SUMMARY
Although post-weaning mortality (PWM) in pig farming is mainly due to the effect of pathogens, farm type or swine management are also directly or indirectly involved. In this work, we used null models and the partial least squares approach (PLS) to structural equation modelling, also known as PLS path modelling (PLS-PM), to explore whether farm type, swine management and pathogens, including porcine circovirus type 2, swine influenza virus, porcine reproductive and respiratory syndrome virus and Aujeszky's disease virus, directly or indirectly influenced PWM in 42 Spanish indoor pig farms. The null model analysis revealed that contact with multiple combinations of viruses could occur by chance. On the other hand, PLS-PM showed that farm characteristics do not influence virus infections, and thus neither farm type nor associated management practices shaped PWM due to pathogens. Accordingly, preventive programmes aimed at controlling PWM in intensive farming should prioritize the control of major pig pathogens.
Key words: Aujeszky's disease virus, co-infection, porcine circovirus type 2, porcine reproductive and respiratory syndrome virus, swine influenza virus
INTRODUCTION
Post-weaning mortality (PWM), including that in nurseries and growing-finishing pigs, is one of the most important economic losses for the porcine industry worldwide. Not only does the reduction in the number of pigs sent to slaughterhouse contribute to this loss, but so do the additional expenses derived from managing non-profitable animals that ultimately die on farm [1]. Different factors such as genetics [2], infectious agents [3], farm characteristics [4] and specific management practices [5] have been linked to PWM. Usually, PWM is the result of the interaction between different factors, and it is rare that PWM is due to a single pathogen infection [6]. A good example of this is porcine circovirus type 2 (PCV2) systemic disease (PCV2-SD, previously known as post-weaning multisystemic wasting syndrome), where the presence of PCV2 and other infectious/non-infectious factors are required for its expression [7]. In fact, diseased pigs affected only by PCV2 are unusual (i.e. 1·9% of cases in a broad survey in the USA [8]), with co-infection with porcine reproductive and respiratory syndrome virus (PRRSV [8, 9]) and swine influenza virus (SIV [10]) or Mycoplasma hyopneumoniae [11] being the main triggers of clinical disease concomitant with PCV2 infection. Another good example of polymicrobial disease is the porcine respiratory disease complex (PRDC [12]), where co-infection with PRRSV, Actinobacillus pleuropneumoniae, M. hyopneumoniae and SIV in combination with a lack of all-in/all-out management, has a major influence on respiratory disease [13].
Furthermore, farm characteristics can also play an important role in influencing PWM either directly or indirectly. In fact, farm design (farrow-to-finish vs. multisite systems) or certain management practices (i.e. early or late weaning) have been introduced in intensive pig farming to reduce PWM [14]. In this sense, a higher mortality has been reported when mean weaning age is ⩽28 days [15], or when piglets are moved to the fattening unit in late autumn (October–December), especially in overcrowded farms [14].
The effect of the above-mentioned factors on PWM have been traditionally analysed under classical statistical approaches, such as generalized linear models [15–17] and mixed models [18] assuming that each risk factor influences the response variable independently. However, no work has disentangled direct and indirect effects of infectious diseases and farm characteristics on PWM, in line with the latest trends in animal health [19, 20]. Moreover, most of the current work has been focused on the effects of pathogens (e.g. [13]) or farm management practices [14] on PWM in an independent manner, and efforts to explore their additive effect are still uncommon. In addition, no work has shown whether or not pathogens causing PWM are random assemblages.
In the present work, two innovative statistical approaches in the field of infectious diseases, null models [21], and partial least squares approach (PLS) to structural equation modelling, also known as PLS path modelling (PLS-PM) [22, 23], have been used to explore direct and indirect effects of farm characteristics (farm type and the associated management practices) and viral infections on PWM in Spanish indoor pig farms.
Null models, a technique originating in the field of community ecology, are based on the principle of the null hypothesis that supports the view that patterns in the data do not reflect biological forces, but represent chance variation or sampling effects [21]. That is to say, certain attributes of the community (species co-occurrence and variations in species dominance, among others) would be strongly dependent on sample size. The null hypothesis can take on a variety of forms, with one of the most common being the independence of species occurrences. In this case, the null hypothesis is that species are distributed randomly and independently from one another, whereas the alternative hypothesis is that species occurrence is non-independent. The null model is used to randomize the occurrence of species and to compare the patterns in these randomized communities with those in the actual data [24].
On the other hand PLS-PM was proposed by Herman Wold [22] as an alternative to the classical covariance-based estimation techniques for structural equation models (SEMs). SEMs meant to estimate a network of causal relationships between two or more unobservable latent variables (LVs) and a number of manifest variables (MVs), i.e. directly measured parameters in the field or in the laboratory. The LVs are those combinations of directly measured parameters defining an abstract concept that would not be defined by a single variable. For example, pig density, continuous animal flow or ‘all-in/all-out’ and number of litters/pen in the fattening unit are measurable traits (MVs) belonging to farm management (LVs).
SEM was first applied in the field of sociology [25], and is very useful in establishing causal relationships in observational studies on natural systems [26]. Several techniques can be used to estimate model parameters in SEM, with PLS-PM one such technique. PLS-PM is an iterative algorithm that separately solves the blocks of the measurement model and then, in a second step, estimates the path coefficients in the structural model. Therefore, PLS-PM is claimed to explain the residual variance of the latent variables and, potentially, that of the manifest variables in any regression run in the model [27]. That is why PLS-PM is considered more an exploratory approach than a confirmatory one. We decided to use this approach because it does not require strong assumptions with respect to the distributions of the MVs, the sample size or the measurement scale.
This work has two goals: (1) to evaluate both direct and indirect effects of farm type, farm management practices and some of the most common viral infections on PWM on 42 farms in Spain; and (2) to apply two new statistical approaches in the field of infectious diseases – null models and PLS-PM – to study causal relationships based on observational data.
METHODS
Farm selection and seroprevalence of selected pathogens
Information was gathered from 42 indoor pig farms (including all production stages: sow gestation/parturition, nurseries and growing-finishing areas) that were included in a previous cross-sectional study on viral infections in intensive pig farming by López-Soria and colleagues [10]. The sample of farms studied herein showed a well-balanced (40–60%) proportion of farms regarding production system (farrow-to-finish vs. multisite operations), pen partitions (solid vs. open) and animal flow during the nursery period (all-in/all-out vs. continuous flow). Proportions were not as well-balanced (1/3 vs. 2/3 of farms) for the remaining discrete variables, with more farms performing all-in/all-out animal flow in the fattening unit and not practising castration. Mortality rate was defined as the percentage of pig deaths per year; definitions for the remaining recorded farm variables are shown in Table 1.
Table 1.
Description of both latent and manifest variables used for fitting the causal model for post-weaning mortality in pigs in 42 farms in Spain
| Latent variable | Manifest variable | Descriptive statistics |
|---|---|---|
| Farm type | Farm system is a categorical variable with two modalities: | |
| 0 = Farrow-to-finish (all productive phases at one site) | 54·7% of farms | |
| 1 = Multisite (productive phases at more than one site) | 45·3% of farms | |
| Farm size, number of reproductive sows | Mean = 924 , s.d. = 1277, min = 45, max = 5186 | |
| Solid partitions between pens (partitions) had two modalities | ||
| 0 = Absence of solid partitions | 40·5% of farms | |
| 1 = Presence of solid partitions | 59·5% of farms | |
| Farm management | Pig density at fattening (density) = pigs/m2 | Mean = 1·4 pigs /m2, s.d. = 0·18, min = 1·2, max = 2·1 |
| Flow nursery | ||
| 0 = All-in/all-out | 59·5% | |
| 1 = Continuous flow | 40·5% | |
| Fattening animal flow | ||
| 0 = All-in/all-out | 66·6% | |
| 1 = Continuous flow | 33·4% | |
| Number of litters/pen in fattening (no. of litters) | Mean = 3·8 litters/pen, s.d. = 2·1, min = 1, max = 13·5 | |
| Age at weaning in days (age weaning) | Mean = 22·6 days, s.d. = 2·5, min = 19·5, max = 30 | |
| Age at fattening in days (age fattening) | Mean = 65·1 days, s.d. = 1·5, min = 45, max = 91·4 | |
| Castration, categorical variable with two modalities: | ||
| 0 = Castration was not practised | 64·3% | |
| 1 = Castration was practised | 35·7% | |
| Antibodies against viruses | SIV = antibodies against swine influenza virus | Mean = 40·6%, min = 0, max = 100 |
| PRRSV = antibodies against porcine reproductive and respiratory syndrome virus | Mean = 67·7%, min = 0, max = 100 | |
| ADV gE = antibodies against glycoprotein E of Aujeszky's disease virus | Mean = 18·3%, min = 0, max = 100 | |
| Post-weaning mortality | Post-weaning mortality (mortality) | Mean = 10%, s.d. = 2·7, min = 5, max = 16·7 |
The study farms did not vaccinate pigs against the studied viruses, precluding the measurement of antibodies elicited by vaccination. In addition, vaccination against Aujeszky's disease virus (ADV) with glycoprotein E (gE)-deleted vaccines (ADV gE) is compulsory in Spain and vaccines for PCV2 were not available in this region at the time of the study. Farms were located in the densest pig-rearing areas in Spain [see: MAPA, 2003 (http://rasve.mapa.es/Publica/Programas/Prevalencias/prevalencias.asp)], the second largest pig producer in the EU. All farms were positive for PCV2, hence seroconversion against this virus was excluded from our PLS-PM analysis.
Thirty pig blood samples per farm (n = 1260) were collected to assess seroprevalence of the studied viruses. Specifically, 15 blood samples were taken from sows and 15 from 20-week-old pigs. Subsequently, serum samples were analysed for the presence of antibodies against PRRSV (indirect ELISA, HerdCheck® PRRSV antibody test kit 2XR, IDEXX Laboratories Inc., USA; cut-off for positivity set at sample/positive ratio ⩾0·40; expected sensitivity (Se) and specificity (Sp) of 97·4% and 99·6%, respectively), SIV (indirect ELISA, CIVTEST™ SUIS Influenza Laboratorios Hipra S.A. Spain; cut-off for positivity set at percentage relative index >20; expected Se and Sp of 94% and 100%, respectively), PCV2 (ORF2-based ELISA test described by [28]; cut-off for positivity set at OD ratio ⩾1·5; expected Se and Sp of 98·2% and 94·5%, respectively) and ADV gE protein (blocking ELISA, HerdCheck® anti-PRV gpI, IDEXX Laboratories Inc.; cut-off for positivity set at sample/negative ratio ⩽0·6; expected Se and Sp of 99% for both parameters). The above-mentioned viral diseases are endemic throughout our study system; therefore, taking into account both Se and Sp of the performed serological tests, 15 pigs per farm were determined to be representative of the sanitary status at the farm level (Survey Toolbox version 1.01). A detailed description on sampling procedures and the observed prevalences of those viruses can be found in López-Soria et al. [10].
Evidence of specific multiple infections by null models
In order to test the co-occurrence of serological patterns against viral infections, a presence/absence matrix [serological combination matrix (SCM)] containing the results of the ADV gE, PRRSV, PCV2 and SIV ELISA tests by farm was performed. Each of the 15 finishing pigs and 15 sows tested per farm represented a row in the matrix. Therefore, the most common viral serological combinations at an individual scale were elucidated by performing a matrix of the observed occurrences of ADV gE, PRRSV and SIV serological responses in the pigs sampled. PCV2 was not considered in this matrix since 100% of the farms showed antibodies. Since measures were established by means of serological tests, timing of co-infection of those viruses could not be assessed.
Regardless of whether the serological combination pattern against the studied pathogens observed in each SCM differed from that expected by chance, they were compared with 10 000 matrices randomized according to a null model. Two types of co-occurrence metrics were used: the C score index [29] and the number of unique species combinations [30].
According to Stone & Roberts [29], the C score index measures the number of ‘checkerboard units’ (CUs) between all possible pairs of viruses. A checkerboard unit is any submatrix of the form: ‘1–0/0–1’ or ‘0–1/1–0’ and the number of CUs for each viral species pair was calculated as:
where S is the number of shared sites (i.e. pigs having both viruses in the present study) and both ri and rj the number of viral species i and j. In the present study, the C score was the average of all possible checkerboard pairs that occurred at least once in the matrix. According to Gotelli [21], in a competitively structured community where a strong inter-specific competition exists (i.e. a specific viral infection would provide protection to a subsequent viral infection), the C score should be significantly larger than expected by chance.
The number of unique species combinations would be the presence of unique viral serological combinations detected in the same pig. For a given assemblage (concomitance of antibodies against viruses in the present study) of n species, there are a maximum of 2n possible species combinations, including the combination of no species being present [30]. In a competitively structured community, the number of unique species combinations should be lower than that expected by chance. For creation of the null models, a randomization algorithm maintaining fixed sums for row and column constraints was used, and each matrix generated had the same number of rows and columns as the original matrix. According to Gotelli & Colwell [24] this algorithm has good type I properties (little chance of falsely rejecting the null hypothesis when it is true), but also has good power for detecting non-random patterns in noisy datasets. The analyses were performed using EcoSim 7.72 software [31].
PLS-PM for PWM
In PLS-PM, an iterative algorithm [32] based on a system of multiple and simple regressions was used to estimate SEM parameters (i.e. the weights linking the MVs to the LVs), as well as the path coefficients in the LVs. Two types of relationships characterizing each SEM were described: the first one involved relationships between the LVs (the structural model), while the other considered the links between each LV and its own block of MVs (the measurement model). These MVs (those directly recorded for each farm) where chosen based on a previous literature revision on the main risk factors associated with PWM in farms, in relation to farm characteristics and associated management practices [15, 16, 18, 33, 34].
In regards to the structural model, it was assumed that management practices directly depended on the farm characteristics and circulation of different viruses depended on both farm characteristics as well as on the management practices. To conclude, all the previous cited LVs should have a direct impact on PWM. For a scheme defining the structural model see Figure 1a. The PWM model was developed in a formative way, where each LV was considered as a linear combination of its own MVs. Thus, the vector of outer weights (Wj) associated at each block of MV was obtained as the vector of the regression coefficients in the multiple regression of the inner estimate of the LV on their associated MVs. Each outer weight can be considered as a proxy for the importance of each MV in the construction of the LV. The path coefficients (β) were obtained by least square regression of connected LV scores, and they were interpreted as standard regression coefficients [27].
Fig. 1.
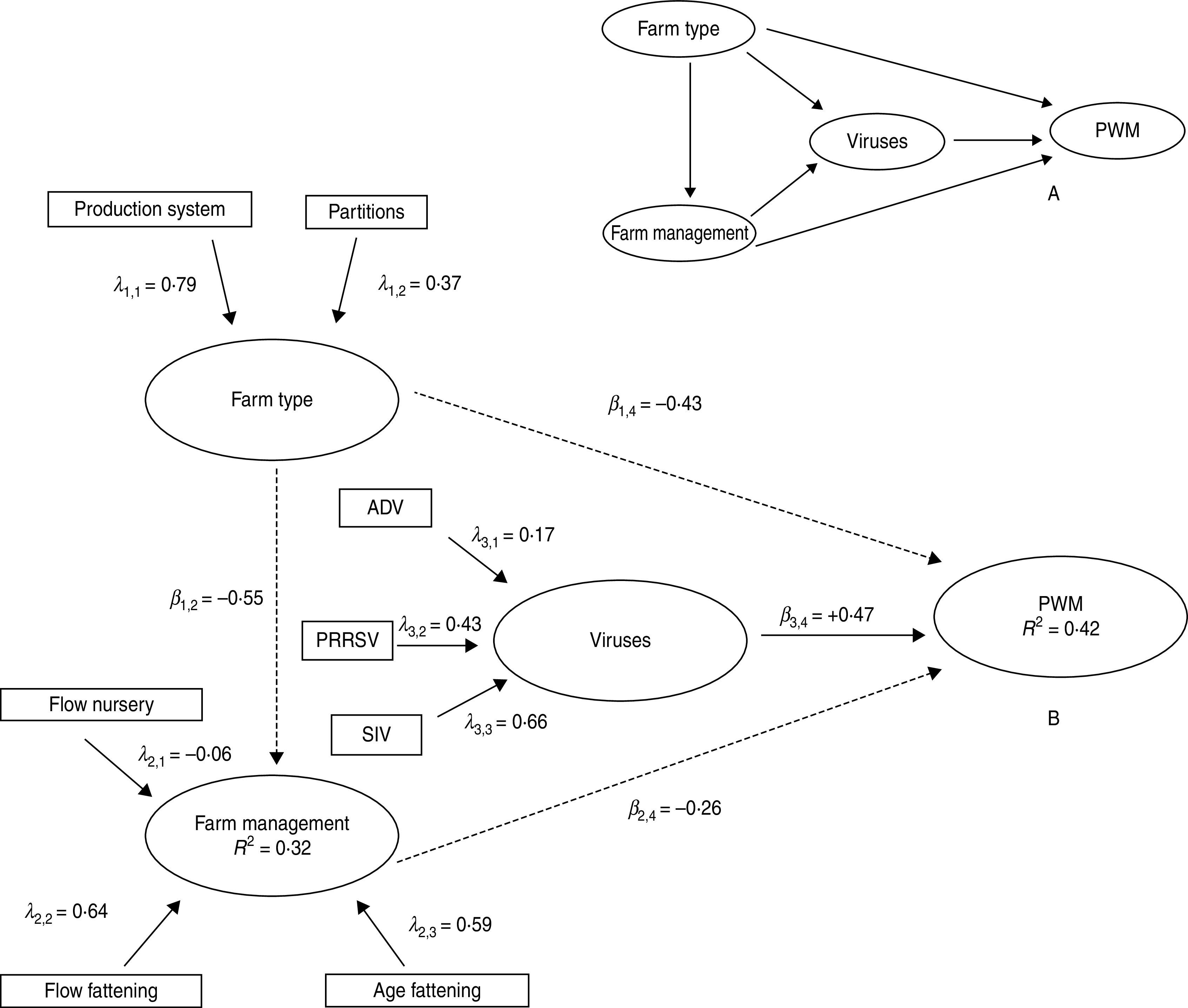
(a) Full initial and (b) final path models describing causes of post-weaning mortality (PWM) in 42 PCV2-infected farms in Spain. Note that the coefficient β1,2 means the path coefficient from the latent variable number 1 (farm type) to latent variable number 2 (farm management). Solid arrows represent positive influences whereas dashed arrows represent negative influences. Viral evidence of infection was studied by serological analysis. ADV, Aujeszky's disease virus; PRRSV, porcine reproductive and respiratory syndrome virus; SIV, swine influenza virus.
Finally, once the full model including all possible relationships between LVs (farm characteristics, farm management, evidence of multiple viral infections; Fig. 1a) was fitted, a model simplification was performed by removing those MVs uncorrelated with their own LVs and later those relationships between LVs with the lowest R2 values. The fit of the final model was measured by the goodness-of-fit index [35]. Model parameters (the path coefficients and the weights of the MVs) and fit indices (the percentages of explained variance and the R2 values) were validated by bootstrapping. This statistical procedure was performed using the package ‘plspm’ version 0.3.7 [36] of the statistical software R version 3.0.0 [37].
RESULTS AND DISCUSSION
Simple descriptive statistics for both categorical and continuous variables that characterized the study farms are shown in Table 1. Patterns of antibody response against viruses and both C scores and the number of viral seroconversions are detailed in Tables 2 and 3, respectively.
Table 2.
Observed frequencies (%) of porcine circovirus type 2 (PCV2), antibodies against glycoprotein E of Aujeszky's disease virus (ADV gE), porcine reproductive and respiratory syndrome virus (PRRSV) and swine influenza virus (SIV) seroconversion in 1260 pigs from 42 Spanish pig farms
| PCV2 | ADV gE | PRRSV | SIV | % of pigs |
|---|---|---|---|---|
| 0 | 0 | 0 | 1 | 0·2 |
| 0 | 1 | 1 | 1 | 0·2 |
| 1 | 1 | 0 | 0 | 0·2 |
| 0 | 1 | 1 | 0 | 0·5 |
| 0 | 1 | 0 | 1 | 0·5 |
| 0 | 0 | 1 | 1 | 1 |
| 0 | 0 | 0 | 0 | 1·3 |
| 0 | 0 | 1 | 0 | 1·3 |
| 1 | 1 | 1 | 0 | 3·2 |
| 1 | 1 | 0 | 1 | 4·2 |
| 1 | 0 | 0 | 1 | 6·2 |
| 1 | 1 | 1 | 1 | 9·8 |
| 1 | 0 | 1 | 1 | 17·9 |
| 1 | 0 | 0 | 0 | 20·3 |
| 1 | 0 | 1 | 0 | 32·5 |
Zero indicates lack of detectable antibodies whereas 1 indicates presence of antibodies against the specific virus. Bold values indicates specific viral seroconversions when they appeared in more than 80% of studied pigs.
Table 3.
Values of observed and simulated C score and number of unique seroconversions against selected viruses in 42 Spanish pig farms
| Farm reference | C score | P value | Unique combinations | P value | ||
|---|---|---|---|---|---|---|
| Observed | Expected | Observed > expected | Observed | Expected | Observed > expected | |
| 1 | 3·33 | 3·52 | 1 | 4·01 | 3·55 | 0·55 |
| 2 | 4·66 | 5·07 | 1 | 5·01 | 5·02 | 1 |
| 3 | 7·33 | 7·27 | 0·56 | 5·01 | 5·02 | 1 |
| 4 | 7·33 | 6·68 | 0·039* | 5·01 | 6·86 | 1 |
| 5 | 3·01 | 3·01 | 1 | 4·01 | 4·01 | 1 |
| 6 | 5·01 | 5·01 | 1 | 4·01 | 4·01 | 1 |
| 7 | 4·01 | 4·01 | 1 | 3·01 | 3·01 | 1 |
| 8 | 8·01 | 8·31 | 1 | 5·01 | 5·01 | 1 |
| 9 | 2·02 | 2·02 | 1 | 4·01 | 4·01 | 1 |
| 10 | 5·33 | 5·33 | 1 | 4·01 | 4·01 | 1 |
| 11 | 7·33 | 7·66 | 0·55 | 6·01 | 6·56 | 0·97 |
| 12 | 1·66 | 1·67 | 1 | 3·01 | 3·01 | 1 |
| 13 | 4·01 | 3·76 | 1 | 4·01 | 5·46 | 1 |
| 14 | 2·66 | 2·71 | 0·57 | 4·01 | 4·01 | 1 |
| 15 | 2·16 | 2·38 | 1 | 5·01 | 4·58 | 0·58 |
| 16 | 1·33 | 1·33 | 1 | 5·01 | 5·02 | 1 |
| 17 | 2·33 | 2·33 | 1 | 4·01 | 4·01 | 1 |
| 18 | 2·33 | 2·32 | 0·43 | 5·01 | 5·56 | 1 |
| 19 | 7·66 | 7·42 | 0·43 | 8·01 | 6·61 | 0·078 |
| 20 | 2·16 | 2·33 | 1 | 5·01 | 5·01 | 1 |
| 21 | 1·01 | 1·01 | 1 | 4·01 | 4·01 | 1 |
| 22 | 1·16 | 1·28 | 1 | 5·01 | 4·51 | 0·51 |
| 23 | 10·01 | 10·11 | 0·62 | 9·01 | 8·43 | 0·51 |
| 24 | 8·66 | 7·62 | 2·01 | 5·01 | 5·73 | 0·99 |
| 25 | 3·01 | 3·01 | 1 | 4·01 | 4·01 | 1 |
| 26 | 12·01 | 10·53 | 0·5 | 5·01 | 5·69 | 1 |
| 27 | 3·01 | 3·51 | 1 | 7·01 | 6·21 | 0·29 |
| 28 | 3·66 | 3·65 | 0·61 | 7·01 | 6·78 | 0·68 |
| 29 | 1·33 | 1·51 | 1 | 6·01 | 5·58 | 0·58 |
| 30 | 1·01 | 1·01 | 1 | 5·01 | 5·02 | 1 |
| 31 | 4·66 | 4·66 | 1 | 4·01 | 4·01 | 1 |
| 32 | 1·33 | 1·33 | 1 | 3·01 | 3·01 | 1 |
| 33 | 2·66 | 2·66 | 1 | 3·01 | 3·01 | 1 |
| 34 | 3·51 | 3·92 | 1 | 6 | 5·75 | 0·75 |
| 35 | 1·51 | 1·73 | 1 | 5·01 | 4·06 | 0·61 |
| 36 | 1·01 | 1·01 | 1 | 3·01 | 3·01 | 1 |
| 37 | 8·16 | 6·34 | 0·04* | 6·01 | 6·42 | 0·95 |
| 38 | 3·33 | 3·37 | 1 | 7·01 | 6·78 | 0·69 |
| 39 | 6·01 | 5·71 | 0·33 | 6·01 | 6·62 | 0·92 |
| 40 | 2·33 | 2·33 | 1 | 3·01 | 3·01 | 1 |
| 41 | 2·01 | 2·01 | 1 | 3·01 | 3·01 | 1 |
| 42 | 11·01 | 10·63 | 0·056 | 6·01 | 6·5 | 0·98 |
Models in which the null hypothesis (the observed C scores or the number of unique combinations of immune response against virus species were more or equally frequent to that expected by chance) was rejected (P values <0·05) are indicated by an asterisk (*). A total of 10 000 simulations were performed.
The mean PWM in the present study was 10% (Table 1), which is consistent with records of PWM in other parts or the world (4–11·9% in USA, [14, 15]; 7% in Canada [38]; or 18·12% in Japan [33]). As previously noted, all farms were seropositive for PCV2. However, prevalence of the remaining viruses was greater for PRRSV, then for SIV or ADV gE. The low prevalence of ADV gE may be attributed to the national Aujeszky's disease eradication scheme implemented in 1995 [39] and in force at the time of the study period.
Pigs that were seropositive for all viruses tested were more common (9·8%) than those fully seronegative (1·3%), a fact that reinforces the endemic nature of the studied pathogens. However, not all viral infections occurred in the same proportion. Co-existence of antibodies against PCV2 and PRRSV was the most frequent combination (32·4%, 204/630 pigs), followed by that of PRRSV, PCV2 and SIV in 17·9% of pigs. Single seroconversion against PCV2 occurred in 20·3% of pigs, while occurring in only 1·3% for only PRRSV and 0·2% for only SIV. No pigs exclusively had antibodies against ADV gE (Table 2). Similar viral prevalence occurrences have been reported in other locations in the world [8, 9], indicating that viral infections in intensive pig farming is a global issue. However, the null model simulation showed that the mean observed number of checkerboard pairs (mean C score = 4·2, min = 1·01, max = 12·01, Table 3) was identical to that obtained from the simulated matrices (mean = 4·1, min = 1·01, max = 10·63), and only two farms (nos. 4 and 37), representing 5% of our population of farms (Table 3), had more checkerboard pairs than expected by chance (significant P values). Along the same lines, the mean number of different observed specific viral infections (mean = 4·9, min = 3·0, max = 9·0, Table 3) was almost identical to that expected by chance (mean = 4·9, min = 3·01, max = 8·43, Table 3). Accordingly, all of these types of viral infections are possible by chance. PRRSV is a well-established triggering factor for the occurrence of PCV2-SD in PCV2-infected pigs, both from the point of view of experimental and natural occurrence of the disease [40]. Moreover, PCV2-SD is one of the main causes of economic loss linked to PWM throughout the world [41]. However, according to the results obtained, past infection does not appear to influence susceptibility to a subsequent infection.
On the other hand, the goodness of fit for the PLS path model used to analyse PWM was 0·91 (more than 90% of the achievable fit), with 42% of PWM variability explained by farm management practices, farm type and viral infection (Table 4, Fig. 1).
Table 4.
Contribution (%) of each latent variable (LV) to global explained observed variability (R2 = 0·42, see Fig. 1) of post-weaning mortality
| LVs explaining mortality | β | Correlation | Contribution to global R2 (%) |
|---|---|---|---|
| Farm type | −0·43*** | −0·29 | 35·7 |
| Farm management | −0·26* | −0·05 | 3·7 |
| EMVI | 0·47*** | 0·45 | 60·6 |
Multiple viral infections (EMVI) were evidenced by immunological tests. Relative goodness-of-fit index = 0·91; β = path coefficient estimated by bootstrapping.
* P < 0·1, ** P < 0·05, *** P < 0·01.
The existence of antibodies against multiple viruses had the greatest impact (60·6%) on the explained PWM variability, followed by farm type (35·7%) and management practices (3·7%, Table 4). However, these LVs had different effects on PWM that increased in farms with evidence of several virus assemblages, and decreased in farms with multisite production, all-in/all-out flow and increased age at fattening (Table 5). Curiously, neither farm type nor associated management affected PWM due to pathogens (no relationship between farm type or management and evidence of multiple virus infections, Fig. 1b), a fact that suggests the low impact of additional management measures aimed to reduce PWM due to infectious diseases. The higher PWMs (14–16·9%) were recorded on farms that were seropositive against PCV2, SIV and PRRSV at the time of slaughter.
Table 5.
Correlation between manifest variables (MVs) and latent variables (LVs) of the partial least squares path modelling of post-weaning mortality
| LVs and MVs | W | Correlation with LVs | |||
|---|---|---|---|---|---|
| Farm type | Farm management | EMVI | Mortality | ||
| Farm type | |||||
| Production system | 0·79 | 0·95 | −0·56 | 0·05 | −0·24 |
| Partitions | −0·37 | −0·52 | 0·2 | 0·156 | 0·24 |
| Farm management | |||||
| Flow nursery | −0·06 | −0·17 | 0·35 | 0·15 | −0·04 |
| Flow fattening | 0·64 | −0·37 | 0·79 | −0·17 | −0·11 |
| Age at fattening | 0·59 | −0·45 | 0·66 | 0·2 | 0·04 |
| EMVI | |||||
| ADV gE | 0·17 | −0·08 | −0·01 | 0·57 | 0·26 |
| PRRSV | 0·42 | −0·15 | 0·09 | 0·64 | 0·29 |
| SIV | 0·66 | 0·13 | −0·13 | 0·84 | 0·38 |
| Post-weaning mortality | |||||
| Mortality | 1 | −0·29 | 0·051 | 0·42 | 1 |
EMVI, Multiple viral infections; ADV gE, antibodies against glycoprotein E of Aujeszky's disease virus; PRRSV, porcine reproductive and respiratory syndrome virus; SIV, swine influenza virus.
W = manifest variables' bootstrapped weights. Only those manifest variables retained in the final path model are shown in Figure 1.
In line with recent surveys [34], early weaning was irrelevant in reducing PWM. However, the fact that most (88%) of the farms in our study showed ages at weaning above the minimum limit recommended in intensive pig farming (21·5 days [5]) would explain its lack of influence in the present study system. Additionally, no evidence of pig density-dependent PWM was observed [17, 33], which probably indicates that farms had a fairly optimum number of pigs per pen.
In conclusion, this study highlights the relevance of infectious diseases in PWM when animals are reared under intensive production systems. Null models and PLS-PM provide a new and clearer view of the network of interactions in risk factors influencing PWM. Farm type and, to a lesser extent, farm management might play an important role in modulating PWM. However, it is evident that pathogens remain the most important challenge limiting intensive pig production. The fact that PWM increases due to certain virus assemblages (evidence of PCV2 + SIV + PRRSV infections) encourages the urgent implementation of specific biosecurity and preventive programmes aimed at controlling some, if not all, major pathogens implicated in PWM.
ACKNOWLEDGEMENTS
E. Serrano was supported by the Beatriu de Pinós programme (BP-DGR 2011) of the Catalan Science and Technology System, Spain.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Miller GY, et al. Productivity and profitability differences between pseudorabies-infected and pseudorabies-non-infected farrow-to-finish swine herds. Journal of the American Veterinary Medicine Association 1995; 206: 446–451. [PubMed] [Google Scholar]
- 2.Rohrer GA, Beattie C W. Genetic influences on susceptibility to acquired diseases. In: Straw BE, D'Allaire S, Mengeling WL, Taylor DJ, eds. Diseases of Swine. Ames, IA: Iowa State University Press, 1999, pp. 977–984. [Google Scholar]
- 3.Dewey CE, et al. Postweaning mortality in Manitoba swine. Canadian Journal of Veterinary Research 2006; 70: 161–167. [PMC free article] [PubMed] [Google Scholar]
- 4.Harris DL. Multi-site Pig Production. Ames, IA: Iowa State University Press, 2007, pp. 271. [Google Scholar]
- 5.Main RG, et al. Increasing weaning age improves pig performance in a multisite production system. Journal of Animal Science 2004; 82: 1499–1507. [DOI] [PubMed] [Google Scholar]
- 6.Ellis J, et al. Porcine circovirus-2 and concurrent infection in the field. Veterinary Microbiology 2004; 98: 159–163. [DOI] [PubMed] [Google Scholar]
- 7.Segalés J, Allan GM, Domingo M. Porcine circovirus diseases. Animal Health Research Reviews 2005; 6: 119–142. [DOI] [PubMed] [Google Scholar]
- 8.Pallarés FJ, et al. Porcine circovirus type 2 (PCV-2) infections in US field cases of postweaning multisystemic wasting syndrome (PMWS). Journal of Veterinary Diagnostic Investigation 2002; 25: 684–693. [DOI] [PubMed] [Google Scholar]
- 9.Wellenberg GJ, et al. The presence of coinfection in pigs with clinical signs of PMWS in the Netherlands: a case-control study. Research in Veterinary Science 2004; 77: 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.López-Soria S, et al. Selected swine viral pathogens in indoor pigs in Spain. Seroprevalence and farm-level characteristics. Transboundary and Emerging Diseases 2010; 57: 171–179. [DOI] [PubMed] [Google Scholar]
- 11.Dorr PM, et al. Epidemiologic assessment of porcine circovirus type 2 coinfection with other pathogens in swine. Journal of the American Veterinary Medicine Association 2007; 230: 244–250. [DOI] [PubMed] [Google Scholar]
- 12.Brockmeier SL, Halbur PG, Thacker EL. Porcine respiratory disease complex. In: Brodgen KA, Gutmiller J, eds. Polymicrobial Diseases. Washington, USA: ASM Press, 2002, pp. 214–231. [Google Scholar]
- 13.Fraile L, et al. Risk factors associated with pleuritis and cranio-ventral pulmonary consolidation in slaughter-aged pigs. Veterinary Journal 2010; 184: 326–333. [DOI] [PubMed] [Google Scholar]
- 14.Maes DA, et al. A retrospective study of mortality in grow-finish pigs in a multi-site production system. Journal of Swine Health and Production 2001; 9: 267–273. [Google Scholar]
- 15.Losinger WC, et al. Mortality attributed to respiratory problems among finisher pigs in the United States. Preventive Veterinary Medicine 1998; 37: 21–31. [DOI] [PubMed] [Google Scholar]
- 16.Losinger WC, et al. An analysis of mortality in the grower/finisher phase of swine production in the United States. Preventive Veterinary Medicine 1988; 33: 121–145. [DOI] [PubMed] [Google Scholar]
- 17.Maes DA, et al. Risk factors for mortality in grow-finishing pigs in Belgium. Journal of Veterinary Medicine. B, Infectious Diseases and Veterinary Public Health 2004; 57: 321–326. [DOI] [PubMed] [Google Scholar]
- 18.Larriestra AJ, et al. Mixed models applied to the study of variation of grower-finisher mortality and culling rates of a large swine production system. Canadian Journal of Veterinary Research 2005; 69: 26–31. [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis FI, et al. Structure discovery in Bayesian networks: an analytical tool for analysing complex animal health data. Preventive Veterinary Medicine 2011; 100: 109–115. [DOI] [PubMed] [Google Scholar]
- 20.Bougeard S, Qabbari EM, Rose N. Multiblock redundancy analysis: interpretation tools ad application in epidemiology. Journal of Chemometrics 2011; 25: 467–475. [Google Scholar]
- 21.Gotelli NJ. Null model analysis of species co-occurrence patterns. Ecology 2000; 81: 2606–2621. [Google Scholar]
- 22.Wold H. PLS path models with latent variables: the nipals approach. In: Blalock HM, Aganbegian A, Borodkin FM, Boudon R, Cappecchi V, eds. Quantitative Sociology: International Perspectives on Mathematical and Statistical Modeling. New York: Academic Press, 1975, pp. 307–357. [Google Scholar]
- 23.Tenenhaus M, et al. PLS path modelling. Computational Statistics and Data Analysis 2005; 48: 159–205. [Google Scholar]
- 24.Gotelli NJ, Colwell RK. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecology Letters 2001; 4: 379–391. [Google Scholar]
- 25.Alwin DF, Hauser RM. The decomposition of effects in path analysis. American Sociological Review 1975; 40: 36–47. [Google Scholar]
- 26.Grace JB. Structural Equation Modeling and Natural Systems. Cambridge, UK: Cambridge University Press, 2006, pp. 378. [Google Scholar]
- 27.Esposito Vinzi V, Trinchera L, Amato S. PLS path modelling: from foundations to recent developments and open issues for model assessment and improvement. In: Esposito VV, Chin WW, Henseler J, Wang H, eds. The Handbook of Partial Least Squares: Concepts, Methods and Applications. New York, USA: Springer, 2010, pp. 47–82. [Google Scholar]
- 28.Blanchard P, et al. An ORF2 protein-based ELISA for porcine circovirus type 2 antibodies in post-weaning multisystemic wasting syndrome. Veterinary Microbiology 2003; 94: 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stone L, Roberts A. The checkerboard score and species distributions. Oecologia 1990; 85: 74–79. [DOI] [PubMed] [Google Scholar]
- 30.Pielou DP, Pielou EC. Association among species of infrequent occurrence: the insect and spider fauna of Polyporus betulinus (Bulliard) Fries. Journal of Theoretical Biology 1968; 21: 202–216. [DOI] [PubMed] [Google Scholar]
- 31.Gotelli NJ, Entsminger GL. EcoSim 7.0: Null models software for ecology (http://garyentsminger.com/ecosim.htm). Accessed 30 September 2012.
- 32.Wold H. Partial least squares. In: Kotz S, Johnson NL, eds. Encyclopedia of Statistical Sciences. New York, USA: Wiley, 1985, pp 581–591. [Google Scholar]
- 33.Koketsu Y. Mortality trends and comparisons between mortality risk and mortality rate of fattening pig operations in farrow-to-finish herds. Journal of Veterinary Epidemiology 2007; 11: 90–95. [Google Scholar]
- 34.Collings CL, Leury BJ, Dunshea FR. Early weaning has minimal effects on lifetime growth performance and body composition of pigs. Animal Production Science 2010; 50: 79–87. [Google Scholar]
- 35.Tenenhaus M, Amato S, Esposito Vinzi V. A global goodness-of-fit index for PLS structural equation modelling. Proceedings of the XLII SIS Scientific Meeting. Padova, Italy, 2004, pp. 739–742. [Google Scholar]
- 36.Sánchez G, Trinchera LR. plspm: Partial Least Squares data analysis methods. R package version 0.3.7 (http://cran.r-project.org/web/packages/plspm/index.html). Accessed 15 April 2013.
- 37.R Development Core Team. Version 3.0.0. (http://www.R-project). Accessed 15 April 2013.
- 38.Van Til LD, et al. A survey of biological productivity of Prince Edward Island swine herds. Canadian Journal of Veterinary Research 1991; 55: 174–179. [PMC free article] [PubMed] [Google Scholar]
- 39.Allepuz A, et al. The role of spatial factors in the success of an Aujeszky's disease eradication programme in a high pig density area (Northeast Spain, 2003–2007). Preventive Veterinary Medicine 2011; 91: 153–160. [DOI] [PubMed] [Google Scholar]
- 40.Segalés J, et al. Quantification of porcine circovirus type 2 (PCV2) DNA in serum and tonsillar, nasal, tracheo-bronchial, urinary and faecal swabs of pigs with and without postweaning multisystemic wasting syndrome (PMWS). Veterinary Microbiology 2005; 111: 223–229. [DOI] [PubMed] [Google Scholar]
- 41.Alarcon P, et al. Assessment and quantification of post-weaning multisystemic wasting syndrome severity at farm level. Preventive Veterinary Medicine 2011; 98: 19–28. [DOI] [PubMed] [Google Scholar]


