SUMMARY
The study characterizes a collection of 67 neonatal septicaemic Escherichia coli isolates on the basis of phylogroup, serotype, virulence, antibiotic resistance and also the association of CTX-M-producing E. coli and the ST131 clone in a developing country. Phylogroups B2 and D were predominant (33% and 19%, respectively). The most prevalent virulence factors (VFs) were traT (69%) and iucC (68%) and most VFs were concentrated in the B2 isolates. High levels of resistance (⩾70%) to cefotaxime, ciprofloxacin and trimethoprim/sulfamethoxazole was recorded but meropenem remained the most active antimicrobial. Six (9%) of the study isolates belonged to the ST131 clone, five of which were from the same hospital, and were either indistinguishable or closely related by pulsed-field gel electrophoresis. Although the prevalence of CTX-M-15-producing isolates was high (81%), the ST131 clone was relatively infrequent (11%) in extended spectrum β-lactamase (ESBL)-producers. The ST131 clone was characterized by the presence of blaCTX-M-15, qnrS, aac(6′)-Ib-cr, IncF plasmids and virulence determinants such as iucC, papC, traT, usp, hlyA, iroNE.coli, cnf, and sat. We conclude that clonal spread of ST131 did not contribute directly to the high prevalence of CTX-M-15 in our settings.
Key words: CTX-M-15, extraintestinal pathogenic Escherichia coli, neonatal sepsis, serotypes, ST131, virulence factors, India
INTRODUCTION
Escherichia coli is one of the most versatile organisms with respect to the diseases it causes, the virulence determinants it possesses and the antibiotic resistance mechanisms that it exhibits [1]. E. coli that have the ability to cause disease outside the intestine are termed extraintestinal pathogenic E. coli (ExPEC). E. coli can be classified on the basis of their O:K:H serotypes, phylogroups (A, B1, B2, C, D, E, F) and sequence types (ST) [2, 3]. Most extraintestinal infections due to E. coli are caused by isolates of phylogroup B2 and D which possess several virulence factors (VFs) and can also be recognized by their characteristic O:K:H serotypes [3]. Phylogroups A and B1 are considered commensals and generally lack potential VFs [4]. Recent studies have shown that phylogroup C is closely related to phylogroup B1, strains belonging to phylogroup F form a sister group to phylogroup B2, and phylogroup E consists of formerly unassigned E. coli isolates. Similarly, when ExPEC are characterized by multilocus sequence typing (MLST) specific lineages such as ST131, ST69, and ST117 appear to be responsible for a large proportion of human extraintestinal infections [3].
Added to the virulence of ExPEC is the increasing degree of antimicrobial resistance, particularly the plasmid-mediated extended spectrum β-lactamases (ESBLs) of which the most prevalent are CTX-M enzymes, especially CTX-M-15, which was first identified from India [5] and are abundant in E. coli worldwide [6, 7]. E. coli strains carrying CTX-M-15 are not only responsible for nosocomial infections, but also are widely disseminated in the community [7, 8].
Historically, antimicrobial-resistant E. coli isolates have been associated with the presence of fewer virulence traits than susceptible E. coli isolates. Of the different lineages of ExPEC, the ST131 clone defies this paradigm by combining extensive resistance with an extensive virulence gene repertoire, exhibiting dual threats to the community [7, 9]. E. coli ST131 associated with serogroup O25b, and of the highly virulent phylogenetic group B2, was isolated from several countries including India and subsequently spread worldwide [8, 9]. Moreover, E. coli ST131 strains positive for CTX-M-15 often co-produce OXA-1, TEM-1 β-lactamases, and harbour plasmid-mediated quinolone resistance (PMQR) genes (qnrA, qnrS, qnrB) [9]. The CTX-M-15-producing ST131 clone is linked to narrow host range IncFII plasmids, while blaCTX-M-15 can be carried by FIA-FIB, FIA-FIB-FII and FIB-FII multi-replicons [9, 10].
E. coli is a common cause of neonatal septicaemia particularly in developing countries [11]. However, there is limited holistic information on such E. coli isolates in terms of phylogroup, serotype, virulence, resistance and clonal type. In addition, although the epidemiology of CTX-M-producing E. coli strains and prevalence of the ST131 clone in adults have already been documented [9, 12], there is a dearth of information on the association of CTX-M-producing E. coli and the ST131 clone in the neonatal population [13, 14].
The present study focused on E. coli blood isolates from septicaemic neonates isolated during 2007–2010 from three different centres in three cities in order to (i) characterize the isolates in terms of phylogroup, serotype, virulence, resistance and clonal group, and (ii) analyse associations of CTX-M-producing isolates and the ST131 clone.
MATERIALS AND METHODS
Bacterial isolates and identification
The E. coli blood isolates from septicaemic neonates were collected from three different centres in India: the Neonatal Intensive Care Unit (NICU) of IPGMER & SSKM Hospital, Kolkata, West Bengal; the Department of Microbiology, Vardhman Mahavir Medical College, and Safdarjang Hospital, New Delhi, and the Sick Newborn Care Unit (SNCU) in Birbhum District Hospital, Seuri, West Bengal.
The blood specimens were collected prior to antibiotic therapy when the neonates were suspected of having sepsis. The isolates were identified by different biochemical tests and further verified by using an ID32E or ID32GN system (bioMérieux, France).
Antimicrobial susceptibility and detection of β-lactamase phenotypes
Antibiotic susceptibility tests for cephalosporins (cefotaxime), fluoroquinolones (ciprofloxacin), carbapenem (meropenem), aminoglycosides (amikacin) and trimethoprim- sulfamethoxazole were performed by the Kirby & Bauer standard disk diffusion method [15] and interpreted according to CLSI guidelines [16]. Isolates exhibiting intermediate susceptibility were recorded as resistant.
All isolates were tested for the production of ESBLs and metallo-β-lactamases (MBLs) using the cephalosporin/clavulanic acid combination disk test (CLSI) and the imipenem-EDTA double-disc combination test, respectively [17].
DNA amplification and sequencing of antibiotic-resistant genes
ESBL-producing and MBL-producing isolates were further tested for the presence of β-lactamase genes (blaSHV, TEM, OXA-1, CTX-M) and carbapenem-resistant genes (blaVIM, IMP, SPM-1, GIM-1, SIM-1, KPC, SME, SPM, NDM, GES), respectively [17]. For the MBL- or CTX-M-producing isolates, all amplified β-lactamase products were further sequenced on both DNA strands in an automated DNA sequencer (Applied Biosystems 3730, DNA Analyzer, PerkinElmer, USA) and aligned with the gene sequences available from Genbank (http://www.ncbi.nlm.nih.gov/genbank).
Phylogenetic grouping and virulence genotyping
ESBL isolates were assigned to one of the seven main E. coli phylogenetic groups (A, B1, B2, C, D, E, F) by a multiplex polymerase chain reaction (PCR) [18] and screened for 11 VFs, namely aerobactin (iucC), invasion of brain endothelium factor (ibeA), P fimbriae (papC), iron acquisition (iroNE.coli), haemolysin (hlyA), type S pili (sfa), cytotoxic necrotizing factor 1 (cnf), serum resistance-associated outer membrane protein (traT), uropathogenic-specific protein (usp), iron-regulated gene homologue adhesin (iha) and afimbrial adhesins (afa) [19]. Serine protease autotransporter toxin (sat) was examined only in ST131 isolates [19].
Serotyping and detection of clonal groups
Fourteen O serogroups (O1, O2a, O4, O6a, O11, O12, O15, O16, O17, O18, O25a, O25b, O75, O157) most frequently observed in ExPEC were sought by PCR with appropriate controls [20]. Group D isolates were evaluated for the presence of antimicrobial resistance-associated E. coli clonal group A (CGA) initially by gene-specific PCR [21] and confirmed by sequence analysis using primers (F: 5′-TGAATACAGTACGCAGCGAA-3′; R: 5′-TTGGCTTTTGTTCACGTCGT-3′) designed in this study, targeting CGA-specific E. coli housekeeping gene fumC single nucleotide polymorphisms.
The B2-O25b isolates were further analysed for the prevalence of the ST131 clone by an allele-specific PCR-based method [12] followed by sequencing using primers (F: 5′-TCCGATTTGCACTTTAACCA-3′; R: 5′-CCAGAATAGGTCGCATGAAAA-3′) designed in this study to detect the ST131-specific alleles within the pabB gene.
Molecular characterization and genotyping of the ST131 clone
For the isolates belonging to the B2-O25b-ST131 clone further detailed analysis was performed. Antimicrobial susceptibility to ciprofloxacin, trimethoprim-sulfamethoxazole, aztreonam, cefpodoxime, ceftriaxone, cefotaxime, ceftazidime, amikacin, and gentamicin were determined (CLSI). Plasmids were assigned to families by a PCR-based replicon typing method [22]. Presence of ampC β-lactamases (MOX, CIT, DHA, ACC, EBC, FOX), 16S rRNA methylase genes (rmtA, rmtB, rmtC, rmtD, armA), PMQR determinants (qnrA, qnrB and qnrS), aminoglycoside modifying enzyme AAC(6′)-Ib-cr and integrons were also evaluated [17, 23].
Pulsed-field gel electrophoresis (PFGE) was performed for ST131 isolates in a CHEF-DR III apparatus (Bio-Rad, USA) with 50 U/plug of the restriction endonuclease XbaI (New England Biolabs, USA). Salmonella Braenderup H9812 was included as a molecular weight marker. The PFGE patterns were analysed and dendrograms were generated by FPQuest™ software, v. 4.5 (Bio-Rad). The levels of similarity were based on the Dice correlation coefficient. For cluster analysis of the patterns the unweighted pair-group matching algorithm (UPGMA) was used with a tolerance and optimization of 1·50%. All isolates with ⩾95% similarity were considered indistinguishable and ⩾85% to <95% similarity as closely related.
Statistics
All data analysis and statistics were performed using SPSS v. 20.0 (SPSS Inc., USA). Data were entered into a standard table, with one row for each sample and columns representing phylogroups, VFs and antibiotic susceptibility patterns. The presence (1) or absence (0) of each characteristic was recorded and their distribution represented in tabular and figure formats. Statistical analysis of the proportions of isolates in different groups was done using χ2 test and all significant differences are reported at 95% confidence intervals.
RESULTS
The 67 E. coli study isolates comprised 28 from IPGMER & SSKM Hospital collected during 2007–2011; 29 from Safdarjang Hospital (2008–2009) and 10 from Birbhum District Hospital during 2009–2010 (Table 1).
Table 1.
Molecular characterization of neonatal septicaemic Escherichia coli isolated from three different hospitals
| Isolate no. | Centre, year of isolation | Phylo-group | Virulence factors | Serotypes | Antibiogram | Extended spectrum β-lactamses (ESBLs) Metallo-β-lactamases (MBLs) |
|---|---|---|---|---|---|---|
| EC1 | SSKM, 2007 | A | traT, hlyA, PapC, IroNEC, iha | O15 | CTX-S, AK-S, CIP-S, SXT-S, IMP-S | ESBL-negative |
| EC2 | Delhi, 2008 | A | iucC, traT, iha | — | CTX-S, AK-R, CIP-R, SXT-S, IMP-S | ESBL-negative |
| EC3 | Delhi, 2008 | C | iucC, traT | — | CTX-R, AK-R, CIP-R, SXT-S, IMP-S | OXA-1, CTX-M-15 |
| EC4 | Delhi, 2008 | A | iucC, iroNE.coli, traT | — | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | OXA-1, CTX-M-15 |
| EC5 | Delhi, 2009 | A | iucC, iha | O2a | CTX-S, AK-S, CIP-S, SXT-S, IMP-S | CTX-M-15 |
| EC6 | Sheuri, 2009 | C | Nil | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | TEM, OXA-1, CTX-M-15 |
| EC7 | Sheuri, 2009 | C | traT | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-R | CTX-M-15, NDM-1 |
| EC8 | SSKM, 2011 | C | traT | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-R | CTX-M-15, NDM-1 |
| EC9 | SSKM, 2011 | C | hlyA, traT, cnf | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-R | TEM-1, CTX-M-15, NDM-1 |
| EC10 | SSKM, 2008 | A | iucC | — | CTX-R, AK-S, CIP-R, SXT-S, IMP-S | ESBL-negative |
| EC11 | SSKM, 2007 | B1 | iucC | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | OXA-1, CTX-M-15 |
| EC12 | Delhi, 2008 | B1 | iucC | — | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | OXA-1, CTX-M-15 |
| EC13 | Delhi, 2008 | F | papC | O1 | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | OXA-1, CTX-M-15 |
| EC14 | Delhi, 2008 | A | iucC, hlyA, papC, iha | O2a | CTX-S, AK-S, CIP-S, SXT-S, IMP-S | ESBL-negative |
| EC15 | Delhi, 2008 | B1 | Nil | O4 | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | OXA-1, CTX-M-15 |
| EC16 | Delhi, 2008 | B1 | iucC | — | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | OXA-1, CTX-M-15 |
| EC17 | Delhi, 2009 | B1 | Nil | — | CTX-S, AK-S, CIP-R, SXT-R, IMP-S | TEM, CTX-M-15 |
| EC18 | Delhi, 2009 | E | iucC, traT | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | TEM, CTXM-15 |
| EC19 | Delhi, 2009 | B1 | traT | — | CTX-S, AK-S, CIP-S, SXT-R, IMP-S | CTX-M-15 |
| EC20 | Delhi, 2009 | B1 | traT | — | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | TEM, OXA-1, CTX-M-15 |
| EC21 | Delhi, 2009 | E | iucC, papC | O18 | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | CTX-M-15 |
| EC22 | SSKM, 2011 | B1 | traT | — | CTX-S, AK-S, CIP-S, SXT-R, IMP-S | SHV, TEM, CTX-M |
| EC23 | Delhi, 2009 | B1 | iucC, usp | O25a | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | CTX-M-15 |
| EC24 | SSKM, 2007 | B2 |
hlyA, sfa, papC, iroNE.coli, cnf, traT, usp |
O6a | CTX-S, AK-S, CIP-S, SXT-S, IMP-S | ESBL-negative |
| EC25 | Delhi, 2008 | B2 | iucC, papC, traT | — | CTX-R, AK-S, CIP-R, SXT-S, IMP-S | OXA-1, CTX-M-15 |
| EC26 | Delhi, 2008 | B2 | iucC, papC, traT, usp | O25b | CTX-R, AK-S, CIP-R, SXT-S, IMP-S | OXA-1, CTX-M-15 |
| EC27 | Delhi, 2008 | B2 | iucC, hlyA, papC, iroNE.coli, sfa, iha, usp | — | CTX-S, AK-S, CIP-S, SXT-S, IMP-S | ESBL-negative |
| EC28 | Delhi, 2008 | B2 | iucC, papC, traT, usp | O25b | CTX-R, AK-S, CIP-R, SXT-S, IMP-S | OXA-1, CTX-M-15 |
| EC29 | Delhi, 2008 | B2 | iucC, hlyA, PapC, iroNE.coli, cnf, traT, usp | O25b | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | OXA-1, CTX-M-15 |
| EC30 | Delhi, 2008 | B2 | iucC, papC, iroNE.coli, usp | O25b | CTX-R, AK-R, CIP-R, SXT-S, IMP-S | OXA-1, CTX-M-15 |
| EC31 | SSKM, 2009 | B2 | iucC, papC | O25b | CTX-R, AK-S, CIP-S, SXT-R, IMP-R | CTX-M-15, NDM-1 |
| EC32 | Delhi, 2009 | B2 | iucC, papC, traT, usp | O25b | CTX-R, AK-R, CIP-R, SXT-S, IMP-S | CTX-M-15 |
| EC33 | Delhi, 2009 | B2 | iucC, hlyA, sfa, papC, iroNE.coli, traT, iha, usp | O25b | CTX-S, AK-R, CIP-S, SXT-R, IMP-S | TEM, CTX-M-15 |
| EC34 | SSKM, 2008 | B2 | iucC, traT, hlyA, cnf, iha, usp | O75 | CTX-R, AK-S, CIP-S, SXT-R, IMP-S | TEM, CTX-M-15 |
| EC35 | SSKM, 2009 | B2 | papC, iroNE.coli, traT, hlyA, sfa, cnf | O6a | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | CTX-M-15 |
| EC36 | SSKM, 2010 | B2 | iucC, hlyA, papC, traT, usp | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | SHV, TEM, CTX-M-15 |
| EC37 | SSKM, 2011 | B2 | iucC, papC, traT, iha, usp | O25b | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | CTX-M-15 |
| EC38 | SSKM, 2011 | B2 | iucC, papC, traT, iroNE.coli, usp | — | CTX-S, AK-S, CIP-S, SXT-S, IMP-S | ESBL-negative |
| EC39 | SSKM, 2008 | B2 | iucC, papC, traT, usp | O75 | CTX-S, AK-S, CIP-S, SXT-R, IMP-S | SHV, TEM |
| EC40 | SSKM, 2008 | B2 | iucC, papC, traT, hlyA, usp | O75 | CTX-S, AK-S, CIP-S, SXT-R, IMP-S | SHV, TEM, CTX-M-15 |
| EC41 | SSKM, 2008 | B2 | iucC, papC, traT, usp | O75 | CTX-S, AK-S, CIP-S, SXT-R, IMP-S | ESBL-negative |
| EC42 | SSKM, 2008 | B2 | iucC, papC, traT, iha | O25b | CTX-R, AK-R, CIP-R, SXT-R, IMP-R | TEM-1, CTX-M-15, NDM-1 |
| EC43 | SSKM, 2009 | E | iucC, traT | — | CTX-R, AK-R, CIP-R, SXT-S, IMP-R | TEM-1, OXA-1, NDM-1 |
| EC44 | SSKM, 2010 | B2 | iucC, papC, traT, usp | O75 | CTX-S, AK-S, CIP-S, SXT-R, IMP-S | TEM, CTX-M-15 |
| EC45 | SSKM, 2010 | B2 | iucC, iroNE.coli, traT, usp | O25b | CTX-R, AK-R, CIP-R, SXT-R, IMP-R | TEM-1, CTX-M-15, NDM-1 |
| EC46 | SSKM, 2007 | D | Nil | — | CTX-R, AK-S, CIP-R, SXT-S, IMP-S | OXA-1, CTX-M-15 |
| EC47 | Delhi, 2008 | D | iucC, papC, traT, hlyA, iha | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | OXA-1, CTX-M-15 |
| EC48 | Delhi, 2008 | D | traT | O15 | CTX-S, AK-S, CIP-S, SXT-R, IMP-S | ESBL-negative |
| EC49 | Delhi, 2009 | E | traT | — | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | CTX-M-15 |
| EC50 | Delhi, 2009 | D | iucC, traT, iha | O157 | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | CTX-M-15 |
| EC51 | Delhi, 2009 | B2 | iucC, traT | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | TEM, CTX-M-15 |
| EC52 | Delhi, 2009 | D | iucC, traT | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | TEM, CTX-M-15 |
| EC53 | Sheuri, 2009 | E | iucC, traT | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | TEM |
| EC54 | Sheuri, 2009 | D | iucC, traT | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | TEM, CTX-M-15 |
| EC55 | Sheuri, 2009 | E | iucC, traT | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | TEM, CTX-M-15 |
| EC56 | Sheuri, 2009 | D | iucC, traT | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | TEM, CTX-M-15 |
| EC57 | Sheuri, 2009 | E | iucC, traT | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | CTX-M-15 |
| EC58 | Sheuri, 2009 | D | iucC, traT | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | TEM, OXA-1 |
| EC59 | Sheuri, 2009 | E | iucC, traT | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-S | CTX-M-15 |
| EC60 | Sheuri, 2010 | F | papC, traT | O1 | CTX-R, AK-R, CIP-R, SXT-R, IMP-R | OXA-1, TEM-1, CTX-M-15, NDM-1 |
| EC61 | SSKM, 2011 | D | papC,hlyA,traT,cnf | O25a | CTX-S, AK-S, CIP-S, SXT-R, IMP-S | CTX-M-15 |
| EC62 | SSKM, 2011 | D | Nil | O12 | CTX-R, AK-S, CIP-S, SXT-R, IMP-S | CTX-M-15 |
| EC63* | SSKM, 2011 | D | iucC, traT | — | CTX-S, AK-S, CIP-S, SXT-R, IMP-S | CTX-M-15 |
| EC64 | SSKM, 2008 | D | iucC, papC, hlyA | — | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | TEM, OXA-1, CTX-M-15 |
| EC65* | SSKM, 2008 | E | iucC, hlyA, papC, cnf, traT, iha | O17 | CTX-R, AK-R, CIP-R, SXT-R, IMP-R | TEM, OXA-1, CTX-M |
| EC66 | SSKM, 2008 | D | Nil | — | CTX-R, AK-S, CIP-R, SXT-R, IMP-S | SHV, TEM, OXA-1, CTX-M-15 |
| EC67 | SSKM, 2011 | A | Nil | — | CTX-R, AK-R, CIP-R, SXT-R, IMP-R | TEM-1, OXA-1, CTX-M-15, NDM-1 |
AK, Amikacin; CIP, ciprofloxacin; CTX, cefotaxime; SXT, trimethoprim-sulfamethoxazole; IMP, imipenem.
Nil, No virulence genes detected; -, targeted O serotypes not present.
Clonal group A.
Phylogroup analysis and VFs
Isolates were distributed into seven phylogroups. Phylogenetic group B2 predominated, accounting for 33% (22/67) of the isolates, followed by phylogroups D (19%, 13/67), B1 and E (each 13%, 9/67), A (10%, 7/67), C (7%, 5/67) and F (3%, 2/67) (Table 2). Approximately half of the strains fell in groups B2 and D, and 24% in the less-virulent, commensal phylogenetic groups A and B1.
Table 2.
Distribution of virulence factors and antibiotic resistance of Escherichia coli isolates by phylogroups
| Virulence/resistance | Isolate no. (n = 67) | Phylogroup A (n = 7) | Phylogroup B1 (n = 9) | Phylogroup B2 (n = 22) | Phylogroup D (n = 13) | Phylogroup C (n = 5) | Phylogroup E (n = 9) | Phylogroup F (n = 2) |
|---|---|---|---|---|---|---|---|---|
| iucC | 46 (68%) | 5 (71%) | 4 (44%)‡∥ | 20 (91%) | 8 (61%)‡ | 1 (20%) | 8 (89%) | 0 (0%) |
| traT | 46 (69%) | 3 (43%)‡∥ | 3 (33%)‡∥ | 18 (82%) | 9 (69%) | 4 (80%) | 8 (89%) | 1 (50%) |
| papC | 28 (42%) | 2 (28%)‡ | 0 (0%)‡ | 19 (86%) | 3 (23%)‡ | 0 (0%) | 2 (22%)‡ | 2 (100%) |
| usp | 18 (27%) | 0 (0%)‡ | 1 (11%)‡ | 17 (77%) | 0 (0%)‡ | 0 (0%) | 0 (0%)‡ | 0 (0%) |
| iha | 12 (18%) | 4 (57%) | 0 (0%)* | 5 (23%) | 2 (15%)* | 0 (0%) | 1 (11%)* | 0 (0%) |
| iroNEC | 10 (15%) | 2 (29%) | 0 (0%)‡ | 8 (36%) | 0 (0%)*‡ | 0 (0%) | 0 (0%)‡ | 0 (0%) |
| hlyA | 15 (22%) | 2 (29%) | 1 (11%) | 7 (32%) | 3 (23%) | 1 (20%) | 1 (11%) | 0 (0%) |
| cnf | 7 (10%) | 0 (0%) | 0 (0%) | 4 (18%) | 1 (7%) | 1 (20%) | 1 (11%) | 0 (0%) |
| sfa | 4 (6%) | 0 (0%) | 0 (0%) | 4 (18%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| ibeA | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| afa | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Presence of 2 VFs | 47 (70%) | 4 (57%)‡ | 2 (22%)‡§∥ | 22 (100%) | 9 (69%)‡ | 2 (40%) | 8 (89%) | 1 (50%) |
| Presence of ⩾4 VFs | 23 (34%) | 1 (14%)‡ | 0 (0%) ‡ | 19 (86%) | 2 (15%)‡ | 0 (0%) | 1 (11%)‡ | 0 (0%) |
| Cefotaxime | 58 (87%) | 4 (57%)†∥ | 9 (100%) | 17 (77%) | 10 (77%) | 5 (100%) | 9 (100%) | 2 (100%) |
| Ciprofloxacin | 47 (70%) | 4 (57%)∥ | 7 (77%) | 11 (50%)∥ | 9 (69%) | 5 (100%) | 9 (100%) | 2 (100%) |
| Trimethoprim/ sulfamethoxazole | 51 (76%) | 2 (29%)†§∥ | 9 (100%) | 13 (59%)†§ | 12 (92%) | 4 (80%) | 8 (88%) | 2 (100%) |
| Amikacin | 29 (43%) | 2 (29%) | 1 (11%)∥ | 8 (36%) | 5 (38%) | 5 (100%) | 6 (66%) | 1 (50%) |
| Meropenem | 10 (15%) | 1 (14%) | 0 (0%) | 4 (18%) | 0 (0%) | 3 (60%) | 2 (22%) | 1 (50%) |
VF, Virulence factor.
Significantly lower than phylogroup A.
Significantly lower than phylogroup B1.
Significantly lower than phylogroup B2.
Significantly lower than phylogroup D.
Significantly lower than phylogroup E.
Difference not significant for all other comparisons, P ⩾ 0·05.
Phylogroups C and F were excluded from the statistical test on account of very small sample size.
On the basis of all 11 studied VFs, the most predominant was traT (69%) followed by iucC (68%), papC, usp, hlyA, iha, iroNE.coli, cnf, and sfa (Table 2). None of the strains possessed ibeA or afa. In general, the B2 phylogroup was associated with a higher prevalence of VFs compared to the other phylogroups and 19 (86%) out of 22 B2 strains harboured ⩾4 virulence genes (Table 2). A number of significant associations between different genes and phylogroups were revealed, namely the increased prevalence of iucC in phylogroup B2 compared to phylogroups B1 and D, and also in phylogroup E compared to B1. Likewise, traT was significantly more prevalent in groups B2 and E than in groups A and B1. Moreover, both papC and usp were more common in group B2, and iha in group A than in groups B1, D and E. Finally, iroNE.coli was significantly more common in B2 compared to groups B1, D and E (Table 2). These results indicate the marked predominance of VFs in phylogroup B2.
Susceptibility profiles and β-lactamase phenotypes
Overall the isolates showed a high level of resistance to all antibiotics tested except to meropenem. Of the 67 isolates, 58 (87%) were non-susceptible (i.e. intermediate or resistant) to cefotaxime, 47 (70%) to ciprofloxacin and 51 (76%) to trimethoprim/sulfamethoxazole. Meropenem was the most-active drug tested with 85% susceptibility followed by amikacin (57%); most meropenem strains were predominantly from Kolkata. The majority (78%, 52/67) of the isolates were resistant to ⩾2 classes of drugs, with the pattern ‘ciprofloxacin-cefotaxime-amikacin-trimethoprim/sulfamethoxazole resistant’ being the most common (24%, 16/67).
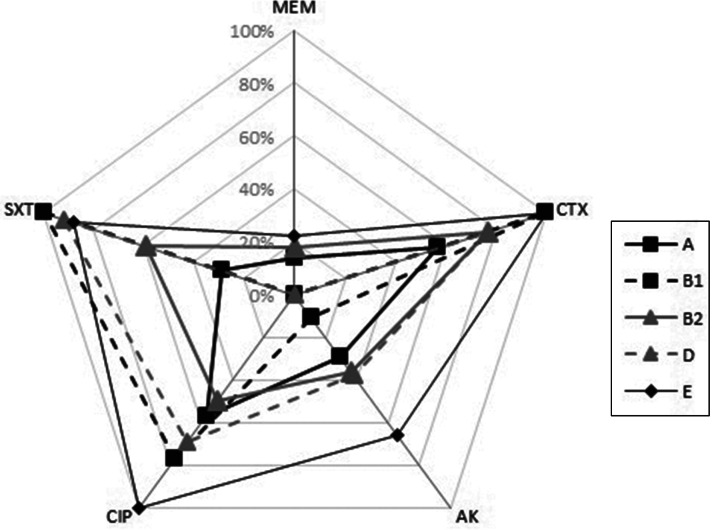
In general, there was no significant difference in the antibiotic resistance pattern of strains across most phylogroups as shown in Figure 1. Nevertheless significant differences were noted: (i) a lower frequency of cefotaxime resistance in group A compared to groups B1 and E phylogroup, (ii) higher ciprofloxacin resistance in group E than in groups A and B2, (iii) lower amikacin resistance in group B1 than in group E and (iv) fewer trimethoprim/sulfamethoxazole resistant strains in group A than in groups B1, D and E, as well as in group B2 compared to groups B1 and D (Table 2).
Fig. 1.
Percentage of resistance to five different classes of antibiotics (CTX, cefotaxime; AK, amikacin; CIP, ciprofloxacin; SXT, trimethoprime/sulphamethoxazole, MEM, meropenem) in isolates of major phylogroups.
β-lactamase gene identification
Evaluation of the ESBL and MBL status of all isolates by phenotypic test revealed that 58 (87%) and 9 (6%) were ESBL- and MBL-producers, respectively. The following genes encoding ESBLs were identified: blaCTX-M (54, 81%), blaTEM (23, 40%), blaOXA-1 (19, 33%) and blaSHV (4, 7%) (Table 1). CTX-M-15 was the only CTX-M-type identified in 54 isolates. The nine isolates resistant to meropenem harboured New Delhi metallo-β-lactamase-1 (NDM-1) which was confirmed by sequencing (Table 1). No other carbapenemases were detected.
Serotyping and detection of CGA
By the PCR-based method only certain O serogroups could be identified. Their distribution in the 22 phylogroup B2 strains in order of prevalence was: O25b (15%, n = 10), O75 (7·5%, n = 5), and O6a (3%, n = 2). Serogroups O12, O15, O157 and O25a were identified in phylogroup D, O25a and O4 in phylogroup B1, and O18 and O17 in phylogroup E. Phylogroup A strains were assigned to serogroups O15 and O2a and both phylogroup F strains were of the O1 serogroup (Table 1). None of the assayed serogroups were represented in phylogroup C.
Ninety percent of the 13 group D and nine group E strains were trimethoprim/sulfamethoxazole resistant but only two strains (EC63 and EC65), each of phylogroups D and E were positive by PCR for CGA and were both isolated from SSKM Hospital, Kolkata.
Identification and molecular characterization of ST131
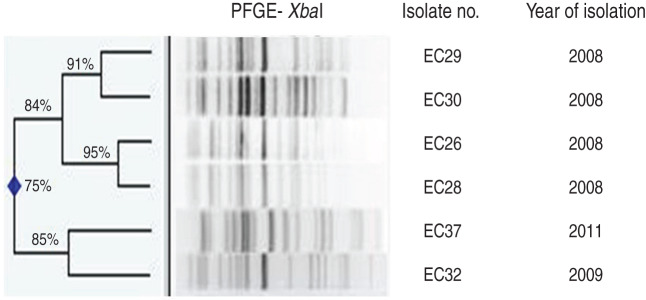
Ten (15%) of the 67 E. coli isolates were classified as phylogroup B2, O25b serogroup and of these, six were typed as the ST131 clone. Five of these latter strains were isolated from Safdarjang Hospital, New Delhi (four in 2008, one in 2009) and the other from IPGMER & SSKM Hospital, Kolkata in 2011 (Table 3). PFGE typing showed that two strains (EC26 and EC28) isolated in New Delhi in 2008 were indistinguishable from each other and the other three strains from the same unit were closely related (Fig. 2). Each of the six ST131 strains were susceptible to meropenem but resistant to other antimicrobials (Table 3).
Table 3.
Characteristics of Escherichia coli ST131 strains from septicaemic neonates
| Isolate no. | Antimicrobial resistance profile | β-lactamases | PMQR determinants | Virulence factors | Replicon typing |
|---|---|---|---|---|---|
| EC26 | CIP, ATM, CPD, CRO, CTX, CAZ, GEM | blaOXA-1, blaCTX-M-15 | qnrS, aac(6′)-Ib-cr | iucC, papC, traT, usp, sat | FIA-FIB |
| EC28 | CIP, ATM,CPD, CRO, CTX, CAZ, GEM | blaOXA-1, blaCTX-M-15 | qnrS, aac(6′)-Ib-cr | iucC, papC, traT, usp, sat | FIA-FIB |
| EC29 | CIP, SXT, ATM, CPD,CRO, CTX, CAZ | blaOXA-1, blaCTX-M-15 | qnrS, aac(6′)-Ib-cr | iucC, hlyA, iroNE.coli, cnf papC, traT, usp, sat | FIB |
| EC30 | CIP, ATM, CPD, CRO, CTX, CAZ, AMK, GEM | blaOXA-1, blaCTX-M-15 | qnrS, aac(6′)-Ib-cr | iucC, papC, iroNE.coli, usp, sat | Non-typable |
| EC32 | CIP, ATM, CPD, CRO, CTX, CAZ, GEM | blaCTX-M-15 | qnrS, aac(6′)-Ib-cr | iucC, papC, traT, usp, sat | FIA |
| EC37 | CIP, SXT, ATM, CPD, CRO, CTX, CAZ, GEM | blaCTX-M-15 | qnrS, aac(6′)-Ib-cr | iucC, papC, traT, usp, iha | FIA-FIB |
PMQR, Plasma-mediated quinolone resistance.
AMK, Amikacin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; CPD, cefpodoxime; CRO, ceftriaxone; CTX, cefotaxime; GEM, gentamicin; SXT, trimethoprim-sulfamethoxazole.
Fig. 2.
Dendrogram of the XbaI-digested patterns of six ST131 Escherichia coli isolates from neonatal septicaemia.
All ST131 strains possessed CTX-M-15 but none harboured ampC genes; all were positive for qnrS and aac(6′)-Ib-cr genes. Resistance to aminoglycosides was variable but 16S rRNA methylase genes were absent. VFs iucC, papC, traT, usp, hlyA, iroNE.coli, cnf, and sat were uniform in all ST131 strains, as well as plasmids of the narrow host range incompatibility group IncF. All but one of the ST131 strains were positive for FIA, FIB and FIA-FIB replicons (Table 3); none possessed integrons.
DISCUSSION
E. coli has long been recognized as an important cause of both early- and late-onset neonatal sepsis particularly in developing countries. This study set out to characterize the phylogeny of neonatal septicaemic E. coli isolates from three Indian hospitals by phenotypic and molecular methods to aid our understanding of the distribution of strain phylogroups and their association with virulence gene content, antibiotic resistance mechanisms and the ST131 clonal type.
Analysis of the phylogenetic background of the strains revealed the predominance of phylogroups B2 and D in neonatal sepsis. This is in agreement with earlier studies which have also shown the prevalence of these groups in isolates from E. coli bloodstream infections and their correspondence with the number of VFs present in the strains [24, 25]. In addition it was noted that a significant proportion (24%) of isolates were of phylogroups A and B1, which are generally considered to have low virulence and are predominantly present in both the adult [6] and neonatal intestinal tract [26]. Studies have shown that E. coli which cause extraintestinal infection often originate in the gut [27] and any breach in the intestinal barrier particularly in preterm neonates may allow commensal E. coli strains such as phylogroups A and B1 to enter the bloodstream. This may explain the finding of approximately one quarter of the isolates studied here being distributed in these phylogroups. The analysis would have been improved by the application of MLST to all isolates but this technique was not available to the study. However, assigning of the isolates to the main phylogenetic lineages recognized in E. coli did provide useful information on the relative distribution of the phylogroups and aided a better understanding of their dynamics in a resource-poor setting.
The prevalent VFs identified in this study were traT and iucC which were present across different phylogroups in good numbers. Gene iucC codes for aerobactin, which is required for iron uptake and confers a significant advantage for bacterial growth in conditions of iron stress [28], while traT inhibits the lysis of sensitized erythrocytes by serum complement [29]. Both of these properties are relevant to septicaemia. By contrast, gene ibeA codes for a factor required for invasion of the brain endothelium and afa mediates adherence to human epithelial cells in enteric or urinary tract infections [30]; neither of these genes were present in the study isolates.
The finding of a significantly higher frequency of virulence-associated genes in phylogroup B2 is in agreement with other studies [4, 31]. We also observed significantly higher frequencies of genes iucC, traT, papC, usp, and iroNE.coli in phylogroup B2 than some other phylogroups. Although the range of VFs is less than might be expected of typical antimicrobial-susceptible pathogenic E. coli from phylogenetic group B2, nonetheless they are more extensive than is usually observed among fluoroquinolone- or extended-spectrum cephalosporin-resistant E. coli [32]. Thus, such strains appear to pose the double threat of multidrug resistance and substantial extraintestinal virulence capability.
Irrespective of the phylogroup, the frequency and extent of antibiotic resistance was markedly high in the septicaemic isolates. It was particularly worrisome that ⩾77% of the strains of all but phylogroup A were resistant to cefotaxime and this corresponded to the high prevalence of ESBLs. The high incidence of ciprofloxacin resistance (⩾70%) was also noted for strains of phylogroups B1, D, C, E, and F which suggests that they may have wider resistance to all available fluoroquinolones – the drug of choice for treatment of infections caused by ESBL-producing strains. Although trimethoprim/sulfamethoxazole resistance was high, this was not linked to the incidence of CGA (3%), indicating that this resistance was not confined to a specific clonal group in our unit. In this study universal susceptibility to any of the antimicrobials tested was not observed which may be due to several factors such as antimicrobial usage, infection control practices and other unrecognized factors including the propensity of these strains to accumulate mutations conferring multidrug-resistant phenotypes. Overall, amikacin and meropenem were found to be superior to cefotaxime, ciprofloxacin and trimethoprim/sulfamethoxazole indicating that these two drugs still retain their potential against ESBL-producing pathogens.
CTX-M-15 was the most common ESBL identified in the isolates which reflects its global dissemination especially in E. coli as reported by others [33]. An association of blaCTX-M-15 with IncFIIK has recently been documented and a high capacity for these plasmids to diffuse and persist over time has been suggested [34]. Moreover, in contrast to a previous report which showed blaCTX-M-15-containing isolates exclusively in phylogroups B2 and D [6], our study showed a nearly similar presence of blaCTX-M-15 in all phylogroups. The appearance of fluoroquinolone- and trimethoprim/sulfamethoxazole-resistant CTX-M-15-producing isolates derived from phylogenetic group B2 with multiple VFs is a worrying development that merits further investigation.
To our knowledge, this study is the first analysis of prevalence of the ST131 clone in septicaemic neonates in a developing country. Detailed molecular characterization of ST131 showed some similarities to and differences from the ST131 clone characterized in earlier studies. Similarities include the presence of blaCTX-M-15 gene, qnrS gene along with aac(6′)-Ib-cr, virulence determinants such as iucC, papC, traT, usp, hlyA, iroNE.coli, cnf, and sat. Variance from other studies includes the absence of plasmid-mediated AmpC-β lactamases and the lack of IncFII plasmids [9]. However, our results are in agreement with Naseer et al. [10] on the high frequency of FIA and FIB replicons in ST131 strains.
The worldwide increase of CTX-M-15-producing E. coli has mainly been attributed to the spread of the ST131 clone [8, 9]. However, our study shows that although CTX-M-15-producing strains were prevalent in septicaemic isolates, the ST131 clone was not. This indirectly indicates that clonal spread of ST131 did not contribute to the high prevalence of CTX-M-15 in our setting. ST131 strains have also been found to be uncommon as colonizers of the neonatal gut [35] and further detailed analysis of the clonal types of the ESBL-producing isolates is warranted to identify whether other E. coli clones are responsible for the spread of the CTX-M enzyme. The dissemination of the blaCTX-M gene in conjunction with clonal spread is an alarming threat, particularly with the emergence of carbapenem-resistant (blaNDM-1) strains in this setting which should not be ignored. A high population density and resource-poor settings may facilitate the rapid spread of the clone and pose a great challenge for clinicians.
ACKNOWLEDGEMENTS
We extend our thanks to Dr Arun K. Singh for his support during the study and Soumya Kanti Sarkar for his assistance with statistical data. We are also grateful to Erick Denamur and James R. Johnson for providing us the strains for PCR controls. The study was funded by an Indian Council of Medical Research (ICMR) intramural fund. Subhasree Roy and Saswati Datta were supported by fellowships from ICMR.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Kaper JB, Nataro JP, Mobley HT. Pathogenic Escherichia coli. Nature Reviews Microbiology 2004; 2: 123–139. [DOI] [PubMed] [Google Scholar]
- 2.Maiden MC, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proceedings of the National Academy of Sciences USA 1998; 95: 3140–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manges AR, Johnson JR. Food-borne origins of Escherichia coli causing extraintestinal infections. Clinical Infectious Diseases 2012; 55: 712–719. [DOI] [PubMed] [Google Scholar]
- 4.Picard B, et al. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infection and Immunity 1999; 67: 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karim A, et al. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiology Letters 2001; 201: 237–241. [DOI] [PubMed] [Google Scholar]
- 6.Coque TM, et al. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerging Infectious Diseases 2008; 14: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicolas-Chanoine MH, et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. Journal of Antimicrobial Chemotherapy 2008; 61: 273–281. [DOI] [PubMed] [Google Scholar]
- 8.Peirano G, Pitout JD. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. International Journal of Antimicrobial Agents 2010; 35: 316–321. [DOI] [PubMed] [Google Scholar]
- 9.Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. Journal of Antimicrobial Chemotherapy 2011; 66: 1–14. [DOI] [PubMed] [Google Scholar]
- 10.Naseer U, et al. Molecular characterization of CTX-M-15-producing clinical isolates of Escherichia coli reveals the spread of multidrug-resistant ST131 (O25:H4) and ST964 (O102:H6) strains in Norway. Acta Pathologica Microbiologica et Immunologica Scandinavica 2009; 117: 526–536. [DOI] [PubMed] [Google Scholar]
- 11.Viswanathan R, et al. Multi-drug resistant gram negative bacilli causing early neonatal sepsis in India. Archives of Disease in Childhood, Fetal and Neonatal Edition 2012; 97: F182–187. [DOI] [PubMed] [Google Scholar]
- 12.Clermont O, et al. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. Journal of Antimicrobial Chemotherapy 2009; 64: 274–277. [DOI] [PubMed] [Google Scholar]
- 13.Oteo J, et al. Extended-spectrum-β-lactamase-producing Escherichia coli as a cause of pediatric infections: report of a neonatal intensive care unit outbreak due to a CTX-M-14-producing strain. Antimicrobial Agents and Chemotherapy 2012; 56: 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahjoub-Messai F, et al. Escherichia coli isolates causing bacteremia via gut translocation and urinary tract infection in young infants exhibit different virulence genotypes. Journal of Infectious Diseases 2011; 203: 1844–1849. [DOI] [PubMed] [Google Scholar]
- 15.Bauer AW, et al. Antibiotic susceptibility testing by a standardized single disk method. American Journal of Clinical Pathology 1966; 45: 493–496. [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 19th informational supplement, M100–S19. CLSI, Wayne, PA, USA, 2009.
- 17.Roy S, et al. Transmission of imipenem resistance determinants during the course of an outbreak of NDM-1 Escherichia coli in a sick newborn care unit. Journal of Antimicrobial Chemotherapy 2011; 66: 2773–2780. [DOI] [PubMed] [Google Scholar]
- 18.Clermont O, et al. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environmental Microbiology Reports 2013; 5: 58–65. [DOI] [PubMed] [Google Scholar]
- 19.Rodríguez-Baño J, et al. Virulence profiles of bacteremic extended-spectrum β-lactamase-producing Escherichia coli: association with epidemiological and clinical features. PLoS ONE 2012; 7: e44238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clermont O, et al. Determination of Escherichia coli O types by allele-specific polymerase chain reaction: application to the O types involved in human septicaemia. Diagnostic Microbiology and Infectious Disease 2007; 57: 129–136. [DOI] [PubMed] [Google Scholar]
- 21.Johnson JR, et al. Rapid and specific detection of Escherichia coli clonal group A by gene-specific PCR. Journal of Clinical Microbiology 2004; 42: 2618–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carattoli A, et al. Identification of plasmids by PCR-based replicon typing. Journal of Microbiological Methods 2005; 63: 219–228. [DOI] [PubMed] [Google Scholar]
- 23.Dhanji H, et al. Molecular epidemiology of fluoroquinolone-resistant ST131 Escherichia coli producing CTX-M extended-spectrum β-lactamases in nursing homes in Belfast, UK. Journal of Antimicrobial Chemotherapy 2011; 66: 297–303. [DOI] [PubMed] [Google Scholar]
- 24.Johnson JR, et al. Epidemiological correlates of virulence genotype and phylogenetic background among Escherichia coli blood isolates from adults with diverse-source bacteremia. Journal of Infectious Diseases 2002; 185: 1439–1447. [DOI] [PubMed] [Google Scholar]
- 25.Russo TA and Johnson JR. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. Journal of Infectious Diseases 2000; 181: 1753–1754. [DOI] [PubMed] [Google Scholar]
- 26.Das P, et al. Composition of Escherichia coli population in the neonatal gut: phylogroups and virulence determinants. Journal of Medical Microbiology 2013; 62: 1680–1687. [DOI] [PubMed] [Google Scholar]
- 27.Bailey JK, et al. Distribution of human commensal Escherichia coli phylogenetic groups. Journal of Clinical Microbiology 2010; 48: 3455–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carbonetti NH, et al. Aerobactin-mediated iron uptake by Escherichia coli isolates from human extraintestinal infections. Infection and Immunity 1986; 51: 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pramoonjago P, et al. Role of TraT protein, an anticomplementary protein produced in Escherichia coli by R100 factor, in serum resistance. Journal of Immunology 1992; 148: 827–836. [PubMed] [Google Scholar]
- 30.Huang SH, et al. Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation and protein expression. Journal of Infectious Diseases 2001; 183: 1071–1078. [DOI] [PubMed] [Google Scholar]
- 31.Johnson JR, et al. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. Journal of Infectious Diseases 2001; 183: 78–88. [DOI] [PubMed] [Google Scholar]
- 32.Johnson JR, et al. Phylogenetic origin and virulence genotype in relation to resistance to fluoroquinolones and/or extended-spectrum cephalosporins and cephamycins among Escherichia coli isolate from animals and humans. Journal of Infectious Diseases 2003; 188: 759–768. [DOI] [PubMed] [Google Scholar]
- 33.Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrobial Agents and Chemotherapy 2004; 48: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coelho A, et al. Characterisation of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. International Journal of Antimicrobial Agents 2010; 36: 73–78. [DOI] [PubMed] [Google Scholar]
- 35.Roy S, et al. Prevalence of ST131 virulence-associated strains among CTX-M-producing Escherichia coli in the gut of hospitalized neonates in India. Diagnostic Microbiology and Infectious Disease 2013; 77: 158–159. [DOI] [PubMed] [Google Scholar]