SUMMARY
This study describes the association between antibiotic resistance of bacteria causing neonatal bloodstream infection (BSI) and neonatal age to inform empirical antibiotic treatment guidelines. Antibiotic resistance data were analysed for 14 078 laboratory reports of bacteraemia in neonates aged 0–28 days, received by the Health Protection Agency's (now Public Health England) voluntary surveillance scheme for England and Wales between January 2005 and December 2010. Linear and restricted cubic splines were used in logistic regression models to estimate the nonlinear relationship between age and resistance; the significance of confounding variables was assessed using likelihood ratio tests. An increase in resistance in bacteria causing BSI in neonates aged <4 days was observed, which was greatest between days 2–3 and identified an age (4–8 days, depending on the antibiotic) at which antibiotic resistance plateaus to almost unchanging levels. Our results indicate important age-associated changes in antibiotic resistance and support current empirical treatment guidelines.
Key words: Antibiotic resistance, bloodstream infections, neonatal infections
INTRODUCTION
Bacterial bloodstream infection (BSI) in neonates (aged 0–28 days) is a serious condition that may result in death or disability unless immediate and effective antibiotic treatment is administered [1–3]. Therefore, treatment is often started empirically prior to identification of the causative organism and determination of its antibiotic susceptibility profile. Current recommendations for empirical treatment of neonatal BSI take into account the likely mode of acquisition of infection, with early-onset sepsis (EOS) attributed to maternal flora and late-onset sepsis (LOS) associated with nosocomial infection. EOS and LOS are therefore thought to be associated with distinct maternal or hospital pathogens, with the latter often assumed to have an increased likelihood of being antibiotic resistant [4, 5].
The criteria used to define EOS and LOS, particularly the time between birth and clinical onset of infection, vary between (or even within) countries, with the timescale for the early-onset period ranging from <48 h to <1 week after birth [6–10]. In the UK, clinical guidelines for the empirical treatment of septic neonates have been issued by the British National Formulary for Children (BNFC) [6], and more recently, the National Institute of Health and Clinical Excellence (NICE) have introduced guidelines for EOS [11]. Both guidelines recommend empirical treatment with penicillin in combination with gentamicin (PEN/GENT) for EOS; however, the timelines for ‘early onset’ differ, with the BNFC using an earlier end-point than NICE (48 h vs. 72 h, respectively). The BNFC also recommends administration of flucloxacillin combined with gentamicin (FLU/GENT) for LOS (i.e. in neonates aged ⩾48 h) or cefotaxime in combination with ampicillin/amoxicillin (CTX/AMOX) in neonates of any age.
Several studies have used arbitrary thresholds of 48–72 h to distinguish between infections of maternal or nosocomial derivation; however, few studies have undertaken data analysis to identify a time threshold [4, 12–14]. Furthermore, review of the published literature did not identify any studies that aimed to correlate definitions of EOS and LOS against antibiotic susceptibility data. This study therefore sought to describe the association between antibiotic resistance of bacteria causing BSI and age of the neonate at the time the blood sample was taken with a view to enhancing the evidence base for guidance on the antibiotic management of neonatal EOS and LOS.
METHODS
Microbiological reports of bacteria isolated from blood cultures taken from neonates aged 0–28 days in England and Wales between January 2005 and December 2010 were extracted from the Health Protection Agency's (now Public Health England; PHE) voluntary surveillance database. Extracted data included information on the patient's age and sex and organism identification and susceptibility to PEN, FLU, AMOX, CTX and GENT. Data on susceptibility to the combinations of antibiotics recommended by the BNFC or NICE for the treatment of EOS and LOS were coded using the criteria outlined in Table 1. Repeat specimens for the same patient within a 14-day period were considered to be part of the same infection episode, therefore data for only the first reported specimen were retained.
Table 1.
Criteria for coding bacterial isolates as susceptible or resistant to dual combinations of antibiotics
| Susceptibility | ||
|---|---|---|
| Antibiotic 1 | Antibiotic 2 | Combination of antibiotics 1 and 2 |
| Susceptible | Susceptible | Susceptible |
| Susceptible | Resistant | Susceptible |
| Susceptible | Not reported | Susceptible |
| Resistant | Resistant | Resistant |
| Resistant | Not reported | Excluded |
| Not reported | Not reported | Excluded |
Susceptible isolates were coded as 0, and resistant or intermediate isolates coded as 1.
Preliminary analysis of the proportion of resistant bacteria plotted against the patient's age at the time the blood sample was taken suggested a nonlinear association for several of the antibiotic combinations under investigation, which could be modelled using linear or restricted cubic splines [linear or cubic functions joined by one or more breakpoints (knots)]. These functions were created in Stata v. 12 (StataCorp., USA; using the user-written command ‘mkspline’) and inserted into a logistic regression model with resistance as the outcome.
Each linear spline incorporated one, two or three knots, whereas restricted cubic splines included three knots (the minimum required for mkspline). Restricted cubic splines generated using mkspline have a linear function before the first knot (knot 1) and after the last knot (knot 3) with a piecewise cubic polynomial between the adjacent knots. All combinations of knot placement between days 1 and 27 were investigated with the condition that knot 1 occurred before knot 2 (for splines with two or three knots) and that knot 2 occurred before knot 3 (for splines with three knots), thus generating 3303 and 2925 different linear and cubic splines, respectively, for each antibiotic combination. Where the association between neonatal age and antibiotic resistance was less complex and did not require splines to be fitted, logistic models incorporating linear or quadratic functions of age were fitted. Akaike's Information Criterion (AIC†) was used to select single variable logistic regression models with the best fit and lowest level of complexity, where models with lower AIC are preferred.
Multivariable logistic regression models incorporating the spline function with the lowest AIC were used to assess whether the association between neonatal age and bacterial resistance to each antibiotic therapy combination was confounded by organism, reporting laboratory, patient's sex, reporting year and calendar quarter of BSI. Organism and reporting laboratory (for laboratories reporting ⩾20 cases, in order to exclude results from laboratories that did not regularly report during the study period) were included in the model as crossed-factor random effects. Calendar year, quarter and patient's sex were included as continuous, factor and binary variables, respectively. Statistical significance of the association of these explanatory variables with outcome was ascertained by means of the likelihood ratio test. Analyses were repeated after excluding BSI with coagulase-negative staphylococci (CoNS), as these bacteria may be contaminants and are associated with less severe morbidity than BSI with other organisms [14, 15]
RESULTS
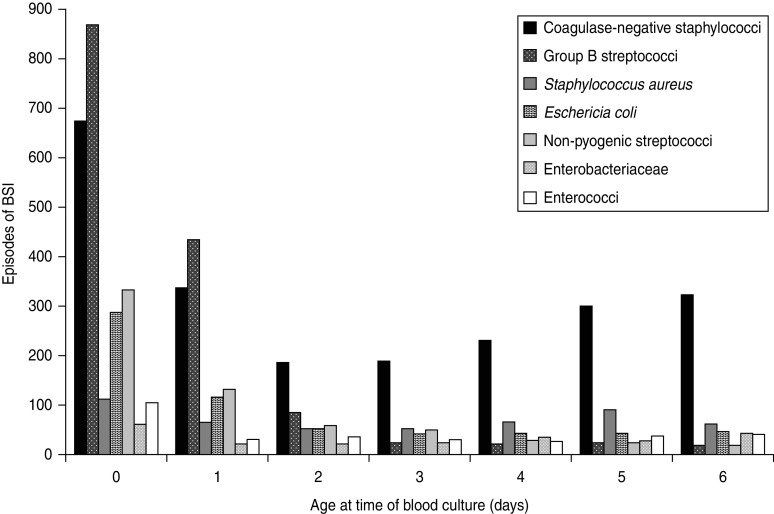
A total of 14 078 episodes of neonatal bacteraemia were reported by 215 laboratories between 2005 and 2010 (of which 137 laboratories reported ⩾20 isolates). The median age when positive blood cultures were taken was 7 days [95% confidence interval (CI) 6–7]. BSI incidence was slightly higher (56%) in males. Seven groups of taxonomically related pathogens accounted for 90% of reports, and comprised: CoNS (38%), group B streptococci (GBS) (14%), Staphylococcus aureus (10%), Escherichia coli (8%), non-pyogenic streptococci (7%), Enterobacteriaceae (6%) and enterococci (6%) (Table 2). The most commonly isolated organisms were GBS for patients with blood cultures taken on day 0 (day of birth) and day 1 and CoNS for all subsequent days (Fig. 1).
Table 2.
Causative organisms and antibiotic susceptibility results reported for bloodstream infection (BSI) in neonates aged 0–28 days
| Organism group | Total | % | PEN/GENT | FLU/GENT | CTX/AMOX | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tested (%) | %R | Tested (%) | %R | Tested (%) | %R | ||||||
| Gram-negative | |||||||||||
| Escherichia coli | 1166 | 8 | 932 | (80) | 1 | 939 | (81) | 1 | 759 | (65) | 3 |
| Enterobacteriaceae | 915 | 6 | 704 | (77) | 0 | 710 | (78) | 0 | 400 | (44) | 20 |
| Pseudomonas/Stenotrophomonas spp. | 253 | 2 | 183 | (72) | 1 | 183 | (72) | 1 | 56 | (22) | 48 |
| Haemophilus influenzae | 112 | 1 | 7 | (6) | 14 | 6 | (5) | 0 | 79 | (71) | 0 |
| Acinetobacter spp. | 106 | 1 | 78 | (74) | 0 | 80 | (75) | 3 | 56 | (53) | 20 |
| Other Gram-negative bacteria | 172 | 1 | 62 | (36) | 0 | 55 | (32) | 2 | 80 | (47) | 10 |
| Gram-positive | |||||||||||
| CoNS | 5419 | 38 | 3248 | (60) | 56 | 3566 | (66) | 54 | 50 | (1) | 26 |
| Group B streptococcus | 1967 | 14 | 1738 | (88) | 0 | 273 | (14) | 0 | 1015 | (52) | 0 |
| Staphylococcus aureus | 1414 | 10 | 1071 | (76) | 2 | 1291 | (91) | 1 | 47 | (3) | 4 |
| Non-pyogenic streptococcus | 954 | 7 | 651 | (68) | 3 | 153 | (16) | 3 | 454 | (48) | 1 |
| Enterococcus spp. | 841 | 6 | 327 | (39) | 17 | 268 | (32) | 10 | 622 | (74) | 0 |
| Micrococcus spp. | 219 | 2 | 156 | (71) | 0 | 154 | (70) | 0 | 4 | (2) | 0 |
| Streptococcus pneumoniae | 157 | 1 | 121 | (77) | 0 | 45 | (29) | 0 | 76 | (48) | 0 |
| Other Gram-positive bacteria | 383 | 3 | 223 | (58) | 4 | 122 | (32) | 3 | 124 | (32) | 2 |
| Total reported BSI | 14 078 | 100 | 9501 | (67) | 20 | 7845 | (56) | 25 | 3822 | (27) | 5 |
| Total BSI excluding CoNS | 8659 | 62 | 6253 | (72) | 2 | 4279 | (49) | 1 | 3772 | (44) | 4 |
PEN, Penicillin; GENT, gentamicin; FLU, flucloxacillin; CTX, cefotaxime; AMOX, ampicillin/amoxicillin; CoNS, coagulase-negative staphylococci.
%R = percentage of isolates tested for a given antibiotic combination that were non-susceptible (resistant or intermediate).
Fig. 1.
Bacteria isolated from blood cultures taken from neonates aged 0–6 days. Reports are shown only for the seven most frequently reported groups, which account for 90% of all reported neonatal bloodstream infections (BSI).
PEN/GENT and FLU/GENT were similar in terms of resistance patterns with increases from 4% and 7%, respectively, for bacteria isolated on day 1 to 35% and 37%, respectively, at day 6. Exclusion of CoNS reduced antimicrobial resistance to <2% over the whole neonatal period for PEN/GENT and FLU/GENT. Resistance to CTX/AMOX was 4–5% and did not change substantially after excluding CoNS. The completeness of reporting of susceptibility test results varied markedly by organism and antibiotic combination ranging from 1% (CTX/AMOX and CoNS) to 91% (FLU/GENT and S. aureus) (Table 2).
Resistance data for all organisms
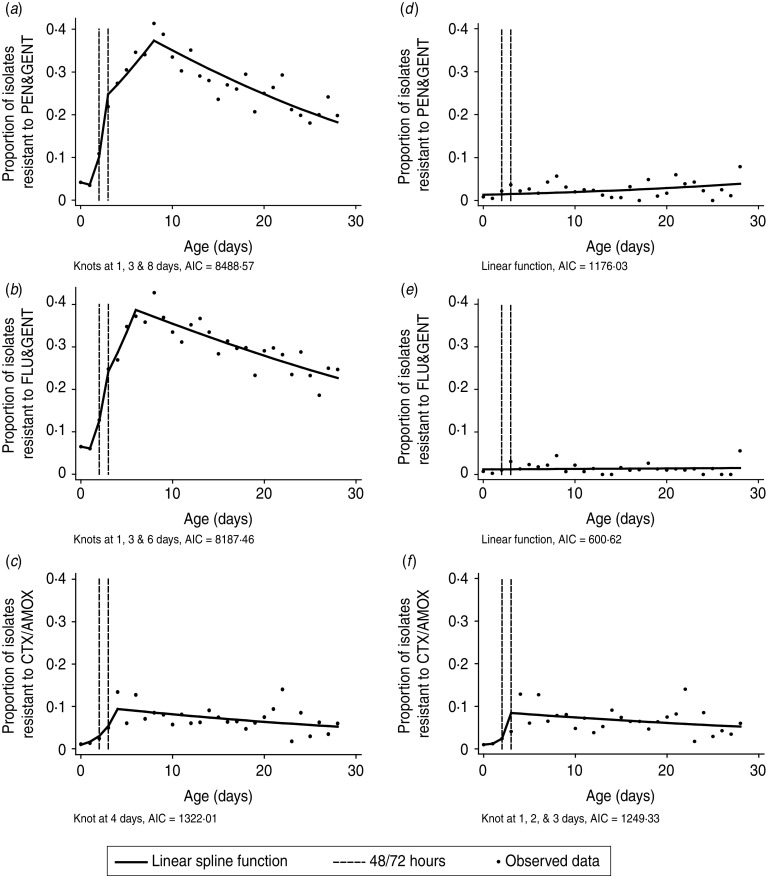
A total of 6228 models incorporating different spline functions were generated for each of the three antibiotic combinations. AICs ranged as follows; 8488·57–9292·17 (PEN/GENT), 8187·46–8988·58 (FLU/GENT) and 1322·01–1389·86 (CTX/AMOX); models with the lowest AIC are outlined in Table 3. The association between PEN/GENT and FLU/GENT resistance in all organisms was best described by a linear spline incorporating three knots placed at 1, 3 and 8 or 1, 3 and 6 days, respectively. By contrast, a linear spline incorporating a single knot at day 4 had the lowest AIC for CTX/AMOX resistance (Fig. 2a–c).
Table 3.
Models incorporating a restricted cubic or linear spline function of age (at which the blood culture was taken) with the lowest AIC
| Spline function | No. knots | PEN/GENT | FLU/GENT | CTX/AMOX | |||
|---|---|---|---|---|---|---|---|
| Knot location (days) | AIC | Knot location (days) | AIC | Knot location (days) | AIC | ||
| All organisms | |||||||
| Linear | 1 | 5 | 8515·68 | 5 | 8197·52 | 4 | 1322·01 |
| 2 | 4, 8 | 8505·14 | 1, 5 | 8196·24 | 4, 5 | 1322·98 | |
| 3 | 1, 3, 8 | 8488·57 | 1, 3, 6 | 8187·46 | 2, 4, 5 | 1323·18 | |
| Cubic | 3 | 2, 3, 9 | 8504·57 | 3, 4, 8 | 8193·88 | 3, 4, 5 | 1323·04 |
| Excluding CoNS | |||||||
| Linear | 1 | 1 | 1281·10 | ||||
| 2 | 1, 2 | 1256·09 | |||||
| 3 | 1, 2, 3 | 1249·33 | |||||
| Cubic | 3 | 1, 2, 3 | 1255·83 | ||||
PEN, Penicillin; GENT, gentamicin; FLU, flucloxacillin; CTX, cefotaxime; AMOX, ampicillin/amoxicillin; AIC, Akaike's Information Criterion; CoNS, coagulase-negative staphylococci.
 , Spline functions not required and hence not fitted;
, Spline functions not required and hence not fitted;  , models with the lowest AIC for each antibiotic combination.
, models with the lowest AIC for each antibiotic combination.
Fig. 2.
Models incorporating a spline or linear function of age (at which the blood culture was taken) to estimate the change in the odds of antibiotic resistance in all organisms (a–c) or excluding coagulase-negative staphylococci (d–f). Models are shown on the logit scale. AIC, Akaike's Information Criterion.
The crude and adjusted odds ratios (ORs) for neonatal age and bacterial resistance for the selected spline models are outlined in Table 4. The crude ORs for resistance to PEN/GENT and FLU/GENT can be interpreted as the odds of resistance being unchanged between birth (day 0) and day 1, then increasing two- to threefold per day from days 1–3. The rate of increasing odds of resistance then slows to 13–25% per day until days 6 or 8 (FLU/GENT and PEN/GENT, respectively) and then decreases slightly (3–5% per day) until day 28. For CTX/AMOX there was an 80% increase in the crude odds of resistance each day between days 0 and 4, followed by unchanging resistance levels thereafter.
Table 4.
Single and multivariable logistic regression models incorporating a spline or linear function of age (at which the blood culture was taken) to estimate the change in the odds of antibiotic resistance
| Antibiotic | Age function | Age (days) | Crude OR | 95% CI | No. obs. | Adjusted OR* | 95% CI | LRT P value for age | No. obs. |
|---|---|---|---|---|---|---|---|---|---|
| PEN/GENT | Linear spline with knots at 1, 3, 8 days | 0–1 | 0·86 | 0·60–1·24 | 9501 | 0·58 | 0·38–0·91 | <0·0001 | 8977 |
| 1–3 | 2·95 | 2·47–3·53 | 2·50 | 1·98–3·16 | |||||
| 3–8 | 1·13 | 1·08–1·17 | 1·18 | 1·11–1·26 | |||||
| 8–28 | 0·95 | 0·94–0·96 | 0·98 | 0·96–0·99 | |||||
| FLU/GENT | Linear spline with knots at 1, 3, 6 days | 0–1 | 0·93 | 0·64–1·36 | 7845 | 0·63 | 0·40–0·97 | <0·0001 | 7460 |
| 1–3 | 2·22 | 1·83–2·70 | 2·28 | 1·79–2·90 | |||||
| 3–6 | 1·25 | 1·16–1·36 | 1·33 | 1·19–1·48 | |||||
| 6–28 | 0·97 | 0·96–0·98 | 0·99 | 0·98–1·00 | |||||
| CTX/AMOX | Linear spline with a knot at day 4 | 0–4 | 1·82 | 1·58–2·11 | 3822 | 1·41 | 1·18–1·69 | 0·0002 | 3533 |
| 4–28 | 0·97 | 0·95–1·00 | 0·99 | 0·96–1·02 | |||||
| PEN/GENT | Linear | – | 1·04 | 1·02–1·06 | 6253 | 1·01 | 0·98–1·04 | 0·5484 | 5705 |
| FLU/GENT | Linear | – | 1·01 | 0·98–1·04 | 4279 | 0·99 | 0·95–1·03 | 0·6816 | 3937 |
| CTX/AMOX | Linear spline with knots at 1, 2, 3 days | 0–1 | 1·20 | 0·44–3·25 | 3772 | 1·73 | 0·57–5·28 | 0·0083 | 3434 |
| 1–2 | 2·06 | 0·57–7·39 | 0·89 | 0·21–3·78 | |||||
| 2–3 | 3·68 | 1·31–10·35 | 2·39 | 0·72–7·92 | |||||
| 3–28 | 0·98 | 0·96–1·00 | 0·99 | 0·96–1·02 |
PEN, Penicillin; GENT, gentamicin; FLU, flucloxacillin; CTX, cefotaxime; AMOX, ampicillin/amoxicillin; OR, Odds ratio; CI, confidence interval; LRT, likelihood ratio test.
Multivariable analysis results for all antibiotic combinations showed strong evidence of an association between age and resistance (P⩽0·0002 for all antibiotic combinations). Crude and adjusted ORs for resistance to PEN/GENT were similar, although estimates for infants aged <3 days were slightly reduced after adjusting for other factors. Conversely, adjusted ORs for resistance in infants aged ⩾3 days were slightly increased relative to the crude estimates. For FLU/GENT, the increase in odds of resistance was slightly inflated in infants aged >1 day after adjusting for other factors. In contrast, for infants aged 0 or 1 day the adjusted OR for change in resistance to FLU/GENT was reduced such that 95% CI no longer included 1. Comparison of crude and adjusted ORs for resistance to CTX/AMOX indicated a relatively decreased rate of resistance prior to day 4 after adjusting for other factors. Adjusted ORs for resistance to all antibiotic combinations suggested either no trend, or a slight decrease in resistance, from the last knot (days 4, 6 and 8 for CTX/AMOX, FLU/GENT and PEN/GENT, respectively) until day 28. Likelihood ratio testing demonstrated that none of the fixed-effect variables included in the models (sex, calendar quarter, reporting year) were significant at the 5% level (P>0·2 in all instances).
Resistance data for all organisms excluding CoNS
The relationship between age and resistance to PEN/GENT or FLU/GENT appeared linear after excluding CoNS. The association between CTX/AMOX resistance in all organisms excluding CoNS and age was best described by a linear spline incorporating three knots placed at 1, 2 and 3 days (Table 3, Fig. 2d–f).
Crude estimates suggested a slight increase (4% per day) in the odds of resistance to PEN/GENT, while resistance to FLU/GENT appeared static (1·01, 95% CI 0·98–1·04). For CTX/AMOX, crude ORs suggested an increase in the odds of resistance from birth until day 3, including a greater than threefold increase between days 2 and 3 that was statistically significant (95% CI 1·31–10·35). The rate of resistance to CTX/AMOX appeared static from days 3–28 of life (Table 4).
Adjusted ORs for resistance to PEN/GENT and FLU/GENT included 1 and were not significantly associated with age (P>0·5 in both instances). After adjusting for other factors there was evidence of an association between CTX/AMOX resistance and age (P = 0·008, age incorporated as a linear spline function with knots at days 1, 2 and 3), although 95% CIs for each age bracket within the spline were wide and included 1 (Table 4). As with the analysis of all organisms, likelihood ratio testing demonstrated that none of the fixed-effect variables included in the models (sex, calendar quarter, reporting year) were significant at the 5% level (P>0·2 in all instances).
DISCUSSION
Current definitions of neonatal EOS and LOS incorporate arbitrary time thresholds that do not consider changes in the antibiotic susceptibility of causative pathogens; the application of these definitions to empirical treatment regimens for sepsis may therefore be suboptimal. We aimed to enhance the evidence base for definitions of EOS and LOS by investigating the association between antibiotic resistance in bacteria causing BSI and the age of the neonate at the time that the blood sample was taken. We observed a rapid increase in antibiotic resistance in bacteria causing BSI in neonates aged <4 days for each of the antibiotic combinations, which may indicate a phased transition from EOS to LOS as maternal or hospital-derived organisms mix in neonates of this age. Our results also identify an age at which antibiotic resistance plateaus to almost unchanging levels (days 4–8, depending on the antibiotic combination).
There is a dearth of studies examining antibiotic resistance across the neonatal period, which limits the scope for comparing results from this study with existing literature. However, several studies have reported evidence of a difference in the organisms and antibiotic resistance occurring in neonates aged 0–1 or ⩾2 days, which is compatible with the observed increase in resistance in bacteria isolated after day 2 of life (relative to <2 days) for the three antibiotic combinations that we investigated [4, 12]. We identified only one study that used data analysis to define EOS, which investigated changes in the frequency of BSI by day of life using finite mixture latent class regression models. BSI caused by all pathogens occurring in two English neonatal units in 2001–2005 were analysed and the results indicated that BSI frequency changed at 2·23 days of life. The study also reported decreased frequencies of CoNS, GBS and Gram-positive organisms after day 2 of life [16]. While these estimates do not describe antibiotic resistance as such, it is likely that a change in the organisms causing BSI will influence levels of resistance and may therefore be compatible with the sharp increases in resistance to PEN/GENT and FLU/GENT observed by this study between days 1–3 of life.
This study used routinely reported surveillance data on microbiologically confirmed BSI in neonates in England and Wales, which captured an estimated 80% of all BSI [17]. However, a substantial proportion of reports did not include susceptibility data for the antibiotics under investigation, which limits the confidence with which resistance levels can be estimated. In particular, data are incomplete for some antibiotic and organism combinations (such as CTX/AMOX and CoNS) that are not routinely tested in UK microbiology laboratories; complete case analysis or imputing values for resistance (the outcome of our study) was considered impractical.
Even though the study's objective was to provide evidence for identifying a time threshold for EOS and LOS by analysing the association between antibiotic resistance of bacteria isolated from neonatal blood and age of the neonate, absence of clinical data in the microbiology surveillance data is a key shortcoming as the severity (or even presence) of disease or patient risk factors cannot be established. The lack of clinical information is particularly important when considering the relevance of CoNS, which are predominantly resistant to PEN/GENT and FLU/GENT but are often considered to be contaminants and are associated with less severe disease [14, 15]. Therefore, we investigated the resistance levels of the empirical antibiotic combinations PEN/GENT, FLU/GENT and CTX/AMOX after excluding CoNS. Resistance to PEN/GENT and FLU/GENT was static and uniformly lower than that for CTX/AMOX. Resistance to CTX/AMOX increased most sharply between days 2–3 of life (thus lending support to both the BNFC and NICE definitions for EOS). It should be emphasized that for all of these combinations the overall resistance rates of relevant pathogens remain low; 2% of isolates tested for PEN/GENT were non-susceptible, 1% were resistant to FLU/GENT and 4% to CTX/AMOX.
It is notable that the highest number of CoNS infections appears to be in the first few days of life, which is also the time that the largest number of blood cultures are obtained in neonates, often not for clinical features of sepsis but because of perceived risk factors and suggesting that these are therefore often contaminants [18]. Risk factors for clinically significant infection with CoNS have been identified for LOS; severe or persistent CoNS infection in neonates aged >2 days is associated with birthweight <1500 g, gestational age ⩽32 weeks and greater exposure to parenteral nutrition, hydrocortisone, antibiotics and mechanical ventilation prior to infection [14, 19, 20]. Infants aged >2 days with any of these risk factors should therefore be considered to be at risk of clinically significant BSI with CoNS.
Although our study did not aim to determine the cause of changes in resistance during the neonatal period, the observed changes in resistance to PEN/GENT and FLU/GENT appear to be driven primarily by CoNS, as exclusion of this organism removed the association between resistance and age. Reported resistance to CTX/AMOX was very low in Gram-positive organisms (although data were incomplete), suggesting that Gram-negative bacteria play a leading role in shaping the association between neonatal age and resistance to this antibiotic combination. This was further confirmed by plotting resistance to CTX/AMOX by day of life for Gram-negative agents (data not shown), as a similar pattern of resistance was seen to that for all organisms. It was not possible to identify which Gram-negative species caused these changes as the total number of isolates resistant to CTX/AMOX in Gram-negative bacteria was small (<10 per day). However, E. coli and Enterobacteriacae account for over 75% of Gram-negative bacteria captured by this study and thus make a substantial contribution to Gram-negative resistance.
Our results indicate important age-associated changes in resistance to combinations of antibiotics recommended for empirical therapy of neonatal sepsis, which appear to be driven by CoNS (for resistance to PEN/GENT and FLU/GENT), and by Gram-negative bacteria for CTX/AMOX. The findings of this study are compatible with both the BNFC and NICE definitions of EOS (thresholds of 48 h and 72 h, respectively) and support empirical treatment with PEN/GENT or FLU/GENT over CTX/AMOX in neonates of all ages. Because of the dominance of GBS on days 0 and 1 of life the preferred combination for this period would therefore be PEN/GENT; after this period S. aureus becomes the dominant Gram-positive pathogen and hence FLU/GENT is preferred. Where CoNS are believed to be true pathogens then vancomycin remains the optimal antibiotic. It is important to note that the data here refer to BSI, the presence of meningitis may indicate a different spectrum of pathogens and a need for antibiotics with optimal cerebrospinal fluid penetration; CTX/AMOX is the standard of care for neonatal meningitis. Additionally, this analysis is based on national data, in individual units local resistance data (i.e. for LOS) may vary significantly from this such that a different empirical antibiotic policy may apply. We emphasize the need for robust microbiological surveillance data on antimicrobial susceptibility to agents used for empirical therapy and the potential benefit of enhancing existing datasets with clinical information.
ACKNOWLEDGEMENTS
We thank Alan Johnson for useful discussions on antimicrobial resistance and empirical antibiotic treatment.
DECLARATION OF INTEREST
Paul Heath was part of the NICE Guideline Development Group for the NICE clinical guideline on Antibiotics for early-onset neonatal infection: ‘Antibiotics for the prevention and treatment of early-onset neonatal infection’.
Footnotes
AIC = –2 × lnL + 2 × k, where lnL is the maximized log-likelihood of the model and k is the number of parameters estimated.
REFERENCES
- 1.Bryce J, et al. WHO estimates of the causes of death in children. Lancet 2005; 365: 1147–1152. [DOI] [PubMed] [Google Scholar]
- 2.Adams-Chapman I, Stoll BJ. Neonatal infection and long-term neurodevelopmental outcome in the preterm infant. Current Opinion in Infectious Diseases 2006; 19: 290–297. [DOI] [PubMed] [Google Scholar]
- 3.Leibovici L, Paul M. Benefit associated with appropriate antibiotic treatment. Clinical Infectious Diseases 2007; 45: 1400–1402. [DOI] [PubMed] [Google Scholar]
- 4.Muller-Pebody B, et al. Empirical treatment of neonatal sepsis: are the current guidelines adequate? Archives of Disease in Childhood – Fetal and Neonatal Edition 2011; 96: F4–8. [DOI] [PubMed] [Google Scholar]
- 5.Newton O, English M. Young infant sepsis: aetiology, antibiotic susceptibility and clinical signs. Transactions of the Royal Society of Tropical Medicine and Hygiene 2007; 101: 959–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paediatric Formulary Committee. BNF for Children 2011–2012 (British National Formulary for Children). London: British National Formulary Publications, 2011. [Google Scholar]
- 7.Christiansen H, Leth H. Breast milk as a cause of group B streptococcal sepsis. Ugeskrift for Laeger 2011; 173: 129–130. [PubMed] [Google Scholar]
- 8.Guerti K, et al. Time to positivity of neonatal blood cultures: fast and furious? Journal of Medical Microbiology 2011; 60: 446–453. [DOI] [PubMed] [Google Scholar]
- 9.van den Hoogen A, et al. Long-term trends in the epidemiology of neonatal sepsis and antibiotic susceptibility of causative agents. Neonatology 2010; 97: 22–28. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Prevention of perinatal group B streptococcal disease: a public health perspective. Centers for Disease Control and Prevention. Morbidity and Mortality Weekly Report. Recommendations and Reports 1996; 45: 1–24. [PubMed] [Google Scholar]
- 11.National Collaborating Centre for Women's and Children's Health. Antibiotics for Early-onset Neonatal Infection: Antibiotics for the Prevention and Treatment of Early-onset Neonatal Infection. London: RCOG Press at the Royal College of Obstetricians and Gynaecologists, 2012. [PubMed] [Google Scholar]
- 12.Vergnano S, et al. Neonatal infections in England: the NeonIN surveillance network. Archives of Disease in Childhood 2011; 96: F9–F14. [DOI] [PubMed] [Google Scholar]
- 13.Parry GJ, Tucker JS, Tarnow-Mordi WO. Relationship between probable nosocomial bacteraemia and organisational and structural factors in UK neonatal intensive care units. Quality & Safety in Health Care 2005; 14: 264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoll BJ, et al. Late-onset sepsis in very low birth weight neonates: a report from the National Institute of Child Health and Human Development Neonatal Research Network. Journal of Pediatrics 1996; 129: 63–71. [DOI] [PubMed] [Google Scholar]
- 15.Isaacs D. A ten year, multicentre study of coagulase negative staphylococcal infections in Australasian neonatal units. Archives of Disease in Childhood 2003; 88: F89–F93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leighton PH. Monitoring blood stream infection in neonatal intensive care units (dissertation). University College London, 2011, 298 pp. [Google Scholar]
- 17.Health Protection Agency (HPA). Voluntary reporting of Staphylococcus aureus bacteraemia in England, Wales and Northern Ireland, 2011 (http://www.hpa.org.uk/webw/HPAweb&Page&HPAwebAutoListName/Page/1191942169769). Accessed 12 December 2012.
- 18.Blackburn RM, et al. Neonatal sepsis – many blood samples, few positive cultures: implications for improving antibiotic prescribing. Archives of Disease in Childhood – Fetal and Neonatal Edition 2012; 97: 487–488. [DOI] [PubMed] [Google Scholar]
- 19.Sohn AH, et al. Prevalence of nosocomial infections in neonatal intensive care unit patients: results from the first national point-prevalence survey. Journal of Pediatrics 2001; 139: 821–827. [DOI] [PubMed] [Google Scholar]
- 20.Russell AB, Sharland M, Heath PT. Improving antibiotic prescribing in neonatal units: time to act. Archives of Disease in Childhood – Fetal and Neonatal Edition 2012; 97; F141–F146. [DOI] [PubMed] [Google Scholar]