SUMMARY
In a 2-year longitudinal study of adult animals on 15 dairy farms and four sheep farms in Lancashire, UK, Arcobacter spp. were isolated from all farms although not at every sampling occasion. Faecal samples were collected and cultured using standard techniques for isolation of campylobacters. Assignment to species was via PCR assays. Apparent prevalence of Arcobacter spp. was higher in dairy cattle compared to sheep (40·1% vs. 8%, P < 0·001) and in housed cattle compared to cattle at pasture (50·1% vs. 20·9%, P < 0·001). This was reflected in the higher prevalence observed in herds that were housed (n = 4) all year compared to herds that grazed cattle on pasture in the summer and housed cattle in the winter (n = 11) (55·5% vs. 36%, P < 0·001). In the case of sheep, peak prevalence was observed in autumn with increased prevalence also being associated with improving pasture quality. There was an apparent inverse association between the faecal pat prevalence of Arcobacter spp. and Campylobacter jejuni although this may in part be an artefact of laboratory test method sensitivity, whereby a relative increase in the frequency of one bacterial species would reduce the sensitivity of detecting the other.
Key words: Arcobacter, Campylobacter, epidemiology
INTRODUCTION
Arcobacter spp. are Gram-negative bacteria belonging to the family Campylobacteraceae first reported by Ellis et al. [1] who isolated a Vibrio-like organism from an aborted bovine foetus and named it Campylobacter cryaerophilus. Arcobacter spp. have been isolated at high levels from poultry carcasses but with much lower isolation rates from poultry intestinal contents suggesting contamination from other sources occurs during processing [2]. High faecal prevalences have been observed in mammals including ruminants [3, 4] and pigs [4]. Arcobacter spp. have been isolated from a wide range of environmental sources such as water [5] and from food products including retail meats and milk [6].
Faecal pat prevalence estimates of Arcobacter spp. in cattle range from 14·3% [3] to 39% [4] suggesting it is a common inhabitant of the cow intestine while a similar prevalence range has been observed in sheep [4, 7]. While Arcobacter spp. have been isolated from cases of abortion [1] and mastitis [8] in cattle, evidence for it being a major pathogen in cattle is scant.
The precise role of Arcobacter spp. in human disease is unclear as a result of its isolation at low frequencies from both diarrhoeic stool samples and from asymptomatic individuals [9–11]. However, there is increasing evidence that it may be associated with diarrhoeal disease in people; there are case reports of the isolation of Arcobacter spp. from the faeces of human cases of diarrhoea [12, 13] and reports of outbreaks of diarrhoea associated with Arcobacter butzleri [14, 15].
In contrast, Campylobacter spp., especially C. jejuni, are well recognized human foodborne pathogens accounting for up to 500 000 cases per annum in the UK [16]. There is a wealth of data implicating poultry as the major source of human campylobacter infections accounting for between 50% and 70% of human cases, including ‘natural experiments’ as occurred in Belgium in 1999 [17], case-control studies [18, 19] and source attribution modelling results [20, 21]. Ruminant and environmental sources are believed to account for much of the remainder. Previous studies have demonstrated seasonality both in human cases [22, 23] and in excretion patterns in ruminants [24] with peak occurrence in both hosts occurring in the spring and summer months. A study in North West England [25] (in which the current study is nested) suggested that in dairy cattle, at least, this apparent seasonality was a reflection of the animals' environment – at pasture or housed; with a lower pat prevalence observed in housed animals irrespective of season.
The objective of the present study is to describe the spatio-temporal epidemiology of Arcobacter spp. on a cohort of dairy and sheep farms in the North West of England and draw comparisons with the concurrently collected C. jejuni epidemiological data from the same cohort [25].
MATERIALS AND METHODS
The data presented here were collected as part of a larger study [25] investigating the epidemiology of C. jejuni in ruminants, thus culture and isolation methods were optimized for the isolation of C. jejuni. However, consequent on the isolation methodology employed, other Campylobacter spp. and Arcobacter spp. were also identified: the focus of this report is the estimation of the faecal pat prevalence of Arcobacter spp.
The study design was a repeated cross-sectional study over a 2-year period starting in January 2006. Fourteen dairy and four sheep farms were recruited with the help of three spatially separated veterinary practices in Lancashire serving the Southern Fylde (zone 1), North Lancashire (zone 2) and South East Lancashire (zone 3). Six dairy farms were recruited in zone 1 while four dairy and two sheep farms were recruited in each of the other zones. One farm in zone 2 ceased trading in December 2006 and was replaced with a neighbouring farm for the rest of the study. Another farm in zone 2 ceased keeping cattle in June 2007 thus sampling on this farm was incomplete. Eligibility criteria for entry to the study were: dairy farms with over 100 adult cows with or without a sheep enterprise; sheep farms with over 150 breeding ewes and no other livestock enterprises.
Each farm was visited on 12 occasions at 8-week intervals when 20 freshly voided faecal samples were collected at random from lactating cows on dairy farms or adult sheep on sheep farms. Samples were only collected from animals observed to defecate by the author. Each faecal pat was sampled from at least three sites within the pat and mixed thoroughly in a sterile sample pot. In the case of sheep faecal pellets, at least three were collected. Samples were transported to the laboratory on ice.
At each visit, current management and production details were obtained via a short questionnaire delivered by the investigator. Average milk yield per cow was calculated by dividing the total production on the day of sampling by the number of cows milked on that day. Farms were coded according to their overall management practice as follows: farms that housed cattle all the year (n = 4) and farms that grazed cattle at pasture during the summer and housed cattle during the winter months (n = 11).
For dairy cows, faecal consistency was scored on a scale of 1–5 [26] and faecal fibre length and presence of partially digested grains assessed by sieving [27]. For analysis purposes, faecal consistency was re-coded as a binary variable ‘loose faeces’ with pats scoring <3 classified as ‘firm’ and pats scoring 4 or 5 classified as ‘loose’.
In the laboratory, 1 g faeces was placed in 9 ml Campylobacter enrichment broth (IDG Ltd, UK) with cefoperazone, vancomycin, trimethoprim and cycloheximide (CVTC supplement: IDG Ltd) and after homogenizing for 30 s in a Colworth 80 stomacher (A. J. Seward & Co. Ltd, UK) was incubated in a plastic universal bottle for 24 h at 37°C in a variable atmosphere incubator (VAIN, Don Whitley Scientific Ltd, UK) maintaining a microaerobic atmosphere (12% CO2, 3% H2, 11% O2, 74% N2). After incubation, 50 μl of the enrichment broth was inoculated onto a Campylobacter blood-free selective agar (CSA) plate (IDG Ltd) enriched with cefoperazone and amphotericin (CA supplement: IDG Ltd). A second CSA plate was inoculated with a 5-μl loopful of enrichment broth. The CSA plates were incubated at 37°C in a microaerobic atmosphere for 60–72 h after which time plates were examined and up to four putative Campylobacter colonies (per faecal sample) were subcultured onto blood agar plates and incubated at 37°C under microaerobic conditions as described earlier. After 72 h incubation, single colonies were subcultured onto two blood agar plates. One plate was incubated for 48 h under microaerobic conditions and the other plate incubated for 48 h at 30 °C in air.
A crude DNA aqueous lysate was prepared by inoculating 200 μl distilled water with a small amount of the culture, heating at 100°C for 15 min followed by centrifugation at 11337 g for 10 min. All putative Campylobacter isolates were frozen in Microbank tubes (Pro-Lab Diagnostics, UK) and stored at −80 °C.
Assignment to species of putative campylobacters was by PCR using the following assays: 16S rRNA PCR for identification of the genus Arcobacter [28]; multiplex PCR for identification of C. jejuni, C. coli and C. lari [29]; duplex PCR for identification of C. fetus and C. hyointestinalis [30] and a monoplex PCR [31] for identification of any C. jejuni that failed to be identified by the colony multiplex PCR.
Data analysis was performed using Stata v. 12 (StataCorp, USA). Covariates recorded at sampling visits and considered for inclusion in statistical analyses are described in Table 1. The sampling period was split into ‘summer’ and ‘winter’ with the winter period defined as being from 1 October to 30 April. The term ‘sampling event’ is defined as ‘a visit to a farm to collect samples’.
Table 1.
Description of variables collected at sampling visits for initial inclusion in statistical analyses
| Variables | Species | Type | Description and coding of variable |
|---|---|---|---|
| Farm identity | Cattle/sheep | Categorical | 0 = housed in winter at pasture in summer (HWPS) |
| Management system | Cattle | Binary | 1 = housed all year (HAY) |
| Purchase policy | Cattle | Categorical | 0 = no purchased stock (closed herd) |
| 1 = occasional purchase of cows | |||
| 2 = frequent purchase | |||
| Group size | Cattle/sheep | Continuous | Number of animals in sampled group (transformed into quintiles) |
| Where sampled | Cattle/sheep | Binary | Inside = 1 or outside = 0 |
| Zone | Cattle/sheep | Categorical | 1 = Southern Fylde |
| 2 = North Lancashire | |||
| 3 = South East Lancashire | |||
| Date of sampling | Cattle/sheep | dd/mm/yy | |
| Average daily milk yield | Cattle | Continuous | Average daily milk yield (litres) on sampling day (transformed into quintiles) |
| Feeding system | Cattle | Categorical | Feeding system used as follows |
| 1 = TMR (total mixed ration) | |||
| 2 = hybrid TMR – TMR and parlour feed | |||
| 3 = grazing and buffer feed and parlour feed | |||
| 4 = grazing and parlour feed | |||
| 5 = silage and parlour feed | |||
| Number of fresh cows | Cattle | Continuous | Number of cows calved within last month (transformed into quintiles) |
| Faecal score | Cattle | Categorical | Score of 1–5 depending on consistency with 1 being very firm and 5 being liquid [26] |
| Binary | Firm = score of 2 or 3; loose = score of 4 or 5 | ||
| Sieve score | Cattle | Categorical | Score of 1–3 being a composite score for presence of grains and long fibre (>1) with 1 = no grains or long fibre, 3 = large amounts of grains and presence of many long fibres |
| Stocking density | Sheep | Continuous | Number of sheep per hectare (transformed into quintiles) |
| Pasture quality | Sheep | Categorical | Quality of pasture scored 1–3 |
| 1 = poor; 2 = mediocre; 3 = lush | |||
| Binary | 0 = rough (poor & mediocre); 1 = lush | ||
| Lambing season | Sheep | Categorical | Were the flock lambing at the time of sampling? |
| 1 = sampled in lambing season | |||
| 0 = sampled out of lambing season |
Multiple logistic regression models were fitted with the binary outcome variable being the Arcobacter spp. test result (presence or absence) of the faecal pat sample. Collinearity and correlation between covariates was investigated using cross-tabulation and Cramer's φ statistic. If present, one of the covariates was discarded taking into account biological plausibility. All remaining covariates (Table 1) were included in the initial models. A backward elimination strategy [32] was employed whereby a full model was built and then each variable removed in turn, a likelihood ratio test performed and the resultant P value noted. The variable with the highest P value was then omitted and the process repeated. This process was repeated until only variables with P < 0·05 remained in the model. The omitted variables were then added back in turn, starting with the lowest P value, a likelihood ratio test performed after each addition, and the variable retained if P < 0·05. This process was continued until no further variables could be added, to produce the final model. Interactions between variables in the final model were considered for inclusion if biologically plausible and retained if they improved model fit as judged by the likelihood ratio test.
The technique of selecting four isolates per plate and testing each independently for Arcobacter spp. and Campylobacter spp. introduces a dependency in the sensitivity of isolation for each species, in that the probability of isolating a specific organism, e.g. Arcobacter spp. from a faecal pat co-colonized by another organism, e.g. Campylobacter spp. will depend on the relative frequency of each organism within the pat. Thus isolation of a Campylobacter would reduce the probability of isolating an Arcobacter and vice versa. To account for this dependency a binary variable ‘Campylobacter present’ was included as a covariate in the multivariable models.
Time was considered as a composite of four sine and cosine functions (harmonic regression) to allow modelling of seasonal periodicity if present [33]. Four time covariates (x1, x2, x3, x4) were generated as follows:
where t = week number with week 1 being the first week in January 2006 when sampling commenced.
Crude prevalence estimates adjusted for clustering at farm level were obtained from univariate logistic regression models; random-effects models in the case of the cattle dataset while logistic models with farm identity as a fixed effect were employed for the sheep dataset.
Separate multiple logistic regression models were fitted for cattle and sheep with the underlying a priori hypothesis being that time of year or season is a primary determinant of the probability of a faecal pat being colonized by Arcobacter spp. with other more proximal covariates also having an effect. A secondary hypothesis was that management practices, namely whether cattle were housed all year or were grazed in the summer, would also have an effect. A mixed-effects modelling strategy was employed for the cattle dataset with farm identity as a random effect while a fixed-effects model was fitted in the case of the sheep dataset due to only four farms being sampled.
Seasonal periodicity in prevalence was described graphically by plotting the logit predictions of isolating Arcobacter spp. estimated from the regression models, using the time covariates (including time interaction terms) only.
RESULTS
The median herd size, defined as total number of lactating and dry cows, was 145 [inter-quartile range (IQR) 104–200, range 71–80] cows. The breed of cattle in 14 of the herds was Holstein Friesian while one herd comprised Ayrshire and Ayrshire × Friesian. Four herds were housed all the time during the study period while one herd was housed for the entire second year of the study. All other herds were housed during the winter months but grazed outside during the summer. Fourteen of the herds were housed in cubicle accommodation while the Ayrshire herd was housed in straw yards. Median annual milk yield was 8000 (IQR 7200–9000) litres. Two of the sheep farms were lowland with one farm grazing on the salt marshes of the River Lune estuary while two were upland with one utilizing summer grazing on moorland. The predominant breeds of sheep kept were Swaledale and North Country Mules. The two upland farms kept 1000 and 700 ewes, respectively, while one lowland farm had 700 ewes and the other kept 150 ewes.
Twenty faecal samples were collected at each sampling visit, yielding a total of 4260 samples. Putative Campylobacter spp. or Arcobacter spp. were isolated from 2724 (64%) faecal pats. Four potential isolates were taken from each faecal sample yielding 17 040 potential bacterial isolates. In total, 9499 putative Campylobacter or Arcobacter spp. isolates were grown and 4535 (47·7%) were identified as Arcobacter spp. (Table 2).
Table 2.
Distribution of the number of isolates and proportion of Campylobacter spp. and Arcobacter spp. isolated by host species
| Cattle | Sheep | |
|---|---|---|
| Number of faecal pat samples | 3300 | 960 |
| Number of potential isolates (4 per pat) | 13 200 | 3840 |
| Number of isolates grown (% of potential isolates) | 7779 (58·9%) | 1720 (44·8%) |
| Arcobacter spp. (% of actual isolates) | 4299 (55·3%) | 236 (13·7%) |
| Campylobacter jejuni (% of actual isolates) | 1857 (23·9%) | 450 (26%) |
| Campylobacter coli (% of actual isolates) | 346 (4·4%) | 815 (47·4%) |
| Campylobacter fetus (% of actual isolates) | 871 (6·8%) | 211 (12·3%) |
| Campylobacter hyointestinalis (% of actual isolates) | 380 (4·9%) | 0 |
| Campylobacter lari (% of actual isolates) | 26 (0·33%) | 8 (0·05%) |
At faecal pat level, assuming 100% specificity of the 16 s rRNA PCR, this equated to an apparent Arcobacter spp. faecal pat prevalence of 40·1% [95% confidence interval (CI) 34·2–46·1] and 8·0% (95% CI 3·7–12·3) for cattle and sheep, respectively, adjusted for clustering at farm level (Table 3). The apparent prevalence is de facto the estimated minimum prevalence due to both the imperfect sensitivity of culture and the dependency introduced by the methodology whereby four isolates were selected per plate and tested independently for Arcobacter spp. and Campylobacter spp. Co-colonization of bovine faecal pats by Arcobacter and Campylobacter spp. occurred relatively frequently with a total of 191 bovine faecal pats (8·6% of culture-positive pats) yielding both species. Co-colonization was less frequent in sheep with only 11 pats (2·2% of culture-positive pats) yielding both species.
Table 3.
Estimated minimum Arcobacter spp. faecal pat prevalence (95% CI) by measured covariates adjusted for clustering at farm level
| Cattle Prevalence, % (95% CI) | Sheep Prevalence, % (95% CI) | |
|---|---|---|
| 40·1 (34·2–46·1) | 8·0 (3·7–12·3) | |
| Geographical location | ||
| Zone 1 | 35·4 (28·1–42·7) | |
| Zone 2 | 46·4 (26·1–66·7) | |
| Zone 3 | 41·3 (29·3–53·4) | |
| Management system | ||
| Housed all year | 55 (43–68)* | |
| Pasture in summer, housed in winter | 35·8 (32·7–39)* | |
| Sampling environment | ||
| Housed | 50·1 (45·3–56·5)* | |
| Pasture | 20·8 (14·7–27)* | |
| Season | ||
| Winter | 45·7 (40·2–51·3)* | 10·5 (7·9–13·1) |
| Summer | 32·6 (27·5–37·6)* | 5·0 (3–7·1) |
| Feeding system (cattle) | Prevalence, % (95% CI) | No. of sampling events |
| TMR | 47·8 (36·2–59·5) | 7 |
| Hybrid TMR | 48·8 (44–53·5) | 95 |
| Grazing and buffer and parlour | 20·5 (17–24·5) | 47 |
| Grazing and parlour | 21·8 (14·7–31) | 10 |
| Silage and parlour | 65·6 (49·3–78·8) | 6 |
CI, Confidence interval; TMR, total mixed ration.
P < 0·01.
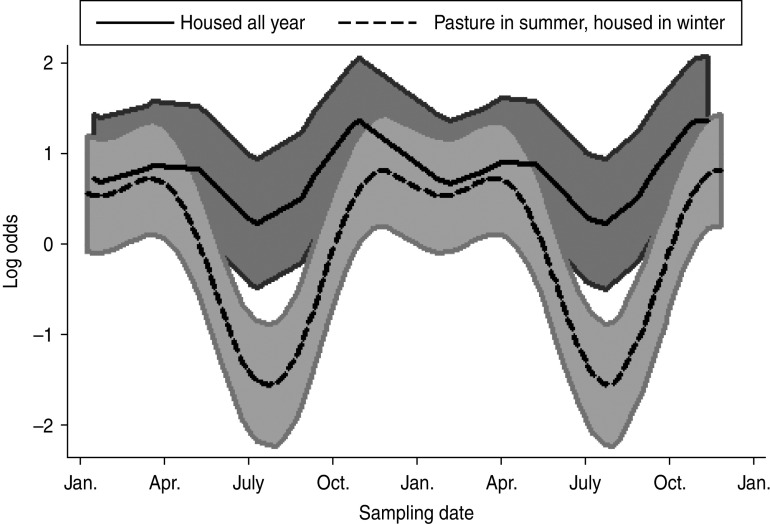
A mixed-effects model with farm identity as a random effect was fitted to the cattle dataset. The final model (Table 4) included an interaction term between the time covariates and the management system employed on the farm, i.e. ‘housed all year’ (HAY) or ‘pasture in summer, housed in winter’ (PSHW). Peak Arcobacter spp. faecal pat prevalence was observed during the winter months when all cattle were housed irrespective of management system. However, faecal pat prevalence was significantly lower in cattle at pasture during the summer months compared to their housed counterparts (Fig. 1).
Table 4.
Mixed effects multivariable logistic regression model including covariates associated with the probability of isolating Arcobacter spp. from cattle faeces on Lancashire dairy farms. Farm is considered as a random effect (n = 15)
| Covariate | Estimate β | 95% CI | OR | 95% CI | Wald test P value |
|---|---|---|---|---|---|
| Proportion of fresh cows (quintiles) | 0·13 | 0·06 to 0·20 | 1·14 | 1·07–1·21 | <0·001 |
| Average herd daily milk yield (quintiles) | −0·17 | −0·30 to −0·50 | 0·84 | 0·74–0·95 | 0·006 |
| Campylobacter isolated | −1·67 | −1·86 to −1·48 | 0·19 | 0·16–0·23 | <0·001 |
| Faeces: loose vs. firm | 0·23 | 0·06 to 0·39 | 1·25 | 1·07–1·48 | 0·006 |
| Management system* | 0·81 | 0·45 to 1·16 | 2·24 | 1·57–3·20 | <0·001 |
| HAY vs. PSHW | |||||
| Time x1 | 0·51 | 0·37 to 0·64 | 1·66 | 1·44–1·91 | <0·001 |
| Time x2 | 0·13 | 0·003 to −0·26 | 1·14 | 1·00–1·23 | 0·045 |
| Time x3 | −0·21 | −0·34 to −0·08 | 0·81 | 0·71–0·92 | 0·001 |
| Time x4 | −0·14 | −0·28 to −0·01 | 0·87 | 0·76–0·98 | 0·028 |
| Management system × x1 | −0·72 | −1·00 to −0·44 | 0·49 | 0·37–0·64 | <0·001 |
| Management system × x2 | −0·38 | −0·64 to −0·13 | 0·68 | 0·53–0·88 | 0·003 |
| Management system × x3 | 0·16 | −0·10 to 0·43 | 1·18 | 0·91–1·53 | 0·217 |
| Management system × x4 | 0·06 | −0·21 to 0·34 | 1·07 | 0·81–1·40 | 0·649 |
| Baseline (PSHW, lowest level of all other covariates) | 0·002 | −0·51 to 0·52 | |||
OR, Odds ratio; CI, confidence interval.
Between-farm variance 0·033 (95% CI 0·013–0·080).
Management system: housed all year (HAY) vs. pasture in summer, housed in winter (PSHW).
Fig. 1.
Seasonal variation of estimated Arcobacter spp. faecal pat prevalence in cattle, adjusted for management system and other covariates. Shaded areas represent 95% confidence intervals.
There was a small positive association between the proportion of freshly calved cows (in quintiles) and Arcobacter spp. faecal pat prevalence [odds ratio (OR) 1·14, 95% CI 1·07–1·21, P < 0·001], while there was a negative association between increasing average daily milk yield and Arcobacter spp. faecal pat prevalence (OR 0·84, 95% CI 0·74–0·95, P = 0·006). The odds of isolating Arcobacter spp. from a faecal pat was increased if the faecal consistency was classified as ‘loose’ (OR 1·25, 95% CI 1·07–1·48, P < 0·006).
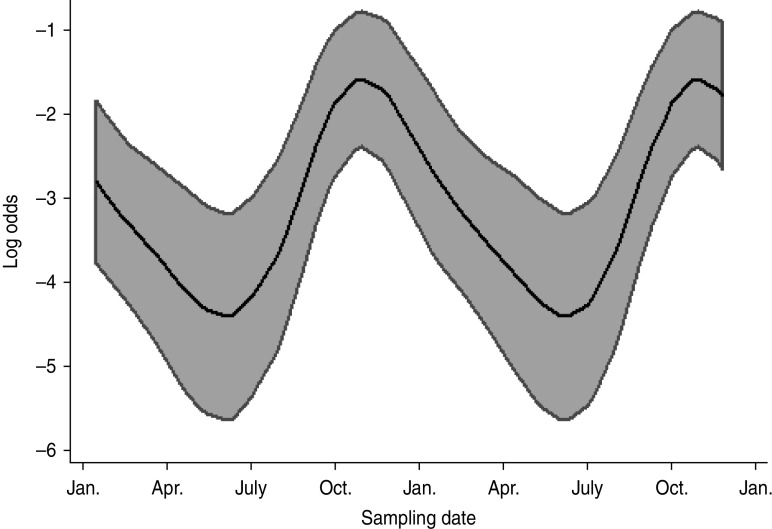
Multivariable analysis of sheep data (Table 5) demonstrated a strong positive association (OR 6·89, 95% CI 2·98–15·90, P < 0·001) between increasing pasture quality (defined as rough or lush pasture) and Arcobacter spp. faecal pat prevalence. As with cattle, there was strong residual seasonal periodicity, after adjusting for pasture quality, with peak faecal pat prevalence observed during the autumn months (Fig. 2).
Table 5.
Multivariable logistic regression model including covariates associated with the probability of isolating Arcobacter spp. from sheep faeces on Lancashire sheep farms. Farm is considered as a fixed effect (n = 4)
| Covariate | Estimate β | 95% CI | OR | 95% CI | Wald test P value |
|---|---|---|---|---|---|
| Pasture: lush vs. rough | 1·93 | 1·09 to 2·77 | 6·89 | 2·98–15·90 | <0·001 |
| Campylobacter isolated | −1·94 | −2·68 to −1·20 | 0·14 | 0·07–0·30 | <0·001 |
| Farm | |||||
| 9 | 0·68 | −0·11 to 1·47 | 1·97 | 0·89–4·33 | 0·092 |
| 15 | 0·58 | −0·34 to 1·50 | 1·79 | 0·71–4·48 | 0·216 |
| 16 | 0·57 | −0·30 to 1·45 | 1·77 | 0·73–4·27 | 0·201 |
| Time x1 | 0·80 | 0·28 to 1·32 | 0·002 | ||
| Time x2 | −1·04 | −1·54 to −0·55 | <0·001 | ||
| Time x3 | −0·28 | −0·75 to 0·19 | 0·243 | ||
| Time x4 | −0·02 | −0·45 to 0·40 | 0·912 | ||
| Baseline: Farm 12, rough | −3·08 | −3·86 to −2·30 | |||
Fig. 2.
Seasonal variation of estimated Arcobacter spp. faecal pat prevalence in sheep, adjusted for pasture type. Shaded areas represent 95% confidence intervals.
As indicated earlier, selecting four isolates per plate and testing each independently for Arcobacter spp. and Campylobacter spp. introduces a dependency in the sensitivity of isolation for each species. The odds ratio for the binary variable ‘Campylobacter present’ was 0·19 (95% CI 0·16–0·23) and 0·14 (95% CI 0·07–0·31) in the cattle and sheep models, respectively, indicating the isolation of Campylobacter spp. was significantly associated with a reduced odds of isolating Arcobacter spp. However, exclusion of the variable ‘Campylobacter present’ in multivariable models had little effect on the magnitude of the estimated coefficients for other covariates (maximum change 10%), suggesting that this dependency did not substantially alter the inferences drawn from the model.
DISCUSSION
In interpreting the results of the present study, it must be borne in mind that the microbiological techniques employed were optimized for the isolation of C. jejuni and not for Arcobacter spp. A recent study [34] suggested that the culture techniques used in the present study are significantly less sensitive than Arcobacter-specific methods. Thus the Arcobacter spp. prevalence estimates obtained in the present study are likely to be an underestimate of the true situation. Similarly the technique of selecting up to four isolates per plate and testing each independently for Arcobacter spp. and Campylobacter spp. introduces a dependency in the sensitivity of isolation for each species. Assuming 100% specificity of the PCR used to determine genus, the results presented are best considered as estimates of the minimum prevalence. Relative changes in the prevalence of one of the genera will affect the probability of detecting the other, i.e. if one becomes relatively more common, the sensitivity of detecting the other would decrease. This is important when interpreting the estimated prevalence and its relationship with covariates, although the effect of including the binary variable ‘Campylobacter present’ on parameter estimates suggests this dependency does not substantially alter the relationship between the measured covariates and the isolation of Arcobacter spp.
The ability of Arcobacter spp. to grow on ‘Campylobacter-specific’ culture media under microaerobic conditions, as demonstrated in this study, together with its very similar colony appearance in culture would suggest that without further testing it is not possible to discriminate between the two microorganisms. This together with the relatively high proportion (9%) of pats co-colonized by both species might suggest that care is needed in studies of Campylobacter spp. or Arcobacter spp. utilizing total bacterial counts since it would be impossible to know if the colonies being counted were actually the organism of interest.
The Arcobacter spp. prevalence estimates reported are in broad agreement with previous studies [3, 4, 7] although with the seasonal and environmental variation observed in the present study, it could be argued that single point prevalence estimates as reported in previous studies are of little value unless the spatial and temporal details are detailed.
In dairy cattle, increasing the proportion of freshly calved cows in the sampled group was associated with increased faecal pat prevalence. This may be associated with cattle being under increased stress at this time. Similar findings were reported for C. jejuni faecal pat prevalence in the same study [25]. The probability of isolating Arcobacter spp. from a faecal pat was increased if the faecal consistency was classified as ‘loose’. However, the direction of the association is unknown, thus causality cannot be inferred.
In this study, four of the dairy herds were housed all the time during the study period while one herd was housed for the entire second year of the study with the remainder being housed during the winter months but grazed outside during the summer. This allowed us to investigate the overall effect of the management system under which cattle were kept, encompassing both environmental and seasonal effects. In both species, the prevalence of Arcobacter spp. was highest in autumn and winter. However, in dairy cattle it appeared that seasonal influences which might be expected to encompass variables such as temperature, rainfall, hours of sunshine, and day length had a greater impact on animals at pasture during the summer months than on their housed counterparts with the lowest prevalence occurring in grazing animals during the summer months.
The North West of England is a major milk-producing area of the UK and the farms recruited in this study may be considered representative of UK dairying both in herd size and management practices. The practice of housing dairy cows throughout the year with no access to pasture is gaining in popularity in the UK due to the greater nutritional demands placed upon the high yielding Holstein dairy cow which are impossible to meet at pasture.
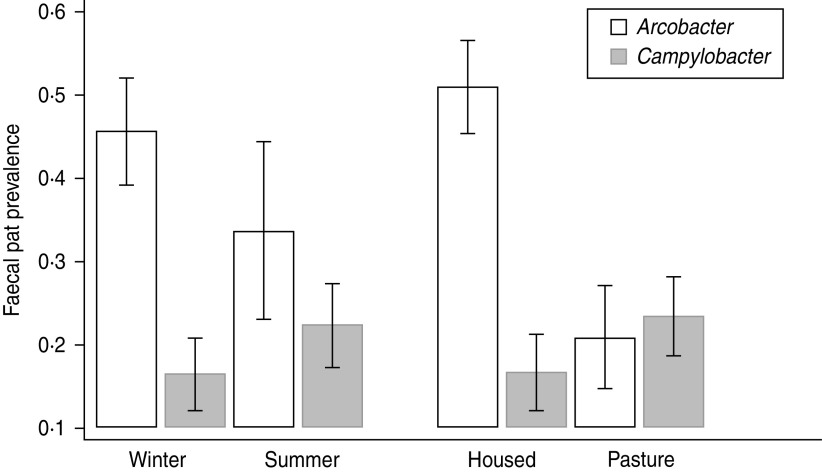
Comparison of Arcobacter spp. and C. jejuni prevalence [25] data by season and sampling environment (Fig. 3) suggest an ‘inverse association; between the two bacterial species in dairy cattle whereby C. jejuni prevalence is highest in animals sampled at pasture and lowest in housed animals whereas Arcobacter spp. prevalence is highest in housed animals and lowest in animals at pasture. We hypothesize that this may be a reflection of the diet consumed rather than environment per se although the current dataset does not allow testing of this hypothesis. At pasture the principal carbohydrates consumed by cattle are short-chain sugars and fructans whereas in housed cattle fed conserved crops and grain products, starches constitute a higher proportion of the dietary carbohydrates [35]. These very different diets will have differing rumen fermentation characteristics such that the composition of ruminal outflow into the small intestine will also be very different. We suggest that this differing intestinal milieu may preferentially favour proliferation of particular species of bacteria. Furthermore, the results of both this and the previous study [25] raise the fascinating question – do Campylobacter and Arcobacter spp. occupy the same ecological niche within the ruminant gastrointestinal tract?
Fig. 3.
The faecal pat prevalence of Arcobacter spp. and Campylobacter jejuni in dairy cattle by environment and season. Error bars represent 95% confidence intervals. C. jejuni data is from Grove-White et al. [25].
While the role of Arcobacter spp. as a human pathogen is unclear, that is not the case with C. jejuni. There is an urgent requirement for the development of interventions to reduce the human campylobacteriosis disease burden and key to this will be efforts to reduce exposure, as are being currently investigated in the poultry industry. While it is increasingly recognized that ruminants are a significant source of Campylobacter infections for people, the exact exposure routes in the UK are unclear but are likely to be via environmental exposure as well as direct exposure to cattle and sheep. The results of our previous study [25] suggest two possible interventions for reducing Campylobacter excretion by dairy cattle, namely housing them all year round or dietary manipulation when they are grazing pasture. While such interventions might result in increased exposure to Arcobacter spp., this might be a cost worth paying in the light of the known harm produced by Campylobacter spp.
ACKNOWLEDGEMENTS
This work is dedicated to the memory of the late Professor Tony Hart who died suddenly in 2007. Before his death he was intimately involved in the work presented here. The study was funded by Defra and HEFCE as part of the Veterinary Training and Research Initiative (VTRI). We thank Ms. J. Sutherst, Ms. P. Houghton and Ms. A. M. Riley for laboratory assistance. We also thank the farmers who participated in the study.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Ellis WA, et al. Isolation of Spirillum/Vibrio-like organisms from bovine foetuses. Veterinary Record 1977; 100: 451–452. [DOI] [PubMed] [Google Scholar]
- 2.Hoa TK, et al. Arcobacter, what is known and unknown about a potential foodborne zoonotic agent. Veterinary Microbiology 2006; 115: 1–13. [DOI] [PubMed] [Google Scholar]
- 3.Wesley IV, et al. Faecal shedding of Campylobacter and Arcobacter spp. in dairy cattle. Applied and Environmental Microbiology 2000; 66: 1994–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Driessche E, et al. Isolation of Arcobacter species from animal faeces. FEMS Microbiological Letters 2003; 229: 243–248. [DOI] [PubMed] [Google Scholar]
- 5.Rice EW, et al. Isolation of Arcobacter butzleri from ground water. Letters in Applied Microbiology 1999; 28: 31–35. [DOI] [PubMed] [Google Scholar]
- 6.Scullion R, et al. Prevalence of Arcobacter spp. in raw milk and retail meats in Northern Ireland. Journal of Food Protection 2006; 69: 1986–1990. [DOI] [PubMed] [Google Scholar]
- 7.De Smet S, et al. Small ruminants as carriers of the emerging foodborne pathogen Arcobacter on small and medium farms. Small Ruminant Research 2011; 97: 124–129. [Google Scholar]
- 8.Logan EF, et al. Mastitis in dairy cows associated with an aerotolerant campylobacter. Veterinary Record 1982; 110: 229–230. [DOI] [PubMed] [Google Scholar]
- 9.Vandenberg O, et al. Arcobacter species in humans. Emerging Infectious Disease 2004; 10: 1863–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prouze-Mauleon VL, et al. Arcobacter butzeleri: underestimated enteropathogen. Emerging Infectious Disease 2006; 12: 307–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houf K, Stephan L. Isolation and characterisation of the emerging foodborne pathogen Arcobacter from human stool. Journal of Microbiological Methods 2007; 68: 408–413. [DOI] [PubMed] [Google Scholar]
- 12.Lerner J, et al. Severe diarrhoea associated with Arcobacter butzeleri. European Journal of Clinical Microbiology and Infectious Disease 1994; 13: 660–662. [DOI] [PubMed] [Google Scholar]
- 13.Wybo I, et al. Isolation of Arcobacter skirrowii from a patient with chronic diarrhea. Journal of Clinical Microbiology 2004; 42: 1851–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandamme P, et al. Outbreak of recurrent abdominal cramps associated with Arcobacter butzeleri in an Italian school. Journal of Clinical Microbiology 1992; 30: 2335–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lappi V, et al. An outbreak of foodborne illness among attendees of a wedding reception in Wisconsin likely caused by Arcobacter butzeleri. Foodborne Pathogens and Disease 2013; 10: 250–255. [DOI] [PubMed] [Google Scholar]
- 16.Tam CC, et al. Longitudinal study of infectious intestinal disease in the UK (IID2 study): incidence in the community and presenting to general practice. Gut 2012; 61: 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vellinga A, Van Loock F. The dioxin crisis as experiment to determine poultry-related Campylobacter enteritis. Emerging Infectious Disease 2002; 8: 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tam CC, et al. Chicken consumption and use of acid-suppressing medication as risk factors for Campylobacter enteritis, England. Emerging Infectious Disease 2009; 15: 1402–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominques AR, et al. Source attribution of human campylobacteriosis using a meta-analysis of case-control studies of sporadic infections. Epidemiology and Infection 2012; 140: 970–981. [DOI] [PubMed] [Google Scholar]
- 20.Wilson DJ, et al. Tracing the source of campylobacteriosis. PLoS Genetics 2008; 4: e1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullner P, et al. Assigning the source of human campylobacteriosis in New Zealand: A comparative genetic and epidemiological approach. Infection, Genetics and Evolution 2009; 9: 1311–1319. [DOI] [PubMed] [Google Scholar]
- 22.Sopwith W, et al. Enhanced surveillance of Campylobacter infection in the North West of England 1997–1999. Journal of Infection 2003; 46: 35–45. [DOI] [PubMed] [Google Scholar]
- 23.Meldrum RJ, et al. The seasonality of human Campylobacter infection and Campylobacter isolates from fresh retail chicken. Epidemiology and Infection 2003: 133: 49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanley KN, et al. The seasonal variation of thermophillic campylobacters in beef cattle, dairy cattle and calves. Journal of Applied Microbiology 1998; 85: 472–480. [DOI] [PubMed] [Google Scholar]
- 25.Grove-White DH, et al. Temporal and farm-management-associated variation in the faecal pat prevalence of Campylobacter jejuni in ruminants. Epidemiology and Infection 2010; 138: 549–558. [DOI] [PubMed] [Google Scholar]
- 26.Hughes J. A system for assessing cow cleanliness. In Practice 2001; 23: 517–524. [Google Scholar]
- 27.Grove-White DH. Rumen healthcare in the dairy cow. In Practice 2004; 26: 88–95. [Google Scholar]
- 28.Gonzalez I, et al. Development of a combined PCR-culture technique for the rapid detection of Arcobacter spp. in chicken meat. Letters in Applied Microbiology 2000; 30: 207–212. [DOI] [PubMed] [Google Scholar]
- 29.Wang G, et al. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis and C. fetus subsp. fetus. Journal of Clinical Microbiology 2002; 40: 4744–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linton D, et al. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Research in Microbiology; 1996: 147: 707–718. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez I, et al. Species Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. Journal of Clinical Microbiology 1997; 35: 759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirkwood BR, Sterne JAC. Essential Medical Statistics, 2nd edn. Oxford: Blackwell Science, 2003, pp. 341–354. [Google Scholar]
- 33.Stolwijk AM, et al. Studying seasonality by using sine and cosine functions in regression analysis. Journal of Epidemiology and Community Health 1999; 53: 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merga JY, et al. Comparison of Arcobacter isolation methods, and diversity of Arcobacter spp. in Cheshire, United Kingdom. Applied and Environmental Microbiology 2011; 77: 1646–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chamberlain AT, Wilkinson. Feeding the Dairy Cow, 1st edn. Lincoln: Chalcombe Publications, 1996, pp. 216. [Google Scholar]