Abstract
We measured the abundance and biovolume of bacteria in intertidal sediments from Tokyo Bay, Japan, by using a dual-staining technique (4′,6-diamidino-2-phenylindole and acridine orange) and several dispersion techniques (ultrasonic cleaner, ultrasonic sonicator, and tissue homogenizer). Dual staining reduced serious background fluorescence, particularly when used for silt-, clay-, and detritus-rich sediments, and allowed us to distinguish bacteria from other objects during both counting and sizing. Within the studied samples, the number of bacterial cells ranged from 0.20 × 109 to 3.54 × 109 g of wet sediment−1. With the cleaner and sonicator treatments, the bacterial numbers for all of the sites initially increased with dispersion time and then became constant. For the homogenizer treatments, the highest bacterial numbers were observed with the shortest (0.5- to 2-min) treatments, and the counts then declined steeply as the homogenization time increased, indicating that cell destruction occurred. The cleaner treatment had the possibility of insufficient dispersion of bacteria for fine-grain sediments. Within the studied samples, the bacterial biovolume ranged from 0.07 to 0.22 μm3. With the cleaner and sonicator treatments, the biovolume peaked during the shorter dispersion time. This pattern was caused not by cell destruction but by the incremental portion of dispersed small cells. We concluded that with the cleaner and sonicator treatments, the longer dispersion time reflected the real size spectrum and was preferable for accurate estimation of mean bacterial biovolumes.
The importance of bacteria in marine and estuarine sediments as a food source and major contributor to biogeochemical processes in benthic ecosystems has been widely recognized (1, 4, 13, 16). The quantification of bacterial roles requires precise measurements of their parameters. A standard procedure used to determine bacterial abundance and biovolume is the microscopic examination of fluorescently stained cells with either 4′,6-diamidino-2-phenylindole (DAPI) or acridine orange (AO). Most benthic bacteria are attached to sediment particles with extracellular polymeric substances (EPS), in contrast to free-living bacteria in water columns. Thus, a direct measurement of the abundance and biovolume of benthic bacteria by epifluorescence microscopy is possible only when bacteria can be detached or segregated from aggregates which include mineral particles and detritus.
Factors affecting the accuracy of the microscopic examination have been reported for the sample dilution and staining procedure (18) and for the efficiency of the bacterial dispersion, including the specification of equipment, treatment time, and dispersing intensity (9). DAPI specifically binds with nucleic acids and emits a brilliant blue light under UV excitation, enabling bacteria to be segregated more easily than with AO, which dyes the protein. In the case of low sample dilution, however, the problems of background fluorescence still remain even with DAPI staining. Several instruments are available to disperse bacterial cells from aggregates. Ultrasonic cleaners and ultrasonicators (9, 18, 21) disperse bacteria by the vibration of individual particles, while tissue homogenizers (1, 7, 12) mechanically break sediments into smaller particles. The dispersing time as well as the dispersing intensity strongly affects cell counts (9) and size distribution. Longer and more intense treatments tend to decrease the aggregate masking effect. This leads to an increase in the bacterial cell counts; however, with the longer and more intense dispersion, the tendency for cell destruction is higher. Moreover, the efficiency of the bacterial dispersion is affected by sediment characteristics, including viscosity and grain size distribution (7).
In this paper, we report a new dual-staining technique using both DAPI and AO for estimating the abundance and biovolume of benthic bacteria. We also explain the effect of dispersion procedures and sediment characteristics on bacterial enumeration and sizing. Intertidal sediments from Tokyo Bay, Japan, were used in the present study.
MATERIALS AND METHODS
Sampling.
Samples were obtained in May 1998 from three sites on the coast of Tokyo Bay, Japan: a sandy beach (35°10.6′N, 139°39.5′E), an intertidal sand flat (35°24.2′N, 139°54.2′E), and a mud flat (35°8.5′N, 139°39.9′E). Core samples were taken to a depth of 5 cm with acrylic core tubes (8.6-cm internal diameter). Each sample was thoroughly mixed and immediately brought back to the laboratory. Sediments for the dispersion procedures were obtained by subsampling from these samples. Subsamples (0.3 g) were mixed with 5 ml of filter-sterilized seawater (particle-free water) in 10 ml of acid-washed polycarbonate tubes and stored at 4°C. The particle-free water was obtained by two filtrations of 10% formalin-seawater solution buffered with sodium tetraborate (final concentration, 3.5 g liter−1) (1) by using Millipore filters (0.22-μm pore size).
Dispersion procedures and bacterial counting.
Prior to dispersion, the samples were incubated for at least 15 min with Tween 80 (final concentration, 1 mg liter−1). This surfactant facilitates even bacterial distribution on the membrane filter (9). Three different devices were used for the bacterial dispersion from the sediments: an ultrasonic cleaner (B-2200; Branson) (60-W output), an ultrasonicator (GE-100; Biomic) (100-W output) equipped with a 3-mm tapered microtip and with the amplitude set at 40% of the maximum, and a homogenizer (PT-2000; Kinematica) set at 20,000 rpm. The dispersion time for samples in the tubes was 5 to 60 min for the cleaner and 0.5 to 8 min for the others. To prevent possible denaturation of nucleic acids caused by overheating, the tubes were placed in ice water during the dispersion treatments (9).
After dispersion, the samples were diluted 50 to 250 times (final dilution, 830 to 4,160) with particle-free seawater. Diluted samples were dual stained with a combination of DAPI to a final concentration of 5 μg ml−1 (18) and AO to a final concentration of 1 mg ml−1 for counterstaining. After more than 30 min of staining, 0.5 to 2 ml of the samples was filtered through polycarbonate black filters (0.2-μm pore size) and then rinsed with particle-free seawater. The filters were immersed in nonfluorescent oil on microscope slides and covered with coverslips. Bacteria retained on the filters were examined within 24 h after dispersion under an Olympus BX-FLA-3 epifluorescence microscope (UV excitation) equipped with a 100× oil immersion objective. On each filter, no fewer than 200 clear-edged cells in 20 microscopic fields were counted.
Measurement of cell volumes.
After dispersion, the samples were centrifuged (100 × g) for 5 min in order to exclude nonbacterial particles as much as possible. Supernatants were dual stained and filtered as described above. A camera (TM-10AK; Olympus) mounted on the microscope was used to take microphotographs on color slide films. Images on the films were scanned with a film scanner (QuickScan 35; Minolta) connected to a computer and digitized, and thresholds were determined by using image analysis software (Image 1.59; NIH). Thresholds were manually adjusted by comparing original color images in each image. Bacterial cell-shaped objects were subsequently segregated from other objects, such as detritus particles and artifacts. Objects having an area of less than 6 pixels were automatically excluded as noise. Dividing cells, which have two well-defined local intensity maxima, were also removed. The pixel size for the resulting image was 0.17 by 0.17 μm. The project area of an object (A), cell length, and cell width (w) were automatically measured by the image analysis software. To compute the cell volume (V), we considered the rod-shaped cells to be cylinders with a hemispherical cap based on the microscopic observations: V = πwA/4 +πw3/6 − π2w3/16. At least 500 bacterial cell-shaped images per sample were analyzed.
Sediment characteristics.
At each study site, water content, sediment granulometry, and EPS were measured for triplicate samples. The water content was determined by the weight loss when wet sediments were dried at 90°C for 24 h. The grain size was measured by sieving the sediment, and the silt and clay components were determined with a Coulter Multisizer. EPS was examined with the phenol-sulfuric acid assay described by Underwood et al. (20) as a parameter of viscosity between the bacterial cells and other objects. The amount of EPS was expressed as micrograms of C per gram (dry weight) of sediment, with glucose as a standard.
Statistical analysis.
Statistical differences in bacterial counts and sizes among dispersing times in each disperser in each sediment were tested by using a one-way analysis of variance. Each analysis of variance was followed by a Student-Newman-Keuls (SNK) multiple-comparison test of means. Data sets were tested for homogeneity of variances (Levene test), and the log-transformed values were used if needed for a normal distribution.
RESULTS
Characteristics of sediments.
The characteristics of the three intertidal sediments are summarized in Table 1. There was a consistent relationship between sediment granulometry and other sediment characteristics. The mud flat sediment exhibited high water and silt contents, with a mean grain size of 92 μm. The mean grain size in the sand flat was almost the same as that in the sand beach (170 μm); however, the proportion of silt was six times higher in the former. The concentration of EPS was higher in the fine sediment.
TABLE 1.
Characteristics of sand beach, sand flat, and mud flat sediments
| Site | Wet density (g cm−3) (mean ± SE) | Dry density (g cm−3) (mean ± SE) | Water content (%) (mean ± SE) | Porosity (%) (mean ± SE) | Grain size distribution
|
EPSa (μg g [dry wt]−1) (mean ± SE) | |||
|---|---|---|---|---|---|---|---|---|---|
| Mean (μm) | Mud (%) | Silt (%) | Sand (%) | ||||||
| Sand beach | 1.75 ± 0.04 | 1.39 ± 0.03 | 20.7 ± 0.2 | 47.7 ± 1.1 | 170 | 0.0 | 0.4 | 99.6 | 17.6 ± 2.1 |
| Sand flat | 1.85 ± 0.01 | 1.38 ± 0.02 | 25.3 ± 0.4 | 48.1 ± 0.7 | 170 | 0.1 | 2.4 | 97.5 | 49.1 ± 0.8 |
| Mud flat | 1.55 ± 0.01 | 0.85 ± 0.01 | 45.3 ± 0.4 | 68.2 ± 0.3 | 92 | 4.6 | 41.2 | 54.2 | 354.3 ± 87.2 |
Measured with glucose as a standard.
Bacterial counts.
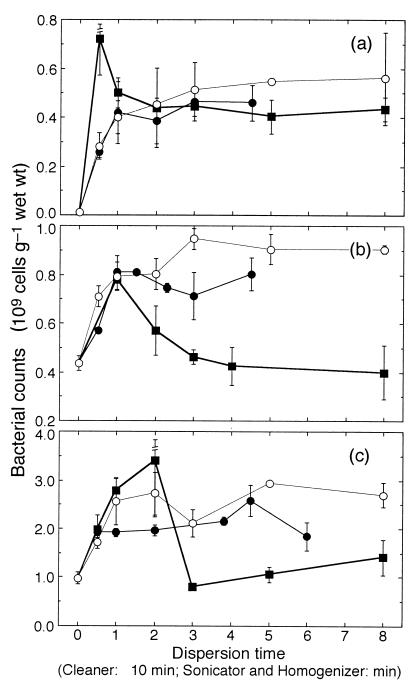
Figure 1 shows the number of dispersed bacteria versus dispersion time for the cleaner, sonicator, and homogenizer techniques for sediments from each site. With both the cleaner and sonicator treatments, the bacterial numbers at all of the sites initially increased with treatment time and then leveled off, resulting in the highest number of bacteria at 15 to 45 min with the cleaner and at 3 to 8 min with the sonicator. However, for the homogenizer treatments, the pattern was totally different from that for the cleaner and sonicator treatments at all of the sites. The highest bacterial numbers were observed for the shortest (0.5- to 2-min) treatments. The bacterial counts then declined steeply as the homogenization time increased, especially in the sand flat and mud flat sediments, where the counts were less than those observed for nondispersed sediments.
FIG. 1.
Comparisons of the numbers of dispersed bacteria with cleaner (●), sonicator (○), and homogenizer (■) treatments in sandy beach (a), sand flat (b), and mud flat (c) sediments. Bars indicate standard errors (n = 3).
We used the SNK test to examine a statistically homogenous subset including the maximum mean bacterial count in each case of the dispersing time series. Table 2 summarizes the average bacterial count in each homogenous subset for each sediment and disperser treatment. The average value was considered the maximum value in each case. For both the cleaner and sonicator treatments, the SNK test revealed that the bacterial counts were homogenous (P > 0.05) among the dispersion times except for several shorter dispersion times. For the sand beach sediment, no statistical differences in the maximum numbers were found among the three dispersion techniques. In the sand flat sediment, the maximum number for the cleaner treatment was significantly lower than that for the sonicator treatment (0.01 < P < 0.05). In the mud flat sediment, the maximum number for the cleaner treatment was significantly lower than those for the sonicator and homogenizer treatments (0.01 < P < 0.05).
TABLE 2.
Maximum numbers of dispersed bacteria at three sites
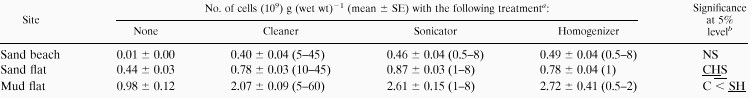 |
The maximum number was calculated by taking the average for a homogenous subset including the maximum mean in the dispersion time series. Values in parentheses indicate the treatment times (minutes) for the homogenous subset.
NS, not significant. C, S, and H, cleaner, sonicator, and homogenizer treatment, respectively. Underlining indicates a statistically homogenous group.
The more dispersed bacteria were observed in the finer-grain sediments. The maximum number for the sonicator treatment ranged from 0.20 × 109 to 0.94 × 109 cells g−1 in the sand beach, 0.69 × 109 to 1.02 × 109 cells g−1 in the sand flat, and 1.67 × 109 to 3.54 × 109 cells g−1 in the mud flat on a wet-sediment basis (Table 2), which corresponded to 0.25 × 109 to 1.18 × 109 cells g−1, 0.93 × 109 to 1.37 × 109 cells g−1, and 3.05 × 109 to 6.46 × 109 cells g−1 on a dry-sediment basis, respectively. The bacterial numbers in the sand flat and mud flat sediments were approximately two- and fivefold higher, respectively, than those in the sand beach sediment. This tendency was more distinct in the nondispersed samples, for which the counts in the sand and mud flats were more than 1 order of magnitude higher than those in the sand beach sediment.
Bacterial cell volume.
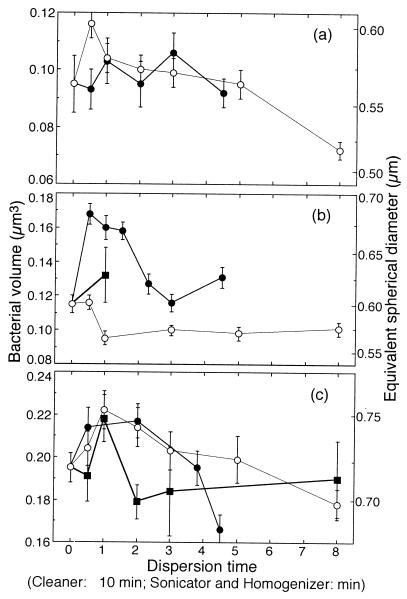
Figure 2 shows the mean bacterial volume of dispersed bacteria versus dispersion time for sediments from each site. In all of the treatments, the biovolumes peaked at shorter times. The biovolumes then dropped as the dispersion time increased.
FIG. 2.
Comparisons of dispersed bacterial volumes with cleaner (●), sonicator (○), and homogenizer (■) treatments in sandy beach (a), sand flat (b), and mud flat (c) sediments. Bars indicate standard errors.
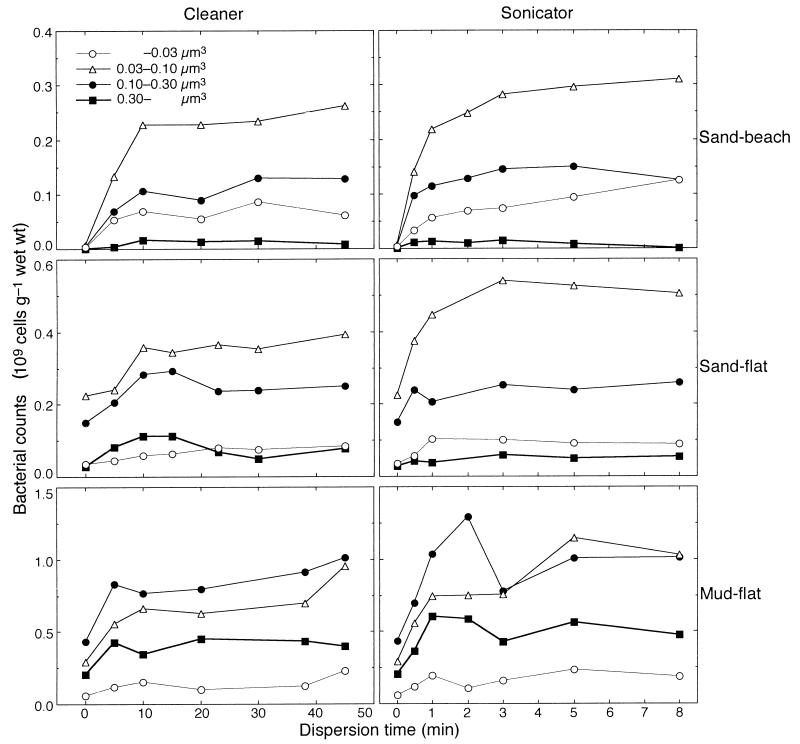
To investigate these fluctuation patterns more thoroughly, we size fractionated the numbers of dispersed bacteria for the cleaner and sonicator treatments. The histogram for each data set showed a Guassian-shaped profile (not shown). When the counted bacterial cells were fractionated into four size classes (Fig. 3), the bacterial cells in the smaller fractions (<0.10 μm3) required a longer dispersion time to become constant than those in the larger fractions (>0.10 μm3) in each data set. No apparent decline was found for either the cleaner or sonicator treatment. These results indicate that the decrease in the mean cell volume with longer dispersion times (Fig. 2) was caused not by cell destruction but by the increased proportion of smaller cells. Consequently, it was concluded that the longer treatment time reflected the real size spectrum and was preferable for the accurate estimation of mean bacterial biovolumes. For the homogenizer treatment, in which only the mud flat sediment was fractionated, all of the bacterial cell fractions dropped steeply after 2 min (not shown). The fraction of 0.03 to 0.1 μm3 dominated in both the sand beach and sand flat sediments, while the fraction of 0.1 to 0.3 μm3 dominated in the mud flat. In the sand flat sediment, the cell counts in the 0.03- to 0.1-μm3 fraction were lower with the cleaner treatment than with the sonicator treatment, which caused the statistical difference in the mean volume between them. In the other sediments, the cell counts of the entire fraction exhibited nearly similar values.
FIG. 3.
Comparison of the numbers of size-fractionated bacteria with cleaner and sonicator treatments at three sites.
The mean bacterial biovolume, equivalent spherical diameter, and cell length/width ratio for the three sediment types and dispersion treatments are summarized in Table 3. The range of bacterial biovolumes was consistent with those previously reported (12, 14, 16, 17). The largest bacterial biovolumes were observed in the finer-grain sediments. Biovolumes ranged from 0.07 to 0.10 μm3 (0.49- to 0.52-μm equivalent spherical diameter) in the sand beach, from 0.10 to 0.13 μm3 (0.53 to 0.56 μm) in the sand flat, and from 0.17 to 0.22 μm3 (0.62 to 0.66 μm) in the mud flat. This tendency was also true for the nondispersed samples. The cell length/width ratios, which ranged from 1.6 to 2.1, were not significantly different among dispersion techniques or sediment types (P > 0.05).
TABLE 3.
Biovolume, equivalent spherical diameter, and cell length/width ratio of dispersed bacteria at three sites
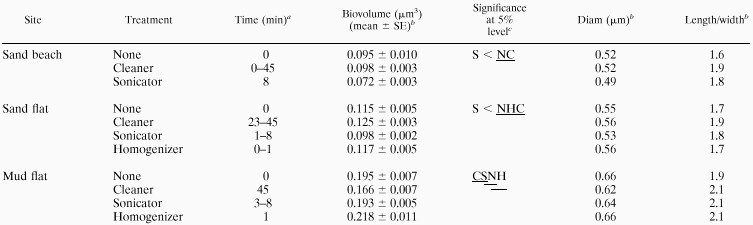 |
Treatment time for a homogenous subset.
Calculated by taking the average for the homogenous subset.
N, C, S, and H, no treatment and cleaner, sonicator, and homogenizer treatments, respectively. Underlining indicates a statistically homogenous group.
DISCUSSION
Dual staining with DAPI and AO.
The dual-staining technique utilized in this study can contribute to a better understanding of the bacterial abundance and size distribution in sediments. Until now, either AO or DAPI alone has been used as a dye for benthic bacteria by other workers. If only AO is used for bacterial staining, all of the aggregate components are stained in similar colors, resulting in the contrast between the bacteria and nonbacterial substances being too low to distinguish between them. Compared with AO staining, DAPI staining provides higher image contrast and is more specific for bacterial staining (12). However, the problems of background fluorescence also occurred when staining with DAPI. In clay- and silt-rich sediments, which are rich in detritus and EPS, the background fluorescence by aggregates containing detritus and minerals as well as bacteria is more intense. When AO and DAPI are used together, under UV light only the bacteria are vividly seen as blue, and the nonbacterial substances are orange. For a counterstaining dye, Epstein and Rossel (9) proposed the use of Evans blue. However, based on our observations, AO is superior to Evans blue in providing contrast to bacteria illuminated blue by DAPI. When the sample is not diluted enough and aggregates accumulate on the filter at a depth greater than that for proper focus, only part of the cells are in constant focus. In this context, some bacteria will be masked by both the nonbacterial substances and background, leading to an underestimation (18). If this occurs, dual staining can reduce the masking effect, as image contrast is improved and background is reduced compared to those with single-staining techniques. Consequently, dual staining simultaneously overcomes the problems of low contrast in the AO technique and background in the DAPI technique.
Moreover, dual staining is also useful for the measurement of cell images. High-color-contrast images allow us to segregate bacteria more easily from nonbacterial substances, which are incorrectly recognized by a computer. Recent progress in the measurement of bacterial size has included automatic threshold determination systems (2, 3), which are faster than the manual threshold determination method used here. The high contrast is also helpful for the problem of dim cells, which have weak fluorescence and are difficult (or almost impossible with the automatic threshold determination system) to distinguish from the background by using only AO or DAPI.
Effect of dispersion and sediment characteristics.
As already mentioned, although the bacterial cell counts increase when the masking effect due to aggregates is decreased by making the treatment time longer, the longer dispersion time can cause cell destruction. This cell destruction can result in an unclear cell edge due to the leaking of protoplasm and lead to the inaccurate measurement of biovolume as well as abundance. In the present study, cell destruction, which was indicated by a decline in the bacterial numbers, was observed with the homogenization treatment but not with the cleaner or sonicator treatment (Fig. 1). The homogenizer mechanically breaks down sediment particles into finer ones, resulting in an increased background. Hence, the counting and sizing for a homogenized sample are relatively time-consuming and somewhat subjective compared to those for the other two treatments. For sonication treatment, no cell destruction has been reported (5, 9, 10). Epstein et al. (10) showed no cell destruction in the samples with optimal dispersing time based on the thorough examination of labeled bacteria with radioisotopes. For cleaner treatment, Ellery and Schleyer (8) reported that cell destruction occurred with a 100- to 200-W output cleaner but not with our 60-W output cleaner. The main reason for this discrepancy is probably the difference in the specifications or intensities of the cleaners. Therefore, there is possible cell destruction in the case of too-long or too-intense dispersion with the sonicator as well as with the cleaner.
For the beach sand sediment, the maximum number of dispersed bacteria was not significantly different among the three dispersion techniques (Table 2). However, for the sand flat and mud flat sediments, the cleaner was inferior to the other dispersers (Table 2). This indicates that for the silt- and clay-rich sediments, where the viscosity (EPS) was also higher (Table 1), the cleaner did not efficiently separate the cells, at least with our low-output equipment. Nonetheless, differences in the maximum yield of bacteria among the dispersion techniques were not as high as those previously reported (9).
For the homogenizer, bacterial counts peaked for the short dispersion time in all of the sediments, while for the cleaner and sonicator, the counts increased as the dispersion time increased and then leveled off (Table 2). The pattern of cell counts during homogenization was similar to those previously reported (7, 9) and somewhat different from the observations of Ellery and Schleyer (8) and Montagna (15), which were similar to those for the cleaner and sonicator. These differences were presumably due to the advantage of dual staining. Direct observation of samples after 0.5 to 2 min of homogenization revealed that there were significant numbers of bacteria in the aggregates. This bacterial retention in the aggregates was also observed by Ellery and Schleyer (8).
The more dispersed bacteria were observed in the finer-grain sediments. This tendency has been found by many workers in relation to sediment grain surface area (5, 19), protected habitat (22), organic content (6), grazer regulation (11), and porosity (19) and need not be discussed further.
In summary, dual staining has advantages over conventional staining techniques, especially for silt-, clay-, and detritus-rich sediments, by reducing serious background fluorescence. With dual staining, bacteria stand out from other objects and can be more easily counted and sized. Cleaner and sonicator treatments are recommended for dispersing bacteria from aggregates, while homogenization treatment has the possibility of cell destruction in the case of dispersion treatments that are too long. The cleaner treatment, however, has the possibility of insufficient dispersion for silt- and clay-rich sediments. Small bacteria (<0.10 μm3) require a longer dispersion time to become constant than large bacteria (>0.10 μm3). It is concluded that studies of bacterial sizing need a sufficient treatment time to disperse small bacteria and to obtain the real size distribution.
ACKNOWLEDGMENTS
The work was supported in part by a grant from the Environment Agency of Japan.
We thank E. Miyoshi, E. Kibe, and Y. Hagimoto for their kind assistance with the physical and chemical analysis; K. Furukawa and M. A. Elzeir for valuable comments, and M. T. Waters for correcting the English text and helpful comments on the manuscript.
REFERENCES
- 1.Alongi D M. Bacterial productivity and microbial biomass in tropical mangrove sediments. Microb Ecol. 1988;15:59–79. doi: 10.1007/BF02012952. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn N, Hagström Å, Wikner J, Cuadros-Hansson R, Bjørnsen P K. Rapid determination of bacterial abundance, biovolume, morphology, and growth by neural network-based image analysis. Appl Environ Microbiol. 1998;64:3246–3255. doi: 10.1128/aem.64.9.3246-3255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloem J, Veninga M, Shepherd J. Fully automatic determination of soil bacterium numbers, cell volumes, and frequencies of dividing cells by confocal laser scanning microscopy and image analysis. Appl Environ Microbiol. 1995;61:926–936. doi: 10.1128/aem.61.3.926-936.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowan J L W, Pennock J R, Boynton W R. Seasonal and interannual patterns of sediment-water nutrient and oxygen fluxes in Mobile Bay, Alabama (USA): regulating factors and ecological significance. Mar Ecol Prog Ser. 1996;141:229–245. [Google Scholar]
- 5.Deflaun M F, Mayer L M. Relationships between bacteria and grain surfaces in intertidal sediments. Limnol Oceanogr. 1983;28:873–881. [Google Scholar]
- 6.Deming J W, Baross J A. The early diagenesis of organic matter: bacterial activity. In: Engel M H, Macko S A, editors. Organic geochemistry. New York, N.Y: Plenum Publishing Corp.; 1993. pp. 119–144. [Google Scholar]
- 7.Dye A H. A method for the quantitative estimation of bacteria from mangrove sediments. Estuar Coast Shelf Sci. 1983;17:207–212. [Google Scholar]
- 8.Ellery W N, Schleyer M H. Comparison of homogenization and ultrasonication as techniques in extracting attached sedimentary bacteria. Mar Ecol Prog Ser. 1984;15:247–250. [Google Scholar]
- 9.Epstein S S, Rossel J. Enumeration of sandy sediment bacteria: search for optimal protocol. Mar Ecol Prog Ser. 1995;117:289–298. [Google Scholar]
- 10.Epstein S S, Alexander D, Cosman K, Dompé A, Gallagher S, Jarsobski J, Laning E, Martinez R, Panasik G, Peluso C, Runde R, Timmer E. Enumeration of sandy sediment bacteria: are the counts quantitative or relative? Mar Ecol Prog Ser. 1997;151:11–16. [Google Scholar]
- 11.Fenchel T. Suspended marine bacteria as a food source. In: Fasham M J R, editor. Flows of energy and materials in marine ecosystems. New York, N.Y: Plenum Publishing Corp.; 1984. pp. 301–305. [Google Scholar]
- 12.Imai I. Size distribution, number and biomass of bacteria in intertidal sediments and seawater of Ohmi Bay, Japan. Bull Jpn Soc Microb Ecol. 1987;2:1–11. [Google Scholar]
- 13.Kuwae T, Hosokawa Y, Eguchi N. Dissolved inorganic nitrogen cycling in Banzu intertidal sand-flat, Japan. Mangr Salt Marsh. 1998;2:167–175. [Google Scholar]
- 14.Meyer-Reil L A. Benthic response to sedimentation events during autumn to spring at a shallow water station in the Western Kiel Bight. Mar Biol. 1983;77:247–256. [Google Scholar]
- 15.Montagna P A. Sampling design and enumeration statistics for bacteria extracted from marine sediments. Appl Environ Microbiol. 1982;43:1366–1372. doi: 10.1128/aem.43.6.1366-1372.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montagna P A. In situ measurement of meiobenthic grazing rates on sediment bacteria and edaphic diatoms. Mar Ecol Prog Ser. 1984;18:119–130. [Google Scholar]
- 17.Rublee P A. Seasonal distribution of bacteria in salt marsh sediments in North Carolina. Estuar Coast Shelf Sci. 1982;15:67–74. [Google Scholar]
- 18.Schallenberg M, Kalff J, Rasmussen J B. Solutions to problems in enumerating sediment bacteria by direct counts. Appl Environ Microbiol. 1989;55:1214–1219. doi: 10.1128/aem.55.5.1214-1219.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt J L, Deming J W, Jumars P A, Keil R G. Constancy of bacterial abundance in surficial marine sediments. Limnol Oceanogr. 1998;43:976–982. [Google Scholar]
- 20.Underwood G J C, Paterson D M, Parkers R J. The measurement of microbial carbohydrate exopolymers from intertidal sediments. Limnol Oceanogr. 1995;40:1243–1253. [Google Scholar]
- 21.Venji M I, Albright L J. Microscopic enumeration of attached marine bacteria of seawater, marine sediment, fecal matter, and kelp blade samples following pyrophosphate and ultrasound treatments. Can J Microbiol. 1986;32:121–126. [Google Scholar]
- 22.Weise W, Rheinheimer G. Scanning electron microscopy and epifluorescence investigation of bacterial colonization of marine sand sediments. Microb Ecol. 1978;4:175–188. doi: 10.1007/BF02015075. [DOI] [PubMed] [Google Scholar]