cloth filtration of water is sustainable and continues to protect villagers from cholera in Matlab, Bangladesh. mBio 2010; 1: e00034–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipp E, Huq A, Colwell R. Effects of global climate on infectious disease: the cholera model. Clinical Microbiology Reviews 2002; 15: 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Islam M, Drasar B, Sack D. Probable role of blue-green algae in maintaining endemicity and seasonality of cholera in Bangladesh: a hypothesis. Journal of Diarrhoeal Diseases Research 1994; 12: 245–256. [PubMed] [Google Scholar]
- 31.Akanda AS, Jutla S, Islam S. Dual peak cholera transmission in Bengal Delta: a hydroclimatological explanation. Geophysical Research Letters 2009; 36: L19401. [Google Scholar]
- 32.Akanda AS, et al. Population vulnerability to biannual cholera outbreaks and associated macro-scale drivers in the Bengal delta. American Journal of Tropical Medicine and Hygiene 2013; 89: 950–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertuzzo E, et al. Hydroclimatology of dualpeak annual cholera incidence: Insights from a spatially explicit model. Geophysical Research Letters 2012; 39: L05403. [Google Scholar]
- 34.Rinaldo A, et al. Reassessment of the 2010–2011 Haiti cholera outbreak and rainfall-driven multiseason projections. Proceedings of the National Academy of Sciences USA 109: 2012; 6602–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Righetto L, et al. The role of aquatic reservoir fluctuations in long-term cholera patterns. Epidemics 2012; 4: 33–42. [DOI] [PubMed] [Google Scholar]
- 36.Righetto L, et al. Rainfall mediations in the spreading of epidemic cholera. Advances in Water Resources 2013; 60: 34–46. [Google Scholar]
- 37.Gatto M, et al. Generalized reproduction numbers and the prediction of patterns in waterborne disease. Proceedings of the National Academy of Sciences USA 2012; 109: 19703–19708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisenberg MC, Robertson SL, Tien JH. Identifiability and estimation of multiple transmission pathways in cholera and waterborne disease. Journal of Theoretical Biology 2013; 324: 84–102. [DOI] [PubMed] [Google Scholar]
- 39.Huq A, et al. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Applied and Environmental Microbiology 1983; 45: 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamplin M, et al. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Applied Environmental Microbiology 1990; 56: 1977–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner JW, et al. Plankton composition and environmental factors contribute to Vibrio seasonality. ISME Journal 2009; 3: 1082–1092. [DOI] [PubMed] [Google Scholar]
- 42.de Magny GC, et al. Role of zooplankton diversity in Vibrio cholerae population dynamics and in the incidence of cholera in the Bangladesh sundarbans. Applied Environmental Microbiology 2011; 77: 6125–6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Islam M, et al. Isolation of Vibrio cholerae O139 synonym Bengal from the aquatic environment in Bangladesh – implications for disease transmission. Applied Environmental Microbiology 1994; 60: 1684–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emch M. Diarrheal disease risk in Matlab, Bangladesh. Social Sciences and Medicine 1999; 49: 519–530. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Moreno D, et al. Cholera seasonality in Madras (1901–4940): Dual role for rainfall in endemic and epidemic regions. EcoHealth 2007; 4: 52–62. [Google Scholar]
- 46.Dick MH, et al. Review of two decades of cholera diagnostics – how far have we really come? PLoS Neglected Tropical Diseases 2012; 6: e1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alam M, et al. Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proceedings of the National Academy of Sciences USA 104: 2007; 17801–17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertuzzo E, et al. On spatially explicit models of cholera epidemics. Journal of the Royal Society Interface 2010; 7: 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]