SUMMARY
The presence of norovirus in shellfish is a public health concern in Europe. Here, we report the results of an investigation into a norovirus gastroenteritis outbreak following a festive lunch which affected 84 (57%) residents and staff members of a nursing home in January 2012 in France. Individuals who had eaten oysters had a significantly higher risk of developing symptoms in the following 2·5 days than those who had not, the risk increasing with the amount eaten [relative risk 2·2 (1·0–4·6) and 3·3 (1·6–6·6) for 3–4 and 5–12 oysters, respectively]. In healthy individuals during those days, 29 (32%) subsequently became ill, most of whom were staff members performing activities in close contact with residents. Genogroup II noroviruses were detected in faecal samples, in a sample of uneaten oysters and in oysters from the production area. Identifying a norovirus's infectious dose may facilitate the health-related management of contaminated shellfish.
Key words: Foodborne infections, gastroenteritis, infectious disease epidemiology, investigation, Norwalk agent and related viruses
INTRODUCTION
Norovirus is the main pathogenic agent involved in acute gastroenteritis outbreaks, especially during the winter season [1]. Although transmission occurs mainly via the faecal–oral route, norovirus foodborne outbreaks are quite frequent. The consumption of shellfish is often implicated as these animals are susceptible to norovirus contamination [2, 3]. In France, oysters are usually consumed raw.
Oyster-related gastroenteritis outbreaks have been documented in several European countries. The prevalence of norovirus in oysters reported in three member states in 2012 varied from <10% to 90% during the winter season [4]. Although noroviruses are highly infectious the relationship between exposure dose and clinical illness has only been described for a few strains [5, 6] and needs to be explored in greater detail.
On 6 January 2012, the French heath authorities were notified about an outbreak of acute gastroenteritis involving about 30 residents and staff members of a nursing home that occurred during the previous night. A festive lunch had been served at the facility on 4 January 2012 during which raw oysters were served. The regional office of the French Institute for Public Health Surveillance (InVS), in collaboration with the French reference laboratory for shellfish microbiology (IFREMER), initiated epidemiological and microbiological investigations on 13 January 2012.
The objectives of the present study were to both describe this outbreak and to contribute to a greater understanding of the risks associated with oyster consumption.
METHODS
Epidemiological survey
The retrospective cohort study of residents and staff members of the nursing home (N = 160 persons) was undertaken between 17 and 19 January 2012. It was conducted using standardized questionnaires which collected demographic data, residents' accommodation units within the institution (five units), staff members' work activities, time of onset of illness, clinical signs and food consumption during the festive lunch on 4 January 2012. The meal itself comprised six foods served successively: raw oysters, shrimps, wild boar, potatoes, cheese and cake.
Case definition
A case was defined as any resident or staff member of the institution having experienced vomiting, diarrhoea, nausea or abdominal pain between 4 and 15 January 2012. Cases were distinguished as follows:
early cases, occurring during the first 60 h after the lunch (until 6 January inclusive);
late cases, occurring after 6 January.
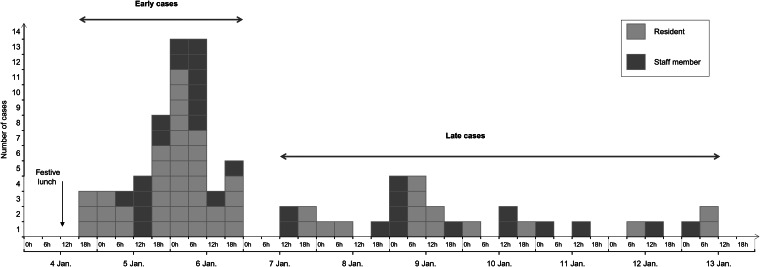
The 60-h cut-off was chosen on the basis of both the incubation period for Norwalk agent infection which ranges from 10 h to 50 h [7] and the two waves of the epidemic curve (Fig. 1). It is consistent with the threshold used by Gotz et al. [8].
Fig. 1.
Epidemic curve of the gastroenteritis outbreak by onset of clinical signs in the nursing home, France, January 2012 (n = 84).
Statistical analysis
Two analyses were performed according to the epidemic wave:
The first was restricted to individuals who had eaten food at the festive lunch. It studied the risk of becoming an early case according to food consumption and, for oysters, the quantity of oysters consumed. This quantity was classified into three categories defined by the two terciles, in order to assess a dose-response relationship using a test for trend. Late cases were classified in the group of non-cases (healthy individuals during this first epidemic wave) in the analysis.
The second analysis studied the risk of a resident and staff member becoming late cases according to, respectively, the resident's accommodation unit and the staff member's work duties involving regular/close contact with residents. In line with Gotz et al. [8], early cases were excluded from this analysis.
Data processing was performed in accordance with pre-established disease outbreak investigation regulations defined by the French regulatory authority on personal data (Commission Nationale de l'Informatique et des Libertés) and guaranteed by InVS (number 341194v42, 16 March 2011).
Data from completed questionnaires were entered using EpiData v. 2.1b (www.epidata.dk). Statistical analyses were conducted using Stata software v. 12.0(StatCorp, USA).
Microbiological analyses
Virological confirmation of cases
Three initial faecal samples from three ill residents were analysed for bacterial pathogens and rotaviruses in a local laboratory.
Other faecal specimens were sampled on 22 and 24 January from five residents who had previously experienced diarrhoea. These specimens were sent to the French National Reference Centre (NRC) for enteric viruses where they were screened for several viruses using molecular methods. Noroviruses were detected by real-time reverse transcriptase–polymerase chain reaction (rRT–PCR), using probes described by Lyman et al. [9] and Da Silva et al. [10] for genogroup I (GI) and genogroup II (GII), respectively. Group A rotaviruses, astroviruses, adenoviruses, noroviruses, sapoviruses and Aichi viruses were all detected by RT–PCR, as described by Sdiri-Loulizi et al. [11]. Enteroviruses were detected with a RT–PCR adapted from Chapman et al. [12], and hepatitis A viruses with a RT–PCR, described by Robertson et al. [13]. Genotyping was then conducted by sequencing a gene fragment, as described previously [11]. For noroviruses, typing was performed both on the gene encoding the RNA polymerase and the gene encoding the capsid.
Shellfish analyses
Four uneaten oysters, kept as a control sample from the lunch on 4 January 2012 were sent to the IFREMER laboratory. Additional oyster samples, comprising at least 12 oysters per sample, were collected from the production area, from the producer's farm, and from the estuary upstream and downstream of the farm (about 2·0 and 0·7 km, respectively).
All oysters were kept at 4 °C during shipment to the laboratory. Analyses were performed as described previously [14]. Briefly, the stomach and digestive diverticula (DT) were removed by dissection (1·5 g portions), homogenized, extracted with chloroform-butanol, and treated with Cat-Floc (Calgon, USA). The virus was then concentrated by polyethylene glycol 6000 (Sigma, France) precipitation. Viral nucleic acids (NA) were extracted with a NucliSens kit (bioMérieux, France), suspended in 100 μl elution buffer and analysed immediately or kept frozen at −80 °C [15]. The efficiency of the virus extraction procedure was determined for each extraction by seeding 104 50% tissue culture-infective doses of mengovirus prior to sample processing and determining the amount of mengovirus recovered by rRT–PCR as described previously [14].
All NA extracts were screened using rRT–PCR, with primers and probes for norovirus and mengovirus, following previously described procedures [10]. Two negative amplification controls (water) were included in each amplification series. Only samples with an extraction efficiency above 10% were considered for quantification. The number of RNA copies present in positive samples was estimated by comparing the Ct values with standard curves for norovirus GI and GII. The final concentration was then determined on the basis of the NA volume analysed (5 μl of 100 μl NA extract) and the measured weight of the DT (1·5 g was analysed) [14]. Similarly to clinical samples, noroviruses were typed by amplification using primers targeting the capsid gene.
RESULTS
Epidemiological results
Characteristics of respondents
One hundred and fifty-two questionnaires were completed. Survey participation rate was 100% for nursing-home residents (98/98) and 87% for staff members (54/62).
One hundred and forty-seven questionnaires (97%) were suitable for analyses (94 residents, 53 staff members). Median age was 87 years for residents [interquartile range (IQR) 84–92] and 40 for staff members (IQR 28–47).
Characteristics of cases and clinical signs
The attack rate in the cohort was 57% (84/147). It was similar in residents (53/94) and staff members (31/53). Diarrhoea and vomiting were experienced by 66% and 55% of the cohort, respectively. Abdominal pain and nausea (without vomiting) were experienced by 44% and 13%, respectively. Nine (6%) individuals experienced nausea or abdominal pain only. Only 4% had fever. The proportion of cases experiencing diarrhoea was significantly higher in residents than in staff members (76% vs. 48%, respectively, P < 0·02).
One person was hospitalized following inhalation of vomit. Another died 2·5 days after the onset of diarrhoea.
The onset of clinical signs occurred over a 10-day period (Fig. 1), with:
55 (65%) early cases occurring within 60 h following the lunch. The peak of the outbreak occurred on 6 January between 00:00 and 06:00 hours.
29 (35%) late cases, occurring after 6 January.
Consumption of food during lunch
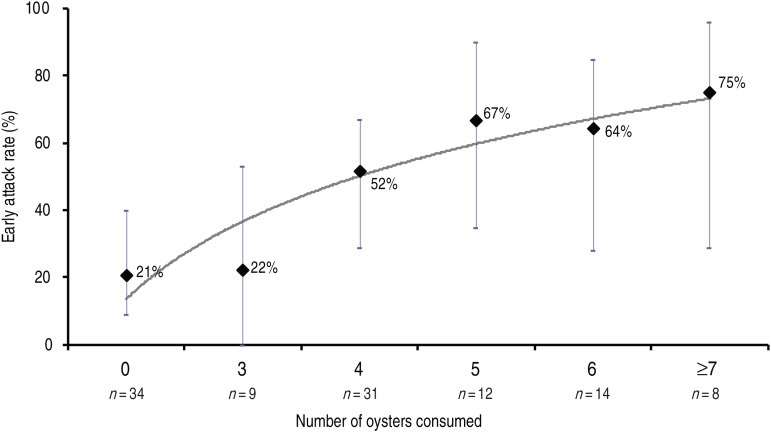
Of the 122 individuals who ate lunch on 4 January, 53 (43%) were defined as early cases. The relative risks for becoming an early case for the six foods served were 2·6 [95% confidence interval (CI) 1·3–5·2] for oysters, 1·6 (95% CI 0·8–3·0) for shrimps, 1·2 (95% CI 0·5–2·7) for wild boar, 2·7 (95% CI 0·4–16·3) for potatoes, 0·9 (95% CI 0·5–1·8) for cheese and 0·9 (95% CI 0·5–1·4) for cake. Consumption of oysters was the only significant risk factor. A dose-response relationship was observed, the risk increasing with the quantity of oysters consumed (test for trend P < 0·001, Table 1 and Fig. 2).
Table 1.
Early attack rate, relative risk and incubation period by quantity of oysters consumed during lunch in the nursing home, France, January 2012
| Total | No. of cases | AR | RR | (95% CI) | Time of incubation (hours) | ||
|---|---|---|---|---|---|---|---|
| Median | (IQR) | ||||||
| No. of oysters consumed | |||||||
| 0 | 34 | 7 | 21% | 1 | 43 | (34–49) | |
| 3–4 | 40 | 18 | 45% | 2·2 | (1·0–4·6) | 38 | (31–44) |
| 5–12 | 34 | 23 | 68% | 3·3 | (1·6–6·6) | 38 | (32–43) |
AR, Attack rate; RR, relative risk; CI, confidence interval; IQR, interquartile range.
Test for trend: P < 0·001.
Fig. 2.
Early attack rate by quantity of oysters consumed during lunch in the nursing home, France, January 2012.
Of the early cases who consumed oysters, the median incubation period was 38 h (IQR 31–44), with no difference according to quantity of shellfish consumed (Table 1).
Secondary transmission
Of the 92 healthy individuals during the 60 h after the lunch, 29 (32%) were defined as late cases.
Staff members with activities involving regular/close contact with residents had a significantly higher secondary attack rate (14/28, 50%) than other staff members (0/6, 0%, Fisher's exact test P = 0·03).
Within the five accommodation units, the late attack rate in residents was inversely correlated to the early attack rate (Table 2). The overall attack rates (early + late) ranged between 47% and 67%.
Table 2.
Early and late attack rates by accommodation unit or type of work activity in the nursing home, France, January 2012
| Total (1) | Early cases | Late cases | All cases | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No. at risk (2)* | No. of cases (3) | AR (3/2) | No. at risk (4)† | No. of cases (5) | AR (5/4) | No. of cases (3 + 5) | AR (3 + 5)/1 | ||
| Residents | |||||||||
| Unit location | |||||||||
| 1 | 19 | 19 | 4 | 21% | 15 | 5 | 33% | 9 | 47% |
| 2 | 19 | 19 | 7 | 37% | 12 | 5 | 42% | 12 | 63% |
| 3 | 19 | 18 | 7 | 39% | 12 | 3 | 25% | 10 | 53% |
| 4 | 18 | 18 | 12 | 67% | 6 | 0 | 0% | 12 | 67% |
| 5 | 19 | 19 | 8 | 42% | 11 | 2 | 18% | 10 | 53% |
| Total | 94 | 93 | 38 | 41% | 56 | 15 | 27% | 53 | 56% |
| Staff members | |||||||||
| Activities in close contact with residents | |||||||||
| Yes | 38 | 17 | 8 | 47% | 28 | 14 | 50% | 22 | 58% |
| No | 13 | 12 | 7 | 58% | 6 | 0 | 0% | 7 | 54% |
| Total | 51 | 29 | 15 | 52% | 34 | 14 | 41% | 29 | 57% |
AR, Attack rate.
Individuals who ate at the festive lunch.
Individuals who were not early cases.
Microbiological results
In all three faecal samples analysed locally, neither bacterial pathogen nor rotavirus was identified. Despite being collected more than 2 weeks after diarrhoeal stage, 4/5 faecal samples sent to NRC for enteric viruses tested positive for norovirus GII. One of these also tested positive for Aichi virus.
Norovirus typing by genetic sequencing identified two strains GII.g/GII.1 and one strain GII.7. The latter related to an early case who attended the festive lunch without consuming oysters.
Food and environmental results
The control sample of four uneaten oysters was collected on 17 January 2012 and sent to the IFREMER laboratory. This sample, with an extraction efficiency of 12%, was contaminated by GII norovirus at a concentration of 105 RNA copies/g DT. Despite several assays, no sequence could be obtained, presumably due to the low concentration.
The implicated batch of oysters were produced on the western coast of France (Brittany) in a small estuary and harvested in November 2011 in a class A area [<230 Escherichia coli organisms/100 g shellfish (meat and liquid), European regulation 54/2004/EC], before relocation 20 m away to an area in front of the shellfish farm in a class B area (<4600 E. coli organisms/100 g). No information could be obtained about purification details before packaging.
All samples collected from the farm and from the estuary showed acceptable extraction efficiencies (>10%) with some variation according to the sampling date (Table 3). The sample collected on 18 January 2012 from the farm tested negative for norovirus, but samples collected 2 days later from the estuary including the area in front of the shellfish farm tested positive for norovirus GII (Table 3). All samples displayed a low concentration varying from below the limit of quantification (70 RNA copies/g DT) to a peak of 540 RNA copies/g DT on 20 January 2012.
Table 3.
Norovirus detected in oysters sampled from three sampling points in the oyster production area, between 20 January and 7 March 2012
| Date | Average extraction efficiency | Downstream class A | Shellfish farm | Upstream class B |
|---|---|---|---|---|
| 20 January | 11% | 540 | +<LQ | 95 |
| 30 January | 10% | <LD | <LD | 430 |
| 7 February | 19% | – | +<LQ | <LD |
| 13 February | 41% | – | +<LQ | <LD |
| 21 February | 11% | – | 100 | <LD |
| 7 March | 10% | – | <LD | <LD |
Number of RNA copies/g DT for positives samples.
+ <LQ, Positive but below limit of quantification (70 RNA copies/g DT).
<LD, Below limit of detection (20 RNA copies/g DT).
Two norovirus GII sequences were obtained from the sample collected on 20 January but did not match the sequence obtained from clinical samples. Sampling was performed until negative results were consistently measured (end of February).
DISCUSSION
Epidemiological and microbiological investigations identified two sources of norovirus infections occurring in the nursing home. The outbreak related first to the consumption of contaminated oysters, and subsequently to transmission from many contagious persons within the establishment.
What makes this gastroenteritis investigation of particular interest is the observed dose-response relationship between the quantity of oysters consumed and acute gastroenteritis which provides an additional and important causal argument to the significant link between consumption of oysters and illness. This relationship raises the question of whether a minimum infectious dose (as estimated by genome copy detection) exists for clinical cases of gastroenteritis. This minimum infectious dose may be <100 viral particles and may differ according to the virus genogroup. Teunis et al. estimated that infected subjects had a dose-dependent probability of becoming ill ranging from 0·1 (at a dose of 103 Norwalk virus genomes) to 0·7 (at 108 virus genomes) [5]. We previously demonstrated that oysters contaminated with <100 RNA copies/g DT were implicated in French gastroenteritis outbreaks [16, 17]. The present investigation found a dose–illness relationship consistent with a model calculated from previous outbreaks [6]. This is important, especially as the European Food Safety Authority recently concluded that it is not currently possible to quantify the public health impact of different established limits or to define thresholds of acceptable risks [4]. Our study provides further evidence that small amounts of virus may result in clinical cases. Data obtained on food directly linked to cases of human illness are still rare and the detailed analysis performed here, which took into account the number of oysters consumed, should prove helpful for future risk analysis model applications [6].
A similar event to the one investigated here occurred in France in 2010 [18], where two distinct foodborne outbreaks involving oysters and mussels led to the temporary withdrawal of these foods, to fishing area closures and cessation of the marketing of shellfish from the contaminated area. In our case, the investigations did not lead to such specific measures. This may have been because of (1) the delay between the date of dispatch from the shellfish farm and the date of confirmation of the origin of the outbreak (28 days), (2) the absence of reports of other outbreaks related to the consumption of shellfish from the same area and (3) the absence of any known sewage treatment plant malfunction affecting the area. Furthermore, the contamination detected in oyster samples from the farm was quite low, both in terms of number of viral RNA copies/g DT and strain diversity, as only norovirus GII was detected. The presence of such low levels of norovirus and one single genogroup has already been demonstrated in some French outbreaks related to shellfish consumption and in marketed French oysters [14, 19]. Multiple-strain contamination is the sign of direct raw sewage contamination, an event becoming increasingly rare in French production areas after large outbreaks occurring in the south of France in 2002 and 2006 led to local authorities and producers implementing stricter safety measures [16, 17]. Oysters are usually produced in the deep sea (class A, EC regulation) and producers usually harvest them twice a month as harvesting in easiest at times of highest tidal coefficient. They are then kept close to the coast in potentially contaminated areas. This present outbreak highlights the need to improve farming practices to avoid contamination, as we know that depuration is not efficient for viral elimination [19, 20].
In the present study, the analysis of secondary transmission within the nursing home (from a large number of infectious people in the days after the lunch) confirms the high risk of secondary transmission between residents and staff members in regular contact with residents. This risk has already been described by Kaplan et al. [21]. Faecal and vomit specimens of ill people infected by norovirus can contain viral concentrations as high as 106 viral particles/ml [22], and droplets projected during vomiting may spread over >1 m. It has recently been shown, in healthcare settings, that transmission of norovirus is mainly caused by symptomatic cases [23]. The maximum attack rate in our cohort was 67% in the accommodation units, consistent with three hypotheses: herd immunity threshold above which the virus transmission stops, the presence of asymptomatic infected people and the presence of people resistant to the infection.
The characteristic of the curve of attack rate as a function of the number of oysters consumed supports the hypothesis of a threshold of clinical response to norovirus exposure, capped at about 70–80%. This finding is consistent with recent studies describing the mechanism of resistance to noroviruses infection [14, 24–26]: norovirus strains bind to glycans of digestive cells belonging to the histo-blood group antigen family whose synthesis depends on the combined polymorphism on loci ABO, FUT2 and FUT3. An individual is deemed a ‘non-secretor’ if he/she does not possess the FUT2 allele. ‘Non-secretor’ volunteers exposed to a viral strain did not experience either clinical signs or antibody response and did not excrete the more common strains of norovirus which were detected in the present study. These individuals who are genetically resistant to infection may represent about 20–30% of the population [27].
Outbreak epidemiological investigations frequently suffer from poor quality data collection. In the present study, the delay between the lunch and the investigations (>10 days) caused memory problems concerning food consumption and time of onset of clinical signs. Most importantly, the use of a cut-off at a fixed point in time may have led to some cases being inaccurately classified as having been infected by the consumption of oysters, when they could have been infected by contact with the first symptomatic cases. These classification errors could have influenced the high attack rate in the group of guests who did not eat oysters (21%). Nevertheless, the dose-response relationship was sufficiently significant to confirm causality of oyster consumption as the original source of the outbreak, confirmed by the detection of norovirus in the uneaten oysters. Although stool specimens were taken more than 2 weeks after the diarrhoeal stage, detection of noroviruses was possible owing to the fact that noroviruses persist in faeces for several weeks [28].
A careful examination of the epidemic curve is needed to identify different sequences of transmission. If the analysis of foods served had included all cases (early + late cases) as is the usual practice, the analytical study would not have identified the oysters as a risk factor for gastroenteritis, the relative risk not being statistically significant [1·3 (95% CI 0·9–1·9) vs. 2·6 (95% CI 1·3–5·2)] in our study of the first epidemic wave. For norovirus the cut-off at 60 h after a punctual exposition seems adequate.
In conclusion, this outbreak highlights the need for swift recognition of foodborne norovirus outbreaks related to shellfish consumption. It is important to react rapidly, especially as shellfish production areas can be promptly investigated and temporarily closed if necessary.
ACKNOWLEDGEMENTS
The authors acknowledge Sylvain Parnaudeau from IFREMER for microbiological analyses of oysters, Jacques Le Pendu from INSERM U707 at the University of Nantes and Nathalie Ruvoën from the National Veterinary School of Nantes for their contribution to the discussion.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Van Beek J, et al. Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Eurosurveillance 2013; 18: 8–9. [PubMed] [Google Scholar]
- 2.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. New England Journal of Medicine 2009; 361: 1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Guyader FS, Atmar RL, Le Pendu J. Transmission of viruses through shellfish: when specific ligands come into play. Current Opinion in Virology 2012; 2: 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Food Safety Authority Panel on Biological Hazards. Scientific opinion on norovirus in oysters: methods, limits and control options. European Food Safety Authority Journal 2012; 10(1), 2500 (39 pp). [Google Scholar]
- 5.Teunis PF, et al. Norwalk virus: how infectious is it? Journal of Medical Virology 2008; 80: 1468–1476. [DOI] [PubMed] [Google Scholar]
- 6.Thebault A, et al. Infectivity of GI and GII noroviruses established from oyster related outbreaks. Epidemics 2013; 5: 98–110. [DOI] [PubMed] [Google Scholar]
- 7.Heymann DL. Control of Communicable Diseases Manual, 18th edn. Washington: APHA, 2004. [Google Scholar]
- 8.Gotz H, et al. Clinical spectrum and transmission characteristics of infection with Norwalk-like virus: findings from a large community outbreak in Sweden. Clinical Infectious Diseases 2001; 33: 622–628. [DOI] [PubMed] [Google Scholar]
- 9.Lyman WH, et al. Prospective study of etiologic agents of acute gastroenteritis outbreaks in child care centers. Journal of Pediatrics 2009; 154: 253–257. [DOI] [PubMed] [Google Scholar]
- 10.Da Silva AK, et al. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Applied and Environmental Microbiology 2007; 73: 7891–7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sdiri-Loulizi K, et al. Acute infantile gastroenteritis associated with human enteric viruses in Tunisia. Journal of Clinical Microbiology 2008; 46: 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman NM, et al. Molecular detection and identification of enteroviruses using enzymatic amplification and nucleic acid hybridization. Journal of Clinical Microbiology 1990; 28: 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson BH, Brown VK, Khanna B. Altered hepatitis A VP1 protein resulting from cell culture propagation of virus. Journal of Virology 1989; 13: 207–212. [DOI] [PubMed] [Google Scholar]
- 14.Le Guyader FS, et al. Comprehensive analysis of a norovirus-associated gastroenteritis outbreak, from the environment to the consumer. Journal of Clinical Microbiology 2010; 48: 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sima LC, et al. Calicivirus removal in a membrane bioreactor wastewater treatment plant. Applied and Environmental Microbiology 2011; 77: 5170–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Guyader FS, et al. Detection of multiple noroviruses associated with an international gastroenteritis outbreak linked to oyster consumption. Journal of Clinical Microbiology 2006; 44: 3878–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Guyader FS, et al. Aichi virus, norovirus, astrovirus, enterovirus, and rotavirus involved in clinical cases from a French oyster-related gastroenteritis outbreak. Journal of Clinical Microbiology 2008; 46: 4011–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillois-Becel Y, et al. Investigation of two foodborne disease outbreaks linked to shellfish consumption in the Loire-Atlantique district, France, 2010 [in French]. Bulletin Epidémiologique Hebdomadaire 2010; 37: 390–392. [Google Scholar]
- 19.Schaeffer J, et al. Norovirus contamination on French marketed oysters. International Journal of Food Microbiology 2013; 166: 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richards GP, McLeod C, Le Guyader FS. Processing strategies to inactivate enteric viruses in shellfish. Food Environmental Virology 2010. Report No.: 2. [Google Scholar]
- 21.Kaplan JE, et al. An outbreak of acute nonbacterial gastroenteritis in a nursing home. Demonstration of person-to-person transmission by temporal clustering of cases. American Journal of Epidemiology 1982; 116: 940–948. [DOI] [PubMed] [Google Scholar]
- 22.French Agency for Food Environmental and Occupational Health and Safety (Anses). Opinion on a risk assessment relating to the reopening of a shellfish growing area closed due to the presence of calicivirus (norovirus and sapovirus) in live shellfish. Maison-Alfort, France: Anses, February 2011.
- 23.Sukhrie FH, et al. Nosocomial transmission of norovirus is mainly caused by symptomatic cases. Clinical Infectious Diseases 2012; 54: 931–937. [DOI] [PubMed] [Google Scholar]
- 24.Hutson AM, et al. Norwalk virus infection and disease is associated with ABO histo-blood group type. Journal of Infectious Diseases 2002; 185: 1335–1337. [DOI] [PubMed] [Google Scholar]
- 25.Larsson MM, et al. Antibody prevalence and titer to norovirus (genogroup II) correlate with secretor (FUT2) but not with ABO phenotype or Lewis (FUT3) genotype. Journal of Infectious Diseases 2006; 194: 1422–1427. [DOI] [PubMed] [Google Scholar]
- 26.Nordgren J, et al. Norovirus gastroenteritis outbreak with a secretor-independent susceptibility pattern, Sweden. Emerging Infectious Diseases 2010; 16: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Pendu J, et al. Mendelian resistance to human norovirus infections. Seminars in Immunology 2006; 18: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atmar RL, et al. Norwalk virus shedding after experimental human infection. Emerging Infectious Diseases 2008; 14: 1553–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]