Abstract
Fusarium subglutinans f. sp. pini (= F. circinatum) is a pathogen of pine and is one of eight mating populations (i.e., biological species) in the Gibberella fujikuroi species complex. This species complex includes F. thapsinum, F. moniliforme (= F. verticillioides), F. nygamai, and F. proliferatum, as well as F. subglutinans associated with sugarcane, maize, mango, and pineapple. Differentiating these forms of F. subglutinans usually requires pathogenicity tests, which are often time-consuming and inconclusive. Our objective was to develop a technique to differentiate isolates of F. subglutinans f. sp. pini from other isolates identified as F. subglutinans. We sequenced the histone H3 gene from a representative set of Fusarium isolates. The H3 gene sequence was conserved and contained two introns in all the isolates studied. From both the intron and the exon sequence data, we developed a PCR-restriction fragment length polymorphism technique that reliably distinguishes F. subglutinans f. sp. pini from the other biological species in the G. fujikuroi species complex.
Fusarium subglutinans f. sp. pini is an important pathogen of pine that causes pitch canker in mature trees (6, 13) and root rot and damping-off in seedlings (2, 34). This fungus can be spread by both infected seedlings and seed (1, 28). The management of F. subglutinans f. sp. pini would be greatly improved if a quick screening method were available for seed and nursery stock.
F. subglutinans f. sp. pini represents one of eight mating populations (i.e., biological species) in the Gibberella fujikuroi species complex (6, 23). Three of these mating populations, B, E, and H (F. subglutinans f. sp. pini), have F. subglutinans anamorphs (5, 14, 19, 20). Strains of Fusarium isolated from pineapple (F. subglutinans f. sp. ananas) and mango, for which a teleomorph is not known, also have F. subglutinans anamorphs (27, 32, 33).
Distinguishing F. subglutinans f. sp. pini from the other species in Fusarium section Liseola usually requires pathogenicity tests or sexual crosses with known tester strains (6, 7, 35). These assays are time-consuming and labor-intensive and do not always yield clear-cut answers. Molecular tools such as random amplification of polymorphic DNA (RAPD) (9, 35, 36), mitochondrial restriction fragment length polymorphisms (RFLP) (7), and ribosomal DNA (rDNA) internal transcribed spacer (ITS1 and ITS2) sequences (25, 37) have been tested for their efficacy in differentiating F. subglutinans f. sp. pini isolates from other isolates of F. subglutinans. Because of the technical difficulties associated with mitochondrial RFLP and the low repeatability of RAPD data, we do not consider these techniques useful for diagnostic purposes. Two different copies of the ITS2 region were identified in the same isolate within some of the species in Fusarium section Liseola (25, 37), and a reliable diagnostic technique based on these sequences could not be developed. Alternative regions such as the histone and β-tubulin genes might be used more effectively.
O’Donnell et al. (26) used the DNA sequences of the nuclear rDNA large subunit, mitochondrial small subunit, and β-tubulin to develop a phylogeny that includes 36 taxa in the G. fujikuroi species complex. These sequences may potentially be useful for diagnostic purposes, but we began our study prior to publication of the phylogeny of O’Donnell et al. (26).
We used an alternative region of the genome, the histone H3 gene, to distinguish F. subglutinans f. sp. pini isolates from other isolates of F. subglutinans. Histone genes encode histone proteins, which are the major constituents of chromatin (16, 21). Four histone proteins, H2A, H2B, H3, and H4, make up the nucleosomal core (17). The gene encoding the H3 protein is well conserved, especially at the amino acid level (12, 31), and the presence of introns enhances its value in taxonomic and phylogenetic studies of closely related organisms (8, 38). Although the histone H4 gene also has these characteristics, it is generally too highly conserved to be suitable for evolutionary studies (30).
Our objectives in this study were (i) to sequence the histone H3 gene from various strains in the G. fujikuroi species complex, (ii) to compare the relationships thus determined with those established by use of other sequences, and (iii) to develop a PCR-RFLP procedure based on the histone H3 gene sequence for the routine identification of F. subglutinans f. sp. pini.
MATERIALS AND METHODS
Fungal isolates.
All isolates were maintained on 2% (wt/vol) malt extract agar (Biolab Diagnostics Ltd., Fedlife Park, Midrand, South Africa) in the culture collections of the Forestry and Agricultural Biotechnology Institute at the University of Pretoria, Pretoria, South Africa, and the Medical Research Council, Tygerberg, South Africa. We examined 42 Fusarium isolates, including F. subglutinans f. sp. pini, pathogenic to pine; F. subglutinans f. sp. ananas, pathogenic to pineapple; F. subglutinans isolates associated with maize and mango; and the mating type tester strains from all eight mating populations in the G. fujikuroi species complex (Table 1). To test the efficacy of the PCR-RFLP technique for use as a species diagnostic technique (see below), we tested 60 strains of the H mating population identified by Britz et al. (5) and 80 strains representing populations A to F identified by Yan et al. (39). These strains were reassorted and then encoded so that the assays were done in a blind manner.
TABLE 1.
Host and origin of the different Fusarium isolates from the G. fujikuroi (Sawada) Wollenw. species complex used in this study
| Mating population | Speciesa | Isolate(s)b | Host and/or origin | Source | GenBank accession no. |
|---|---|---|---|---|---|
| A | F. moniliforme Sheldon | MRC 6191, KSU 0999, PEN-M3703 | Maize, United States | J. F. Leslie | AF150859 |
| A | F. moniliforme | MRC 6155, KSU 0149, PEN3125 | Maize, United States | J. F. Leslie | AF150858 |
| B | F. subglutinans (Wollenw. and Reinking) Nelson, Toussoun, and Marasas | MRC 6524, KSU 3852, PEN-M6865 | Laboratory cross | J. F. Leslie | AF150861 |
| B | F. subglutinans | MRC 6525, KSU 3853, PEN-M6866 | Laboratory cross | J. F. Leslie | AF150860 |
| C | F. proliferatum (Matsushima) Nirenberg | MRC 6570, KSU 4921 | Rice, Taiwan | J. F. Leslie | AF150873 |
| C | F. proliferatum | MRC 6571, KSU 4922 | Rice, Taiwan | J. F. Leslie | AF150872 |
| D | F. proliferatum | MRC 6568, KSU 4853 | Laboratory cross | J. F. Leslie | AF150871 |
| D | F. proliferatum | MRC 6569, KSU 4854 | Laboratory cross | J. F. Leslie | AF150870 |
| E | F. subglutinans | MRC 6483, KSU 0990, PEN-M3696 | Maize, United States | J. F. Leslie | AF150845 |
| E | F. subglutinans | MRC 6512, KSU 2192, PEN-M3693 | Maize, United States | J. F. Leslie | AF150844 |
| F | F. thapsinum Klittich et al. | MRC 6536, KSU 4092 | Laboratory cross | J. F. Leslie | AF150857 |
| F | F. thapsinum | MRC 6537, KSU 4093 | Laboratory cross | J. F. Leslie | AF150856 |
| G | F. nygamai Burgess and Trimboli | MRC 7548, KSU 5111 | Laboratory cross | J. F. Leslie | AF150854 |
| G | F. nygamai | MRC 7549, KSU 5112 | Laboratory cross | J. F. Leslie | AF150855 |
| H | F. subglutinans f. sp. pini Correll et al. | MRC 6209, BBA 69854 | Pine, South Africa | A. Viljoen | AF150846 |
| H | F. subglutinans f. sp. pini | MRC 6211 | Pine, South Africa | A. Viljoen | AF150847 |
| H | F. subglutinans f. sp. pini | MRC 6213 | Pine, South Africa | A. Viljoen | AF150849 |
| H | F. subglutinans f. sp. pini | MRC 6228, PEN-M1290 | Pine, United States | P. E. Nelson | AF150850 |
| H | F. subglutinans f. sp. pini | MRC 7437, FL 103 | Pine, United States | T. R. Gordon | AF150848 |
| H | F. subglutinans f. sp. pini | MRC 7438 | Pine, United States | A. Viljoen | AF150851 |
| H | F. subglutinans f. sp. pini | MRC 7439, FL 15 | Pine, United States | T. R. Gordon | AF150852 |
| H | F. subglutinans f. sp. pini | MRC 7440, FSP 9 | Pine, United States | T. R. Gordon | AF150853 |
| F. subglutinans | MRC 2730 | Mango, South Africa | W. F. O. Marasas | AF150865 | |
| F. subglutinans | MRC 3477 | Mango, South Africa | W. F. O. Marasas | AF150868 | |
| F. subglutinans | MRC 3478 | Mango, South Africa | W. F. O. Marasas | AF150869 | |
| F. subglutinans | MRC 3479 | Mango, South Africa | W. F. O. Marasas | AF150867 | |
| F. subglutinans | MRC 7034 | Mango, United States | W. F. O. Marasas | AF150864 | |
| F. subglutinans | MRC 7035 | Mango, United States | W. F. O. Marasas | AF150866 | |
| F. subglutinans | MRC 7037 | Mango, United States | W. F. O. Marasas | AF150863 | |
| F. subglutinans | MRC 7038 | Mango, United States | W. F. O. Marasas | AF150862 | |
| E | F. subglutinans | MRC 115 | Maize, South Africa | W. F. O. Marasas | AF150843 |
| E | F. subglutinans | MRC 620 | Maize, South Africa | W. F. O. Marasas | AF150842 |
| E | F. subglutinans | MRC 714 | Maize, South Africa | W. F. O. Marasas | AF150841 |
| E | F. subglutinans | MRC 756 | Maize, South Africa | W. F. O. Marasas | AF150839 |
| E | F. subglutinans | MRC 837 | Maize, South Africa | W. F. O. Marasas | AF150840 |
| E | F. subglutinans | MRC 1077 | Maize, South Africa | W. F. O. Marasas | AF150837 |
| E | F. subglutinans | MRC 1084 | Maize, South Africa | W. F. O. Marasas | AF150838 |
| F. subglutinans f. sp. ananas Ventura, Zambolim, and Gilb. | MRC 6782 | Pineapple, Brazil | J. A. Ventura | AF150834 | |
| F. subglutinans f. sp. ananas | MRC 6783 | Pineapple, Brazil | J. A. Ventura | AF150833 | |
| F. subglutinans f. sp. ananas | MRC 6784 | Pineapple, Brazil | J. A. Ventura | AF150836 | |
| F. subglutinans f. sp. ananas | MRC 6785 | Pineapple, Brazil | J. A. Ventura | AF150835 | |
| F. oxysporum Schlecht. emend. Snyd. and Hans. | MRC 6212 | Pine, South Africa | A. Viljoen | AF150832 |
Synonyms for F. moniliforme, F. subglutinans f. sp. pini, and F. subglutinans f. sp. ananas are F. verticillioides Gerlach and Nirenberg (11), F. circinatum Nirenberg and O’Donnell (24), and F. guttiforme Nirenberg and O’Donnell (24), respectively. The proposed synonyms for F. subglutinans from mating population B and F. proliferatum from mating population C are F. sacchari O’Donnell and Cigelnik (25) and F. fujikuroi Gerlach and Nirenberg (11), respectively.
MRC, W. F. O. Marasas, Programme on Mycotoxins and Experimental Carcinogenesis (PROMEC), Medical Research Council, Tygerberg, South Africa; KSU, J. F. Leslie, Department of Plant Pathology, Kansas State University, Manhattan; BBA, Biologische Bundesanstalt für Land- und Forstwirtschaft, Berlin, Germany; FL and FSP, T. R. Gordon, Department of Plant Pathology, University of California, Davis; PEN, P. E. Nelson Culture Collection, Department of Plant Pathology, Pennsylvania State University, University Park, Pa.
DNA isolation.
Flasks containing 100 ml of malt extract broth (2% [wt/vol]) (Biolab) were inoculated with 1-ml spore suspensions (>1,000 spores/ml). After 2 weeks of static incubation at room temperature (20 to 25°C), mycelium was harvested by filtration through no. 1 filter paper (Whatman BioSystems Ltd., Maidstone, Kent, United Kingdom). Harvested fungal tissue was ground to a powder in liquid nitrogen with a mortar and pestle and homogenized in extraction buffer containing 5% (wt/vol) CTAB (N-cetyl-N,N,N-trimethylammonium bromide), 1.4 M NaCl, 0.2% (vol/vol) 2-mercaptoethanol, 20 mM EDTA, 100 mM Tris-HCl (pH 8.0), and 1% (wt/vol) polyvinylpyrrolidone. This homogenate was incubated at 60°C for 1 h and centrifuged (16,000 × g) at room temperature. We performed phenol-isoamyl alcohol-chloroform (25:1:24) extractions and removed residual phenol with an additional chloroform extraction. Nucleic acids were precipitated by adding 0.1 volume of 3 M sodium acetate (pH 5.2) and 0.6 volume of 2-propanol, followed by incubation at 4°C overnight. Precipitated DNA was centrifuged (16,000 × g), washed with 70% ethanol, and resuspended in deionized water. This protocol is a variation of the one developed by Murray and Thompson (22).
PCR amplification.
PCR amplification was performed as described by Glass and Donaldson (12) with primers H3-1a (5′-ACTAAGCAGACCGCCCGCAGG-3′) and H3-1b (5′-GCGGGCGAGCTGGATGTCCTT-3′). These primers were constructed to flank at least one intron and amplify approximately 450 bp of the Neurospora crassa histone H3 gene. Each PCR mixture contained 1 mM deoxynucleotide triphosphates (0.25 mM each), 2.5 mM MgCl2, 0.2 μM H3-1a, 0.2 μM H3-1b, 0.25 ng of DNA per μl, 0.05 U of Super-Therm DNA polymerase [Southern Cross Biotechnology (Pty.) Ltd., Cape Town, South Africa] per μl, and 1× Super-Therm reaction buffer. PCR mixtures were overlaid with mineral oil, and reactions were performed on an Omnigene thermocycler (Hybaid, Middlesex, United Kingdom) with an initial denaturation step of 1 min at 92°C. This step was followed by 30 cycles of denaturation at 92°C (1 min), annealing at 68°C (1 min), and elongation at 72°C (1 min). A final extension was performed at 72°C for 5 min.
DNA sequencing.
PCR products were purified with a QIAquick PCR Purification Kit (Qiagen GmbH, Hilden, Germany). Histone H3 gene fragments from the 42 Fusarium isolates included in this study, were sequenced (see Table 1 for GenBank accession numbers) in both directions with primers H3-1a and H3-1b. Reactions were performed on an ABI PRISM 377 automated DNA sequencer with an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer, Warrington, United Kingdom).
Sequence Navigator version 1.0.1 (Perkin-Elmer Applied BioSystems, Inc., Foster City, Calif.) was used for translation of DNA sequences to amino acid sequences. DNA sequences were aligned manually by inserting gaps, and phylogenetic analyses were performed with PAUP (Phylogenetic Analysis Using Parsimony) version 4.0b1 (29). Each gap was treated as a fifth character (NEWSTATE) in heuristic searches, with tree-bisection-reconnection branch swapping and Mul Trees (saving of all optimal trees) effective. Bootstrap analyses were based on 1,000 replications. Fusarium oxysporum (MRC 6212) was used as an outgroup.
Sexual compatibility tests.
The seven F. subglutinans isolates recovered from maize in South Africa (Table 1) were crossed with mating population E tester strains and with one another in all possible pairwise combinations (5, 18). Crosses were scored as positive when ascospores were observed exuding from perithecia.
PCR-RFLP technique.
Amplified DNA was digested with two restriction enzymes, CfoI and DdeI (Boehringer Mannheim South Africa Pty. Ltd.). Digestions were performed consecutively by adding 5 U of CfoI to 15 μl of unpurified PCR product (3); after 3 h of incubation at 37°C, 5 U of DdeI was added and the sodium chloride concentration was adjusted to 100 mM. These digestion reaction mixtures were then incubated at 37°C for an additional 5 h. We resolved PCR-RFLP profiles on 3% (wt/vol) agarose gels (Promega Corporation, Madison, Wis.; molecular biology-grade agarose) containing ethidium bromide (0.2 μg/ml). Electrophoresis was performed at 3 V/cm (room temperature) with 0.5× electrophoresis buffer containing 4.5 mM Tris, 4.5 mM boric acid, and 1 mM EDTA (pH 8.0). Nucleic acids were visualized with a UV transilluminator (302 nm).
Verification of technique.
To test the efficacy of the PCR-RFLP technique described here, histone H3 gene PCR products from 60 strains representing mating population H and 80 strains representing mating populations A to F were amplified, digested, and electrophoresed as described above. We compared the resulting PCR-RFLP profiles to those generated from the representatives of the G. fujikuroi species complex.
RESULTS
DNA sequencing.
The Fusarium histone H3 gene fragments ranged from 519 to 527 bp in length and contained two introns (intron 1 and intron 2) whose positions within the sequences were conserved. Intron 1 was 83 bp long for strains from mating population H, F. oxysporum, and F. subglutinans f. sp. ananas; 81 bp long for mating populations C and D and F. subglutinans isolated from mango; 85 bp long for mating populations A and G; 82 bp long for mating populations E and F and F. subglutinans isolated from maize; and 77 bp long for mating population B. Intron 2 was 57 bp long for all of the isolates, except for F. oxysporum, for which it was 58 bp long.
The coding regions of the Fusarium histone H3 genes were highly conserved, and we observed no deletions or insertions. We detected no differences in amino acid sequence, and coding sequence variation within the Fusarium genes was generally limited to the third position within the codon. The Fusarium histone H3 amino acid sequence differed from that of N. crassa (GenBank accession no. CAA25761) only at position 91 (A→L) (38), whereas that of Aspergillus nidulans (GenBank accession no. CAA39154) differed at two positions, 29 and 99 (both S→A) (10). N. crassa has a single intron at the same position as Fusarium intron 2, but its sequence was quite different from that of intron 2.
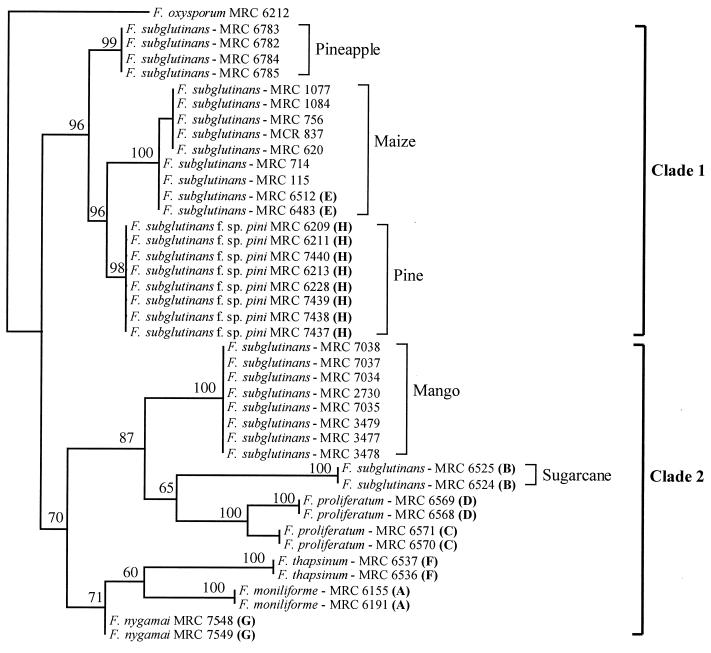
Phylogenetic analysis with PAUP 4.0b1 generated a single most-parsimonious tree from 469 bp of aligned DNA sequence (Fig. 1). This tree was comprised of two distinct clades. Clade 1 included isolates from mating populations H and E as well as isolates of F. subglutinans f. sp. ananas and F. subglutinans isolates from maize. The bootstrap value for this clade indicated 96% unity. Clade 2 included isolates from mating populations A, B, C, D, F, and G as well as F. subglutinans isolates from mango. The support for the unity of this clade was 70%.
FIG. 1.
Phylogram generated with histone H3 gene sequence data from the isolates included in this study by use of PAUP 4.0b1. Bootstrap values based on 1,000 replications are indicated as percentages. Bold letters in parentheses refer to the G. fujikuroi mating populations. This dendrogram is rooted to F. oxysporum MRC 6212. The length of the tree was 201 steps, and the values for homoplasy index and the retention index were 0.24 and 0.94, respectively.
Two subgroups made up clade 1 (Fig. 1). The first subgroup included F. subglutinans f. sp. ananas. The second subgroup included F. subglutinans f. sp. pini and isolates from mating population E, clustering together with 96% certainty. Clade 2 was subdivided into two smaller subgroups, one of which included isolates from mating populations B, C, and D as well as the F. subglutinans isolates from mango, with 87% support. The second subgroup in clade 2 contained isolates from mating populations A, F, and G, with 71% support.
Sexual compatibility tests.
Three of the F. subglutinans isolates associated with maize (MRC 1077, MRC 837, and MRC 714) were sexually compatible with one of the mating type tester strains for mating population E (MRC 6483). The remaining four isolates did not cross with one another or either of the tester strains.
PCR-RFLP technique.
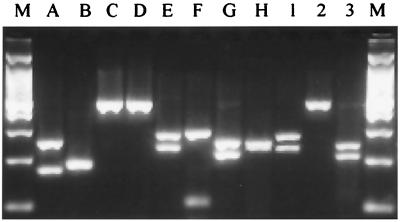
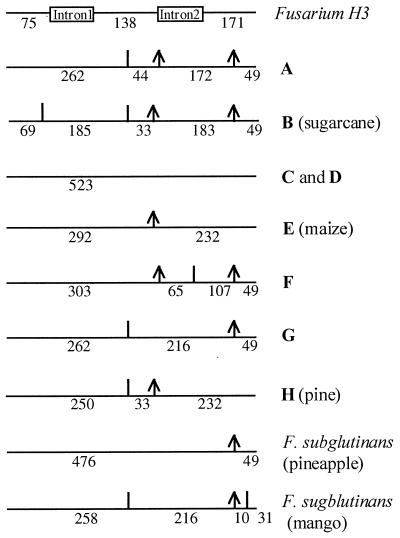
PCR-RFLP analysis of the amplified histone H3 gene products with DdeI and CfoI enabled us to distinguish F. subglutinans f. sp. pini from the rest of the isolates included in this study (Fig. 2). Unique PCR-RFLP profiles were generated for each group included in this study, except for mating populations C and D, mating population G, and F. subglutinans isolated from mango. From the restriction enzyme profiles, we constructed restriction maps for all host-specific groups of F. subglutinans as well as F. moniliforme, F. proliferatum, F. thapsinum, and F. nygamai (Fig. 3).
FIG. 2.
PCR-RFLP profiles generated by digestion of Fusarium histone H3 gene amplification products with DdeI and CfoI. Electrophoresis were performed on 3% agarose gels at 3 V/cm. Lanes M, 100-bp ladder (1,500, 1,000, 900, 800, 700, 600, 500, 400, 300, 200, and 100 bp); lane A, mating population A; lane B, mating population B (F. subglutinans associated with sugarcane); lane C, mating population C; lane D, mating population D; lane E, mating population E (F. subglutinans associated with maize); lane F, mating population F; lane G, mating population G; lane H, F. subglutinans f. sp. pini (mating population H); lane 1, F. subglutinans from maize; lane 2, F. subglutinans from pineapple (F. subglutinans f. sp. ananas); lane 3, F. subglutinans from mango.
FIG. 3.
Restriction maps of the histone H3 genes from the different isolates of Fusarium, generated with restriction enzymes DdeI and CfoI. The Fusarium introns are indicated as boxes, and the exons are indicated as horizontal lines. Bold letters refer to the G. fujikuroi mating populations. An arrow indicates a CfoI restriction site, and a vertical line indicates a DdeI restriction site. Exon and fragment sizes are indicated in base pairs.
Verification of technique.
All 60 mating population H strains were positively identified as F. subglutinans f. sp. pini in a blind test of the PCR-RFLP technique. We identified none of the strains from the collection of Yan et al. (39) as F. subglutinans f. sp. pini, and the expected profiles were generated for each of their representatives of mating populations A, B, E, and F. The blind test of 140 samples was 100% successful, providing 95% confidence that the error rate for this test is less than 2%.
DISCUSSION
In this study, we were able to distinguish F. subglutinans f. sp. pini (mating population H) from F. subglutinans isolates associated with mango, maize (mating population E), sugarcane (mating population B), and pineapple and F. moniliforme (mating population A), F. proliferatum (mating populations C and D), F. thapsinum (mating population F) and F. nygamai (mating population G). The PCR-RFLP technique has been used successfully by the Tree Pathology Co-operative Programme diagnostic clinic to identify isolates of F. subglutinans f. sp. pini for the last year. Seven outbreaks of root rot in South African nurseries have been correctly diagnosed as being caused by F. subglutinans f. sp. pini (4). We thus have confidence that this technique is robust and can be used with a high degree of certainty.
Phylogenetic analyses with the Fusarium histone H3 gene sequence data generated a phylogram (Fig. 1) that was similar to those produced by O’Donnell et al. (26). The results presented here and those based on β-tubulin and mitochondrial small-subunit DNA sequences (26) are similar to those obtained with isozymes (15) in two aspects. First, mating populations C and D form a closely related group in all cases. Second, mating population E is phylogenetically distinct from mating populations A, B, C, D, F, and G.
There are, however, two major differences between DNA-based phylogenies and those based on isozymes. With isozymes, Huss et al. (15) showed mating populations C and D to be most closely related to mating population G. The DNA-based phylogenies (26; this study), however, indicate that mating population G is most closely related to mating populations A and F and that these three mating populations form a distinct cluster separate from both mating populations C and D. Also, in contrast to the results from the isozyme study (15) both DNA-based phylogenies (26; this study) indicate that mating populations C and D are most closely related to mating population B.
F. subglutinans f. sp. pini has previously been reported to belong to mating population B (29), but our results and those presented by Britz et al. (5) and O’Donnell et al. (26) suggest otherwise. Nirenberg and O’Donnell (24) elevated this fungus to species level and provided the name F. circinatum (teleomorph = G. circinata) for it. Although our results are consistent with those of O’Donnell et al. (26) and support the placement of F. subglutinans f. sp. pini in a distinct taxon, the distinguishing morphological characters reported by Nirenberg and O’Donnell (24) appear to be inadequate to make definite identifications of the fungus (5).
F. subglutinans f. sp. pini, F. subglutinans f. sp. ananas, mating population E, and F. subglutinans isolated from maize are closely related to each other and are included in clade 1. Although some of the F. subglutinans isolates from maize and those belonging to mating population E appeared in two separate but closely related groups, this separation was caused by only two nucleotide base-pair differences. Since some individuals from both of these groups could cross with one of the mating type E tester strains, we do not believe that the second cluster of isolates from maize represents a separate mating population. The overall appearance of clade 1 corresponds to that of the so-called American clade described by O’Donnell et al. (26). This similarity suggests an equivalence of F. subglutinans f. sp. pini and F. circinatum as well as of F. subglutinans f. sp. ananas and F. guttiforme.
The two subgroups that constitute clade 2 in our study correspond to the African and Asian clades of O’Donnell et al. (26). The African clade includes mating populations A, F, and G, whereas the Asian clade includes mating populations B, C, and D. The latter clade also includes F. subglutinans isolates associated with mango, which are phylogenetically separate from F. subglutinans isolates associated with maize, pineapple, and pine but phylogenetically more closely related to F. subglutinans from mating population B (Fig. 1).
The results of this study and those of O’Donnell et al. (26) have identified a number of conserved genes that are useful for phylogenetic and taxonomic studies among species of Fusarium. The H3 gene, as well as the β-tubulin gene, allows for a higher degree of resolution than rDNA ITS1 and ITS2. Species previously considered too closely related for separation into distinct groups can now be separated based on histone or β-tubulin gene sequence. Moreover, rapid identification of fungi such as the pitch canker pathogen is now possible with a PCR-RFLP technique based on the histone H3 gene sequence.
ACKNOWLEDGMENTS
We thank the Foundation for Research Development (FRD) and the members of the Tree Pathology Co-operative Programme (TPCP) for financial support.
REFERENCES
- 1.Anderson R L, Belcher E, Miller T. Occurrence of fungi inside slash pine seeds produced in seed orchards in the United States. Seed Sci Technol. 1984;12:795–799. [Google Scholar]
- 2.Barnard E L, Blakeslee G M. Pitch canker of slash pine seedlings: a new disease in forest tree nurseries. Plant Dis. 1980;64:695–696. [Google Scholar]
- 3.Blanck A, Glück B, Wartbichler R, Bender S, Pöll M, Brandl A. Activity of restriction enzymes in a PCR mix. Biochemica. 1997;3:25. [Google Scholar]
- 4.Britz, H. Personal communication.
- 5.Britz H, Coutinho T A, Wingfield M J, Marasas W F O, Gordon T J, Leslie J F. Fusarium subglutinans f. sp. pini represents a distinct mating population in the Gibberella fujikuroi species complex. Appl Environ Microbiol. 1999;65:1198–1201. doi: 10.1128/aem.65.3.1198-1201.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correll J C, Gordon T R, McCain A H, Fox J W, Koehler C S, Wood D L, Schultz M E. Pitch canker disease in California: pathogenicity, distribution and canker development on Monterey pine (Pinus radiata) Plant Dis. 1991;75:676–682. [Google Scholar]
- 7.Correll J C, Gordon T R, McCain A H. Genetic diversity in California and Florida populations of the pitch canker fungus Fusarium subglutinans f. sp. pini. Phytopathology. 1992;82:415–420. [Google Scholar]
- 8.Donaldson G C, Ball L A, Axelrood P E, Glass N L. Primer sets developed to amplify conserved genes from filamentous ascomycetes are useful in differentiating Fusarium species associated with conifer. Appl Environ Microbiol. 1995;61:1331–1340. doi: 10.1128/aem.61.4.1331-1340.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DuTeau N M, Leslie J F. RAPD markers for Gibberella fujikuroi (Fusarium section Liseola) Fung Genet Newsl. 1991;38:37. [Google Scholar]
- 10.Ehinger A, Denison S H, May G S. Sequence, organization and expression of the core histone genes of Aspergillus nidulans. Mol Gen Genet. 1990;222:416–424. doi: 10.1007/BF00633848. [DOI] [PubMed] [Google Scholar]
- 11.Gerlach W, Nirenberg H I. The genus Fusarium—a pictorial atlas. Mitt Biol Bundesanst Land Forstwirtsch Berlin Dahlem. 1982;209:1–406. [Google Scholar]
- 12.Glass N L, Donaldson G C. Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hepting G H, Roth E R. Pitch canker, a new disease of some southern pines. J For. 1946;44:742–744. [Google Scholar]
- 14.Hsieh W H, Smith S N, Snyder W C. Mating groups in Fusarium moniliforme. Phytopathology. 1977;67:1041–1043. [Google Scholar]
- 15.Huss M J, Campbell C L, Jennings D B, Leslie J F. Isozyme variation among biological species in the Gibberella fujikuroi species complex (Fusarium section Liseola) Appl Environ Microbiol. 1996;62:3750–3756. doi: 10.1128/aem.62.10.3750-3756.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igo-Kemenes T, Horz W, Zachau H G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- 17.Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- 18.Klittich C J R, Leslie J F. Nitrate reduction mutants of Fusarium moniliforme (Gibberella fujikuroi) Genetics. 1988;118:417–423. doi: 10.1093/genetics/118.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhlman E G. Varieties of Gibberella fujikuroi with anamorphs in Fusarium section Liseola. Mycologia. 1982;74:756–768. [Google Scholar]
- 20.Leslie J F. Gibberella fujikuroi: available populations and variable traits. Can J Bot. 1995;73:S282–S291. [Google Scholar]
- 21.McGee J D, Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- 22.Murray M G, Thompson W F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson P E, Toussoun T A, Marasas W F O. Fusarium species: an illustrated manual of identification. University Park: Pennsylvania State University Press; 1983. [Google Scholar]
- 24.Nirenberg H I, O’Donnell K. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia. 1998;90:434–458. [Google Scholar]
- 25.O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7:103–117. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- 26.O’Donnell K, Cigelnik E, Nirenberg H I. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 1998;90:465–493. [Google Scholar]
- 27.Rohrbach K G, Pfeiffer J B. Susceptibility of pineapple cultivars to fruit disease incited by Penicillium funiculosum and Fusarium moniliforme. Phytopathology. 1976;66:1386–1390. [Google Scholar]
- 28.Storer A J, Gordon T R, Clark S L. Association of the pitch canker fungus, Fusarium subglutinans f. sp. pini, with Monterey pine seeds and seedlings in California. Plant Pathol. 1998;47:649–656. [Google Scholar]
- 29.Swofford D L. PAUP phylogenetic analysis using parsimony version 4.0b1. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 30.Tabata T, Kimiko S, Iwabuchi M. The structural organization and DNA sequence of wheat H4. Nucleic Acids Res. 1983;11:5865–5875. doi: 10.1093/nar/11.17.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thatcher T H, MacGaffey J, Bowen J, Horowitz S, Shapiro D L, Gorovsky M A. Independent evolutionary origin of histone H3.3-like variants of animals and Tetrahymena. Nucleic Acids Res. 1994;22:180–186. doi: 10.1093/nar/22.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varma A, Lele V C, Raychaudhuri S P, Ram A, Sang A. Mango malformation: a fungal disease. Phytopathol Z. 1974;79:254–257. [Google Scholar]
- 33.Ventura J A, Zambolim L, Gilbertson R L. Pathogenicity of Fusarium subglutinans to pineapples. Fitopatol Bras Supl. 1993;18:280. [Google Scholar]
- 34.Viljoen A, Wingfield M J, Marasas W F O. First report of Fusarium subglutinans f. sp. pini on pine seedlings in South Africa. Plant Dis. 1994;78:309–312. [Google Scholar]
- 35.Viljoen A, Wingfield M J, Marasas W F O. Characterization of Fusarium subglutinans f. sp. pini causing root disease of Pinus patula seedlings in South Africa. Mycol Res. 1997;101:437–445. [Google Scholar]
- 36.Voigt K, Schleier S, Brückner B. Genetic variability in Gibberella fujikuroi and some related species of the genus Fusarium based on random amplification of polymorphic DNA (RAPD) Curr Genet. 1995;27:528–535. doi: 10.1007/BF00314443. [DOI] [PubMed] [Google Scholar]
- 37.Waalwijk C, de Koning J R A, Baayen R P, Gams W. Discordant groupings of Fusarium spp. from the sections Elegans, Liseola and Dlaminia based on ribosomal ITS1 and ITS2 sequences. Mycologia. 1996;88:361–368. [Google Scholar]
- 38.Woudt L P, Pastink A, Kempers-Veenstra E, Jansen A E M, Mager W H, Planta R J. The genes encoding histone H3 and H4 in Neurospora crassa are unique and contain intervening sequences. Nucleic Acids Res. 1983;11:5347–5360. doi: 10.1093/nar/11.16.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan K, Dickman M D, Xu J R, Leslie J F. Sensitivity of field strains of Gibberella fujikuroi (Fusarium section Liseola) to benomyl and hygromycin B. Mycologia. 1993;85:206–213. [Google Scholar]