SUMMARY
We describe a foodborne outbreak in Italy caused by enteroinvasive Escherichia coli (EIEC), an enteric pathogen uncommon in industrialized countries. On 14 April 2012 a number of employees of the city of Milan Fire Brigade (FB) were admitted to hospital with severe diarrhoea after attending their canteen. Thirty-two patients were hospitalized and a total of 109 cases were identified. A case-control study conducted on 83 cases and 32 controls attending the canteen without having symptoms identified cooked vegetables to be significantly associated with the disease. Stool samples collected from 62 subjects were screened for enteric pathogens using PCR-based commercial kits: 17 cases and two asymptomatic kitchen-workers were positive for the Shigella marker gene ipaH; an ipaH-positive EIEC strain O96:H19 was isolated from six cases. EIEC may cause serious dysentery-like outbreaks even in Western European countries. Microbiologists should be aware of microbiological procedures to detect EIEC, to be applied especially when no common enteric pathogens are identified.
Key words: Enteroinvasive Escherichia coli (EIEC), Escherichia coli, foodborne infections, Italy, outbreaks
INTRODUCTION
Enteroinvasive Escherichia coli (EIEC) are a group of diarrhoeagenic E. coli biochemically, genetically, and pathogenetically related to Shigella spp. [1–3]. Both pathogens have the ability to invade the colonic epithelium due to the presence of genes located on large invasive plasmids and on the bacterial chromosome [1–3].
The pathogenesis of EIEC infection comprises colonic epithelial cell penetration, lysis of the endocytic vacuole, intracellular multiplication and extension into adjacent epithelial cells [2–4]. This sequence of events elicit the characteristic dysentery syndrome, characterized by the presence of blood, mucus, and leukocytes in the stools [2, 5], even if EIEC infections may often result in watery diarrhoea [2].
EIEC are human pathogens occurring worldwide [2, 5–7]. They usually belong to specific E. coli O serogroups [3, 5, 6] and are considered, with few exceptions [7], as uncommon agents of diarrhoea [2, 6, 7]. EIEC are spread primarily by the faecal–oral route and infections have been mainly reported in countries where sanitation and socioeconomic status are poor [2, 6]. In industrialized countries, EIEC infections are typically sporadic and travel related [6, 8, 9]. Nonetheless, outbreaks characterized by mild clinical symptoms have been reported in the USA [10–13], Japan [14] and Israel [15]. In Europe, EIEC infections appear to be endemic in some eastern countries [6]. In Western Europe, to the best of our knowledge, EIEC have never been associated with outbreaks and also sporadic cases are rare [6, 9]. In Italy, EIEC infections have never been documented to date, although it is notable that diagnostic methods specifically for their detection have been included in only two studies on childhood diarrhoea conducted in two regions in the 1980s [16, 17].
In this paper we describe, for the first time, an outbreak of EIEC in Italy. The episode occurred in 2012 and involved employees of the city of Milan Fire Brigade (FB).
METHODS
Background
On 14 April 2012, the Local Public Health Service (LPHS) of Milan was notified of over 50 persons, all employees of the Milan FB, who had been admitted to the emergency unit of different city hospitals due to severe gastroenteric symptoms, including bloody diarrhoea.
Preliminary investigations revealed that patients had fallen ill between 13 and 14 April 2012, and that the only common exposure was the consumption of meals at the FB canteen before developing symptoms. Information on other possibly linked cases of gastroenteritis was collected from all the public hospitals and emergency units of the city, but no other cases could be found. The hypothesis of a foodborne outbreak linked to the canteen of the FB was formulated. The canteen was inspected on 15 April 2012 by the LPHS which reported gross failures in the hygiene conditions of the equipment, including refrigerators and freezers, and in the procedures of preparation and storage of food (e.g. the habit of serving food leftovers from a meal during subsequent days). The activity of the canteen was suspended and an investigation was undertaken to establish the magnitude of the outbreak and to identify its aetiological agent, with its source and vehicle.
Epidemiological investigation
The staff of Milan FB includes about 400 employees, some of whom live in the fire station. Most of them regularly consumed meals at the FB canteen, which served the FB exclusively. Neither the list, nor the exact number of people who had consumed meals before 15 April 2012 was available. On 15 April 2012 FB employees who had experienced any gastroenteric symptom from 9 April 2012 onwards were invited via a public announcement to contact the LPHS of Milan for an interview using a semi-structured questionnaire. Besides basic demographic elements, information was collected for each patient on the clinical course of illness (date and time of onset of illness, type of symptoms, whether or not he/she had been hospitalized), and which meals and food items he/she had consumed at the FB canteen between 9 and 14 April 2012. To assess possible secondary cases, patients were asked whether any episode of gastroenteritis occurred in their household after the start of their illness. The same epidemiological questionnaire was administered to the six kitchen staff members, regardless of their clinical status, and to 33 FB employees who had also eaten at the canteen in the same period without developing symptoms. These latter were enrolled as healthy controls in a case-control study undertaken to investigate the association between illness and the consumption of food items. A case was defined as a FB employee who had consumed at least one meal prepared in the FB canteen between 9 and 14 April 2012 and developed diarrhoea within the following 6 days.
Controls were defined using the same criteria, except for clinical symptoms, as it was necessary that no signs of gastroenteritis were reported within 6 days after consuming the last meal.
Microbiological investigation
Stool samples
All the case-patients and the kitchen staff members were requested to submit a stool sample. These were examined at the laboratories of the LPHS or of the admitting hospitals for the presence of enteric pathogens. The Seeplex® Diarrhoea B1 (Salmonella spp., Shigella spp., Vibrio spp., Campylobacter spp., Clostridium difficile) and B2 [Yersinia enterocolitica, Aeromonas spp., E. coli O157:H7, Shiga toxin-producing E. coli (STEC), and Clostridium perfringens] ACE detection panels (Seegene Diagnostics, South Korea) were used at LPHS level. The xTAG® Gastrointestinal Pathogen panel (xTAG GPP; Luminex Corp., USA), which includes (in addition to the pathogens listed for the previous panels) viruses (adenovirus 40/41, rotavirus A, norovirus GI/GII) and parasites (Giardia, Entamoeba histolytica, Cryptosporidium), was employed in the hospital laboratories. Samples positive for the presence of the ipaH gene, encoding one of the invasion plasmid antigen proteins [3], were plated onto SS agar for the isolation of Shigella spp.
Aliquots of the stool specimens were stored at −20 °C until transportation to the National Reference Laboratory (NRL) for E. coli for the detection of diarrhoeagenic E. coli. This was accomplished as described previously [18]: stools were streaked onto MacConkey agar plates and sweeps of both lactose-fermenting and non-fermenting colonies were tested by PCR amplification for the presence of genes encoding verocytotoxins (vtx1 and vtx2), intimin (eae), and the antiaggregation protein transporter gene (aat) of enteroggregative E. coli. The ipaH gene was amplified using the primer pair shig1/shig2, as described by Lüscher & Altwegg [19]. For the isolation and characterization of EIEC strains, single colonies were picked up from the ipaH PCR-positive colony sweeps and tested again by PCR. The ipaH-positive strains obtained were biochemically identified as E. coli and examined by a cell culture invasion assay [20] using HEp-2 cells instead of HeLa cells [17]. Serotyping was performed by F. Scheutz at the WHO International E. coli and Klebsiella Centre of the Statens Serum Institut, Copenhagen (Denmark). The antimicrobial susceptibility testing was performed by disk diffusion as described previously [21], using the following antibiotic disks: nalidixic acid, ampicillin, cefotaxime, ciprofloxacin, chloramphenicol, gentamicin, kanamycin, streptomycin, sulfonamide, tetracycline, trimethoprim-sulfamethoxazole. Subtyping of the isolated strain was performed by pulsed-field gel electrophoresis (PFGE) as described previously [22], using 100 U of XbaI (New England Biolabs Inc., USA) for overnight restriction. A strain of Salmonella enterica serotype Braenderup was used as standard molecular-weight marker.
Food, water and environmental samples
Leftovers of food from the kitchen, samples of drinking water and swabs of the kitchen's working surfaces were collected on 15 April 2012 by the LPHS, and examined at the LPHS laboratory for the presence of S. enterica, Shigella spp., E. coli O157 and Y. enterocolitica using the international standard methods. Enrichment cultures of the samples in buffered peptone water were also tested with Seeplex Diarrhoea ACE (panels B1 and B2) for the presence of ipaH-positive bacteria.
Statistical analysis
Data were analysed using Stata v. 12·1 (StataCorp, USA). Categorical variables are described using counts and percentages, whereas quantitative variables are reported with medians and ranges. Differences in proportions were assessed for statistical significance using the χ2 test or, when appropriate, Fisher's exact test. Differences in median values were tested with the Mann–Whitney U test. P values <0·05 were considered statistically significant.
The incubation period of the disease was calculated in hours, based on cases who had consumed one meal only and with information available for the time of symptom onset.
In the case-control study, only cases and controls remembering the food items consumed each day were included. Because not all cases attended the canteen on every day considered, and some fell ill during the period considered at risk, we decided to analyse the exposure to food items each day independently. Food-specific odds ratios (OR) and 95% confidence intervals (CI) were calculated for each item using exact logistic regression. This approach was chosen because some of the cells formed by the outcome (i.e. case-control) and predictor variables (i.e. food consumption) had no observations.
RESULTS
Epidemiological investigation
A total of 117 patients replied to the LPHS invitation and 109, including three kitchen staff members, met the case definition. The median age of cases was 41 years (range 25–60 years) and most (n = 93) were males. Dates of onset of symptoms ranged from 10 to 17 April 2012, with the peak occurring on 14 April 2012 (Fig. 1).
Fig. 1.
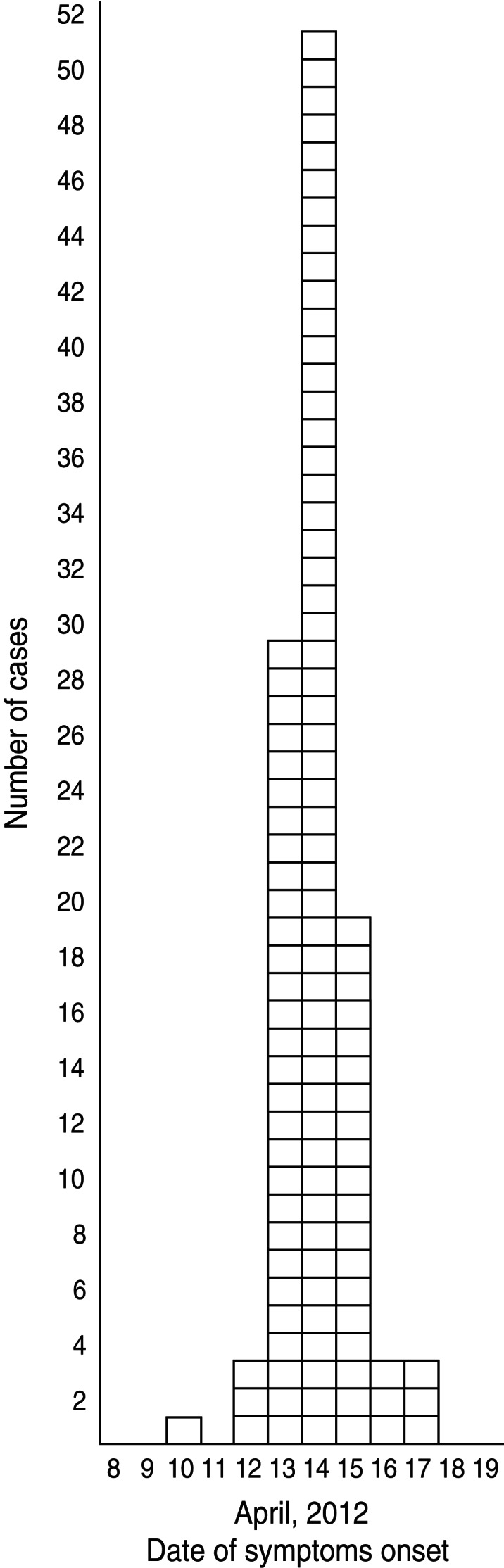
Distribution of cases (n = 109) by date of symptom onset during an outbreak of gastroenteritis, Milan, Italy, April 2012.
The estimate of the median incubation time was 21 h (range 10–37 h), based on the history of ten cases.
Besides diarrhoea (no distinction was drawn between watery and bloody diarrhoea), the symptoms most commonly reported were fever (n = 82, 75·2%), abdominal cramps (n = 66, 60·6%) and vomiting (n = 46, 41·8%). Some cases reported headache, fatigue and nausea (Table 1). Overall, 74 cases were examined at an emergency department, of which 32 were hospitalized, one with a diagnosis of septic shock syndrome (Table 1).
Table 1.
Clinical symptoms and healthcare-seeking in cases involved in an outbreak of gastroenteritis, Milan, Italy, April 2012 (n = 109)
| Symptoms | No. | % |
|---|---|---|
| Diarrhoea* | 109 | 100 |
| Fever | 82 | 75·2 |
| Abdominal cramp | 66 | 60·6 |
| Vomiting | 46 | 41·8 |
| Headache† | 6 | 5·5 |
| Nausea† | 4 | 3·7 |
| Malaise† | 3 | 2·8 |
| Emergency department visit | 74 | 67·9 |
| Hospitalized | 32 | 29·4 |
Includes both, watery and bloody diarrhoea.
Symptom non-systematically enquired.
Seven cases reported a household member experiencing gastroenteric symptoms after the start of their disease, making seven the number of possible secondary cases identified.
The median number of days of canteen attendance was two (range 1–5 days), according to the 108 cases who remembered on which days they attended the canteen before developing clinical symptoms. Eighty-three cases and 32 controls were capable of recalling the food items eaten and were included in the case-control study (Table 2).
Table 2.
Distribution of cases (n = 108) attending the canteen, number of cases (n = 83) and healthy controls (n = 32) with complete information on food consumption, and number of food items served in the canteen for each day in an outbreak of gastroenteritis, Milan, Italy, April 2012
| Day of canteen attendance | Number of cases consuming meals in the canteen (n = 108) | Fire Brigade employees included in the case-control study | Overall number of food items served in the Fire Brigade canteen | |
|---|---|---|---|---|
| Cases (n = 83) | Controls (n = 32) | |||
| 9 April | 15 | 7 | 6 | 12 |
| 10 April | 29 | 16 | 5 | 17 |
| 11 April | 35 | 22 | 9 | 20 |
| 12 April | 69 | 50 | 19 | 26 |
| 13 April | 79 | 59 | 23 | 26 |
| 14 April | 21 | 15 | 4 | 18 |
The results of the case-control study showed that, of the 119 food items served in the canteen, beet greens served on 12 April 2012 (OR 10·64, P = 0·017) and green beans and potatoes served on 13 April 2012 (OR 10·33, P = 0·008 and OR 8·77, P = 0·032, respectively) were those significantly associated with the illness. In addition, peppers and beet greens served on 13 April 2012 had OR values >4, which although not significant (P = 0·098 and P = 0·180, respectively), indicate stronger chances of illness among the exposed persons. Peppers and potatoes were also served on 14 April 2012, and retained an OR >2 (Table 3).
Table 3.
Food exposure: univariate odds ratio (OR) of being exposed to food items served at the canteen of Milan Fire Brigade from 9 to 14 April 2012. Only food items with an OR >2 for at least 1 day are reported
| Day of consumption | Food items | Exposed cases (total cases) | Exposed controls (total controls) | OR | 95% CI | P value |
|---|---|---|---|---|---|---|
| 12 April | Beet greens | 16 (48) | 0 (16) | 10·64 | 1·63–∞ | 0·017 |
| 13 April | Green beans | 19 (55) | 1 (21) | 10·33 | 1·42–460·11 | 0·008 |
| 13 April | Potatoes | 17 (55) | 1 (21) | 8·77 | 1·20–392·29 | 0·032 |
| 13 April | Peppers | 8 (55) | 0 (21) | 4·74 | 0·68–∞ | 0·098 |
| 13 April | Beet greens | 7 (55) | 0 (21) | 4·04 | 0·57–∞ | 0·180 |
| 14 April | Peppers | 5 (15) | 0 (4) | 2·25 | 0·23–∞ | 0·296 |
| 14 April | Potatoes | 4 (15) | 0 (4) | 1·63 | 0·16–∞ | 0·530 |
CI, Confidence interval.
None of these vegetables alone could explain more than the 22% of cases included in the analytical study. However, 57 (69·9%) of the 83 case-patients consumed at least one of the aforementioned vegetables, compared to one (3·1%) of the 32 controls.
Laboratory investigation
Stool samples were collected between 15 April and 4 June 2012 from 59 case-patients and three asymptomatic kitchen staff members.
All stool samples were negative for common enteric pathogens, but the molecular screening assay showed the presence of the ipaH gene in specimens from 17 cases and two asymptomatic kitchen employees, one of whom had recently travelled to South East Asia.
This gene is considered as a marker for the presence of Shigella spp., but no Shigella strains were isolated from the ipaH-positive samples.
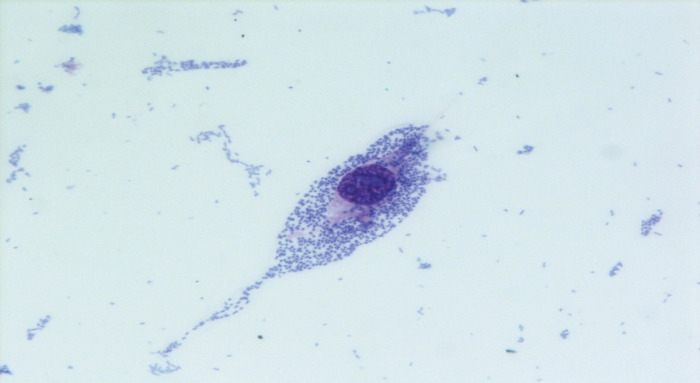
Fifteen ipaH-positive samples were sent to the NRL for E. coli detection. All were negative for the vtx1, vtx2, eae and aat genes, and were confirmed as ipaH-positive. ipaH-positive E. coli strains were isolated from six samples: all were motile, lactose-fermenting, lysine-positive, negative by indole test, and resistant to streptomycin and sulphonamides; they did not agglutinate with antisera against the typical EIEC serogroups and with Shigella antisera, gave a strong positive result when tested by the HEp-2 cell invasion assay (Fig. 2), and shared the same PFGE profile (not shown). A representative strain was identified as E. coli O96:H19 at the World Health Organization International E. coli and Klebsiella Centre.
Fig. 2.
[colour online]. Invasive pattern of the Escherichia coli O96:H19 epidemic strain in the HEp-2 cells invasion assay (400x, May–Grunwald Giemsa staining), outbreak of gastroenteritis, Milan, Italy, April 2012.
For the ipaH-positive cases, the interval between onset of symptoms and first collection of stool samples (median 13 days, range 6–18 days) was significantly shorter than that of ipaH-negative cases (median 19 days, range 20–30 days) (P = 0·0014). Seven ipaH-positive cases were repeatedly sampled. The median interval between onset of symptoms and the last positive assay for ipaH before two consecutive negative molecular screening tests was 13 days (range 1–46 days).
The two ipaH-positive asymptomatic kitchen employees were also repeatedly tested. The one returning from South East Asia was still positive 42 days after the first sampling and became negative only after 3 weeks of antimicrobial treatment.
All the food and water samples and the surface swabs collected in the kitchen were negative for ipaH and for the other enteropathogens.
DISCUSSION
This investigation illustrates some peculiarities with respect to the epidemiology of EIEC, including the occurrence of such a large outbreak in Italy, the severity of the infection, and the unusual serotype and biochemical profile of the EIEC outbreak strain.
EIEC infections are considered uncommon in industrialized countries [6, 8, 9]. To date in Italy EIEC have never been detected, although it should be noticed that studies investigating EIEC as possible cause of diarrhoea in humans are limited [1] The aetiological role of the EIEC O96:H19 strain in this outbreak is supported by the demonstration of evidence for EIEC infection, i.e. the isolation of the strain or a positive ipaH screening test in 29% of the subjects submitting a stool sample, together with the negative results for the other enteric pathogens in all the stool samples. The negative results obtained in 71% of the cases might be explained by the delay in collecting the stool samples, since the interval between the onset of symptoms and stool collection was significantly shorter for EIEC-positive samples.
The knowledge that EIEC are human pathogens transmitted by the faecal–oral route [2], the significant association between cooked vegetables served at the FB canteen and illness, as well as the identification of two asymptomatic EIEC carriers among the kitchen workers, support the hypothesis that the primary source of the outbreak could have been one of these workers who could have contaminated the cooked vegetables during post-cooking handling procedures. In particular, it is likely that the primary source was the asymptomatic kitchen worker who recently returned from South East Asia, where EIEC infections appear to be endemic [23, 24]. This individual also reported not having eaten cooked vegetables in the period of exposure, contrary to his two colleagues (one symptomatic and one asymptomatic carrier) who remembered the food items consumed each day. Finally, his long shedding period (i.e. he was still positive for the ipaH gene test 42 days after the first positive sample) might also have facilitated the contamination process. EIEC healthy carriers with median shedding periods comparable to those observed in this outbreak have been reported in another outbreak that occurred in the USA [11], while long-lasting carriage has been described in countries where EIEC infections are endemic, with asymptomatic persons excreting the pathogen for over 1 year [25].
Once the contamination had occurred, the poor hygiene conditions of the kitchen (i.e. no separation between meat, fish and vegetables, working areas and equipment), the improper storage of foodstuffs (i.e. because firefighters must respond to emergencies, canteen schedules were flexible, and foods could be kept out of the fridge for long periods and/or removed from and returned to the fridge several times a day), in addition to the unhygienic practice of serving leftover vegetables, could have amplified the EIEC bacterial load. This might have been exacerbated, moreover, by the absence of a competing microbial flora in cooked food, and the spread to other foodstuffs.
Transmission of EIEC infection through ready-to-eat or contaminated cooked food has been documented in most of the EIEC outbreaks reported in industrialized countries, and the implicated food items included French Camembert cheese [10], cold potato salad [12], guacamole [13] and sasayuki tofu [14]. In this case the EIEC outbreak strain was not isolated from foods nor from the kitchen environment. However, there was no evidence that the food items tested were those served during the exposure period, with the kitchen having already been cleaned by the time the LPHS inspection was conducted.
Because consumption of the suspected vegetables does not explain all the cases, it can be argued that other food items possibly contaminated to a lower degree, or other transmission routes might have played a role. Indeed we cannot exclude that in some cases EIEC infection might have been acquired by contact with a contaminated environment (e.g. the FB toilets) or by direct contact with other cases, as demonstrated by the occurrence of episodes of diarrhoea within the households of seven cases. The short incubation period of EIEC infection (between 10 h and 37 h in the present episode), coupled with the exposure to contaminated food for at least two and possibly three consecutive days, might have allowed the onset of symptoms of possible secondary cases to occur together with those of primary cases. Although the EIEC infectious dose is considered to be as high as 106–108 organisms, rendering the probability of person-to-person transmission low [26], this route has been reported [11].
An unusual feature of the present episode with respect to other reported EIEC outbreaks [10, 12, 14] was the severity of the clinical illness. The clinical symptoms were not thoroughly investigated, but the attendance of emergency departments and the hospitalization rates (70% and 30% of the cases, respectively) were markedly higher than in other episodes [10, 12]. One patient was admitted to the intensive care unit with a diagnosis of septic shock syndrome, and some patients reported mild physical discomfort for weeks. These findings are also impressive considering the exposed population consisted of relatively young and healthy adults. The factors that could have influenced the severity of the illness might include high EIEC dose in the food, the naive immunological status of the exposed population to this unusual pathogen, as well as the virulence of the EIEC strain involved. In fact, the outbreak strain showed features that are unusual for EIEC: it was motile, lysine decarboxylase positive [3], belonged to a serotype, E. coli O96:H19, which has never been reported for EIEC (F. Scheutz, personal communication), and showed a very strong capacity to invade cultured cells. Further studies are needed to assess the potential virulence of this EIEC strain.
This episode confirms the possibility that EIEC infections may be under-reported in non-endemic countries because of the limited awareness of their role as a cause of diarrhoeal illness and because EIEC can be mistaken for non-pathogenic E. coli and/or Shigella. Commercial diagnostic kits for enteric pathogens based on molecular screening may include genetic targets (e.g. the ipaH gene) that may be 100% specific for both Shigella and EIEC, but this does not allow discrimination between them [3]. In the present outbreak, stool specimens were tested for EIEC because the positive molecular screening for Shigella was not confirmed by the stool cultures and because the samples were submitted to a reference laboratory for E. coli.
Finally, some limitations affect this investigation. Failure in tracing back, interviewing and testing all FB employees, in particular, was identified as the main drawback. This possibly impeded the identification of all the cases and the estimation of epidemiological measures such as the attack rate. Moreover, the systematic collection of further details on symptoms (e.g. characteristics of diarrhoea and duration of symptoms) might have allowed a more accurate description of the severity of the epidemic.
In conclusion, this episode indicates that EIEC may represent a public health hazard even in non-endemic countries, causing serious dysentery-like disease and outbreaks and that long-lasting asymptomatic carriers may favour the entry of these unusual pathogens into the food chain. Therefore strict compliance with good hygiene practices and HACCP plans, as well as attention to the health status of the employees handling food, is needed to avoid EIEC foodborne transmission in community settings. Importantly, the prompt suspension of the canteen activity and its thorough sanitization, together with the suspension from service of the ipaH-positive kitchen workers resulted in an effective control measure in limiting the spread of the present outbreak.
From a diagnostic point of view, clinical microbiologists should be aware of the procedures of classical and molecular microbiology that allow the laboratory diagnosis of EIEC infections, which should be applied in particular to stool specimens in which common enteric pathogens cannot be identified.
ACKNOWLEDGEMENTS
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
The authors sadly report that one of the co-authors, Roberta Casa, died suddenly during the revision process of this paper. This paper is dedicated to her memory.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Kopecko DJ, Baron LS, Buysse J. Genetic determinants of virulence in Shigella and dysenteric strains of Escherichia coli: their involvement in the pathogenesis of dysentery. Current Topics in Microbiology and Immunology 1985; 118: 71–95. [DOI] [PubMed] [Google Scholar]
- 2.Nataro JP, Kaper JB. Diarrhoeagenic Escherichia coli. Clinical Microbiology Reviews 1998; 11: 142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van den Beld MJC, Reubsaet FAG. Differentiation between Shigella, enteroinvasive Escherichia coli (EIEC) and noninvasive Escherichia coli. European Journal of Clinical Microbiology & Infectious Diseases 2012; 31: 899–904. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien AD, et al. Shigellosis and Escherichia coli diarrhoea: relative importance of invasive and toxigenic mechanisms. American Journal of Clinical Nutrition 1979; 32: 229–233. [DOI] [PubMed] [Google Scholar]
- 5.Taylor DN, et al. Clinical and microbiological features of Shigella and enteroinvasive Escherichia coli infections detected by DNA hybridization. Journal of Clinical Microbiology 1988; 26: 1362–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutin L, et al. Origin and characteristics of enteroinvasive strains of Escherichia coli (EIEC) isolated in Germany. Epidemiology and Infection 1997; 118: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vieira N, et al. High prevalence of enteroinvasive Escherichia coli isolated in a remote region of Northern Coastal Ecuador. American Journal of Tropical Medicine and Hygiene 2007; 76: 528–533. [PMC free article] [PubMed] [Google Scholar]
- 8.Wanger AR, et al. Enteroinvasive Escherichia coli in travelers with diarrhoea. Journal of Infectious Diseases 1988; 158: 640–642. [DOI] [PubMed] [Google Scholar]
- 9.Svenungsson B, et al. Enteropathogens in adult patients with diarrhoea and healthy control subjects: a 1-year prospective study in a Swedish clinic for infectious diseases. Clinical Infectious Diseases 2000; 30: 770–778. [DOI] [PubMed] [Google Scholar]
- 10.Tulloch EF Jr., et al. Invasive enteropathic Escherichia coli dysentery. An outbreak in 28 adults. Annals of Internal Medicine 1973; 79: 13–17. [DOI] [PubMed] [Google Scholar]
- 11.Harris JR, et al. Person-to-person transmission in an outbreak of enteroinvasive Escherichia coli. American Journal of Epidemiology 1985; 122: 245–252. [DOI] [PubMed] [Google Scholar]
- 12.Snyder JD, et al. Outbreak of invasive Escherichia coli gastroenteritis on a cruise ship. American Journal of Tropical Medicine and Hygiene 1984; 33: 281–284. [DOI] [PubMed] [Google Scholar]
- 13.Gordillo ME, et al. Molecular characterization of strains of enteroinvasive Escherichia coli O143, including isolates from a large outbreak in Houston, Texas. Journal of Clinical Microbiology 1992; 30: 889–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamura K, et al. Food poisoning caused by enteroinvasive Escherichia coli (O164:H–) – a case in which the causative agent was identified [in Japanese]. Kansenshōgaku zasshi 1992; 66: 761–768. [DOI] [PubMed] [Google Scholar]
- 15.Ozbonfil D, et al. An outbreak of enteritis associated with enteroinvasive E. coli in an Israeli military base. Public Health Reviews 1991. –1990; 18: 171–177. [PubMed] [Google Scholar]
- 16.Capano G, et al. Enteric infections, cow's milk intolerance and parenteral infections in 118 consecutive cases of acute diarrhoea in children. European Journal of Pediatrics 1984; 142: 281–285. [DOI] [PubMed] [Google Scholar]
- 17.Bisicchia R, et al. Toxin production and haemagglutination in strains of Escherichia coli from diarrhoea in Brescia, Italy. Journal of Hygiene 1985; 95: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scavia G, et al. Enteroaggregative Escherichia coli associated with a foodborne outbreak of gastroenteritis. Journal of Medical Microbiology 2008; 57: 1141–1146. [DOI] [PubMed] [Google Scholar]
- 19.Lüscher D, Altwegg M. Detection of shigellae, enteroinvasive and enterotoxigenic Escherichia coli using the polymerase chain reaction (PCR) in patients returning from tropical countries. Molecular and Cellular Probes 1994; 8: 285–290. [DOI] [PubMed] [Google Scholar]
- 20.Mehlman IJ, et al. Detection of invasiveness of mammalian cells by Escherichia coli: collaborative study. Journal of the Association of Official Analytical Chemists 1982; 65: 602–607. [PubMed] [Google Scholar]
- 21.Busani L, et al. Antibiotic resistance in Salmonella enterica serotypes Typhimurium, Enteritidis and Infantis from human infections, foodstuffs and farm animals in Italy. Epidemiology and Infection 2004; 132: 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morabito S, et al. Detection and characterization of Shiga toxin-producing Escherichia coli in feral pigeons. Veterinary Microbiology 2001; 82: 275–283. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen TV, et al. Detection and characterization of diarrhoeagenic Escherichia coli from young children in Hanoi. Journal of Clinical Microbiology 2005; 43: 755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hien BTT, et al. Diarrhoeagenic Escherichia coli and Shigella strains isolated from children in a hospital case-control study in Hanoi, Vietnam. Journal of Clinical Microbiology 2008; 46: 996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kétyi I. Epidemiology of the enteroinvasive Escherichia coli. Observations in Hungary. Journal of Hygiene, Epidemiology, Microbiology, and Immunology 1989; 33: 261–267. [PubMed] [Google Scholar]
- 26.DuPont HL, et al. Pathogenesis of Escherichia coli diarrhoea. New England Journal of Medicine 1971; 285: 1–95. [DOI] [PubMed] [Google Scholar]