SUMMARY
In March 2012, a second outbreak of Cryptosporidium parvum affected children following a stay at a holiday farm in Norway; the first outbreak occurred in 2009. We studied a cohort of 145 schoolchildren who had visited the farm, of which 40 (28%) were cases. Cryptosporidium oocysts were detected in faecal samples from humans, goat kids and lambs. Molecular studies revealed C. parvum subtype IIa A19G1R1 in all samples including human samples from the 2009 outbreak. A dose–response relationship was found between the number of optional sessions with animals and illness, increasing from two sessions [risk ratio (RR) 2·7, 95% confidence interval (CI) 0·6–11·5] to six sessions (RR 8·0, 95% CI 1·7–37·7). The occurrence of two outbreaks 3 years apart, with the same subtype of C. parvum, suggests that the parasite is established in the farm's environment. We recommend greater emphasis on hand hygiene and routines related to animal contact.
Key words: Cryptosporidium, gastrointestinal infections, hand hygiene, outbreaks, zoonoses
INTRODUCTION
Cryptosporidium is an intestinal protozoan parasite considered an important cause of diarrhoeal illness worldwide, particularly in young children and immunocompromised patients. The robust transmission stage of Cryptosporidium, along with its high excretion rate and low infectious dose, means that it lends itself to waterborne transmission and waterborne outbreaks of cryptosporidiosis occur regularly worldwide [1]. Foodborne outbreaks of cryptosporidiosis have also been identified [2], as well as infections associated with contact with animals and farm visits [3, 4]. About 25 species of Cryptosporidium are currently recognized, along with various genotypes, several of which are known to have zoonotic potential. Cryptosporidium hominis and C. parvum are the major species infecting humans [5].
There has been considerable interest in identifying those animal species that may be reservoirs for Cryptosporidium spp. with zoonotic potential, in order to limit faecal contamination of drinking-water sources [6]. C. parvum is the major zoonotic species of Cryptosporidium and is common in calves; an infected calf may excrete millions of infective oocysts on a daily basis [6].
In Norway, cryptosporidiosis first became a notifiable infection in 2012. Awareness of this parasite among healthcare professionals in Norway is low. Testing for Cryptosporidium is not included in routine parasite testing of stool specimens, and has to be specifically requested. A laboratory survey showed that between 1998 and 2002 most laboratories performed fewer than 10 tests annually; and only 0–2 positive tests per year were reported [7]. Nevertheless, some outbreaks of cryptosporidiosis have previously been documented in Norway. The first was a small outbreak among veterinary students examining calves at a dairy farm [8], and the second was at a hotel, where a water dispenser or ice-cubes were suspected sources of infection [9]. In 2009, an outbreak of cryptosporidiosis at a holiday farm affected 74 individuals, the majority of whom were schoolchildren [10]. Additionally, during a large waterborne outbreak of giardiasis in Norway in 2004, 115 diarrhoeic patients were also diagnosed with Cryptosporidium [11]. There have been various outbreaks of cryptosporidiosis in other Scandinavian countries, with the largest outbreak recorded in Europe occurring in Østersund, Sweden in 2010 [12]. Cryptosporidium infections have been reported from a range of different animals in Norway, and are considered relatively widespread in dairy calves in Norway [13].
On 25 March 2012, the Norwegian Institute of Public Health (NIPH) was notified about several cases of gastroenteritis in schoolchildren following a stay at a Norwegian holiday farm. Cryptosporidium oocysts were identified in faecal samples from some of the children. Pupils from four different schools visiting the farm during 5–23 March were affected. This is the same holiday farm at which an outbreak occurred during March 2009, and for which no source or vehicle of infection could be definitively identified [10]. An outbreak investigation for the current outbreak was initiated to determine its extent and to identify the transmission vehicle and contamination source, in order that appropriate control measures could be implemented.
METHODS
Epidemiological investigation
A retrospective cohort study was conducted in schoolchildren, teachers and farm staff who had been on the holiday farm during the period 5–23 March 2012. For the descriptive analysis, a case was defined as a person who had been on the holiday farm during that period and had developed gastrointestinal symptoms during or within 2 weeks of staying at the farm. For the analytical study, the case definition was restricted to a person who had been on the farm in the relevant period, and had experienced diarrhoea or at least two of the following symptoms within 2 weeks of the visit: vomiting, nausea, abdominal pain, fever, with duration of illness >1 day. The cohort was restricted to schoolchildren.
A standardized web-based questionnaire was adapted to obtain information on demographic data, clinical symptoms and illness characteristics, contact with other symptomatic people, consumption of food, beverages including drinking water and animal contact during the stay. On 29 March, a link to the questionnaire was sent to the contact persons of the relevant schools by email. Pupils answered the questionnaire under the guidance of their teachers. Pupils with symptoms were asked to contact their doctors and provide faecal samples.
The cohort for the analytical study was restricted to schoolchildren. Persons reporting illness, but not fulfilling the case definition, were excluded. We calculated risk ratios with 95% confidence intervals for all food, drink and animal exposures. Stata v. 12 (StataCorp LP, USA) was used for the statistical analysis.
Environmental analysis
The premises of the holiday farm were inspected by the Norwegian Food Safety Authority on 28 March 2012 and 26 animal faecal samples, from 17 goat kids, two goats, two calves, three horses and two lambs, were collected for analysis. Food samples were not available.
Eleven litres of raw tap water were collected and stored at 4°C overnight before transportation to the Norwegian school of veterinary science (NVH).
Microbiological analysis
Human faecal samples
Initially, faecal samples were investigated for Salmonella, Shigella, Campylobacter, and Yersinia by culture and biochemical identification according to standard clinical microbiological methods at Vestfold Hospital [14] and by real-time RT–PCR for norovirus [15, 16] and sapovirus [17]. They were then fixed in formaldehyde, concentrated (Para-Pak SpinCon, Meridian Bioscience, USA) and examined for Cryptosporidium oocysts and Giardia cysts by immunofluorescence antibody test (IFAT, MeriFluor, Meridian Biosciences).
Cryptosporidium-positive samples were concentrated by replacing formaldehyde with distilled water in the standard concentration protocol and oocysts opened by boiling 0·2 ml faecal concentrate for 1 h, followed by one freeze-thawing cycle and then incubation with proteinase K for 3 h, and automated DNA extraction (Nuclisens easyMAG, bioMérieux France). A quadruplex real-time PCR (LightCycler 2·0, Roche Diagnostics, Switzerland) was performed at the SSU rRNA and LIB 13 locus using primers from a recent publication [18]. The TaqMan probes were modified from that publication by using LNA-binding TaqMan probes instead of MGB-TaqMan probes; the probe sequences for binding C. parvum and C. hominis DNA were otherwise identical to those used by Hadfield et al. [18]. Furthermore, LightCycler DNA Master Hybridization probes mastermix (Roche Diagnostics) was used and the PCR protocol was modified from two-step PCR to three-step PCR, with the following thermocycling conditions: 55 cycles; 15 s at 95 °C, 30 s at 60 °C and 60 s at 72 °C. An in-house internal control (lambda phage) was included, using primers (5′–3′) ‘LF4’ (AATCGGACTTGCCTGCAAAGAT), ‘LR2.2’ (TCCTGCGGTAGTGGTAACACC) and probe ‘L’ (Dyomics-TAGCTCGTACCATGTCCTGATACAG-BHQ3).
Selected samples were amplified by nested PCR at the GP60 gene [19], with separation of the product by gel electrophoresis on 2% agarose and visualized with GelGreen (Biotium Inc., USA). DNA products were purified (Zymoclean Gel DNA Recovery kit, Zymo Research, USA) and sequenced on both strands at a commercial laboratory (GATC Biotech, Germany). Sequences were analysed with LaserGene software (DNASTAR, USA). Comparisons with sequences in GenBank were conducted via BLAST and GP60 allele type (‘subtype’) determined by the common nomenclature [20].
Animal faecal samples
The animal faecal samples were examined by light microscopy after standard concentration procedures, involving homogenization of weighed subsamples in water, sieving and centrifugation, and individual subsamples of each sample concentrate examined by IFAT (Aquaglo, Waterborne Inc., USA) for Cryptosporidium oocysts and Giardia cysts. From Cryptosporidium-positive samples, the oocysts were isolated by modified immunomagnetic separation (IMS) [21], and DNA isolated using a QIAamp DNA mini kit (Qiagen GmbH, Germany). Standard (not real-time) PCR at the SSU rRNA gene and at the actin gene was performed as described previously [6], with product visualization by electrophoresis on 1% agarose and staining with SYBR Safe (Invitrogen, USA). For four positive samples (two from lambs, two from goat kids), the DNA products were purified (High Pure PCR product purification kit, Roche Diagnostics) and sequenced on both strands at a commercial laboratory (MWG Biotech, Germany). Sequences were examined using Chromas version 2.01 and DNA Baser 3.5 software. Comparisons with sequences in GenBank were conducted via BLAST. For oocysts from four animals PCR and sequence analyses were conducted at the GP60 gene to obtain information on species subtype, as described for Cryptosporidium oocysts found in human samples.
Water samples
The water samples were analysed using the NVH internal method for Cryptosporidium oocysts and Giardia cysts, which is based on ISO method 15 553 [22] and US EPA method 1622 [23] and US EPA method 1623·1 [24], in which the water is filtered and the eluate purified using IMS, before examination of the total sample concentrate by IFAT with additional staining with 4′,6-diamidino-2-phenylindole. The laboratory undertaking the water analyses is accredited by the national accreditation body for analysis of water for Cryptosporidium and Giardia, and, at this laboratory, the recovery efficiency of this method for Cryptosporidium oocysts is generally in excess of 60%.
RESULTS
Epidemiological investigation
Descriptive analysis
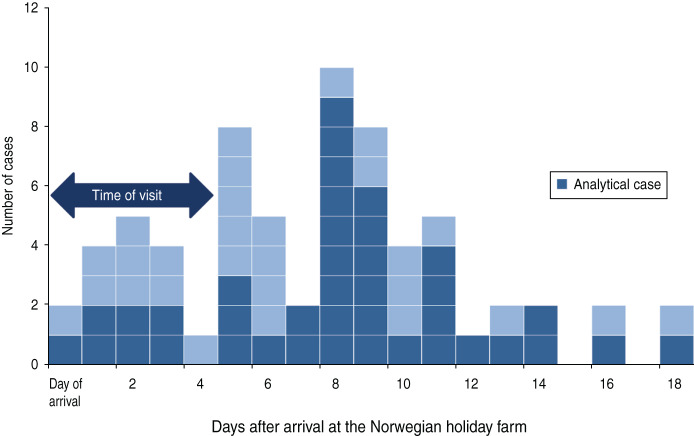
A total of 290 people visited the holiday farm between 5 and 23 March 2012, including five school groups comprising 266 pupils and teachers, and 24 farm staff. The questionnaire was sent to all of them of which 209 responded (181 pupils, eight teachers, 20 farm staff) (response rate 72%). The male:female ratio of the respondents was 1:1 and 89% were in the 10–14 years age group, reflecting the main age distribution of the visitors. Of the 209 respondents, 78 reported gastrointestinal illness following the visit (75 children, three adults). Two children had illness onset >2 weeks after the visit and were therefore excluded from further analyses. Onset of symptoms, with respect to day of arrival at the farm, is shown in Figure 1 for all cases included in the descriptive study; the 40 cases also included in the analytical study are shown in a different colour. The cases were distributed over an 18-day period; however, there is a peak between 5 and 11 days after arrival. The first case reported symptoms on the evening of 5 March and the last on 18 April. Illness onset of the farm staff was between 15 and 21 March. The most common symptoms were abdominal pain, nausea and headache (Table 1). Median duration of symptoms was 4–5 days, ranging from a few hours to >2 weeks. Most cases included in the analytical study (43%, 17/40) reported symptoms 7–9 days after arrival at the farm (Fig. 1).
Fig. 1.
[colour online]. The distribution of cryptosporidiosis cases in schoolchildren visiting a holiday farm in Norway, 5–23 March 2012 with respect to day of arrival (n = 67*). All cases fulfilled the descriptive case definition. Those also fulfilling the analytical case definition are shown in a darker colour. [* n = 67, six respondents stated illness without giving precise onset of symptoms (one of which was included in the analytical study), three were farm staff.]
Table 1.
Clinical features of respondents reporting gastrointestinal symptoms due to Cryptosporidium infection in schoolchildren visiting and farm staff working on a holiday farm in Norway, 5–23 March 2012 (n = 72*), by testing history†
| Type of symptom | Self-reported symptoms only (n = 64) | Attack rate (%) | Self-reported symptoms (positive Cryptosporidium) (n = 8) | Attack rate (%) |
|---|---|---|---|---|
| Abdominal pain | 52 | 81 | 8 | 100 |
| Nausea | 45 | 70 | 6 | 75 |
| Headache | 38 | 59 | 5 | 63 |
| Fever | 30 | 47 | 7 | 88 |
| Diarrhoea | 29 | 45 | 7 | 88 |
| Watery diarrhoea | 11 | 17 | 5 | 63 |
| Vomiting | 25 | 39 | 4 | 50 |
| Sore throat | 18 | 28 | 2 | 25 |
| Blood in faeces | 1 | 1·6 | 1 | 13 |
Four respondents stated illness without specifying symptoms.
Cryptosporidium oocysts were detected in faecal samples from eight cases in the cohort.
Of the pupils answering the questionnaire and reporting illness, Cryptosporidium oocysts were detected in faecal samples from eight cases. Most laboratory-diagnosed cases (100%, 8/8) expressed abdominal pain, diarrhoea (7/8), fever (7/8), nausea (6/8) and headache (5/8). Half of the cases reported vomiting (50%, 4/8) (Table 1). Only three of the cases reported contact with ill persons before symptom onset.
Analytical study
Only 40 matched the case definition of the analytical study (attack rate 19%), and all were aged between 10–14 years. A total of 40 cases and 105 non-cases, all pupils, were included in the risk-factor analysis. No specific food, or beverages including drinking water were significantly associated with illness. However, pupils who had had contact with lambs or drank milk directly from the nanny goat had a higher risk of developing symptoms (Table 2). All the pupils had had contact with the animals through the holiday programme arranged by the farm (Table 2). However, the pupils also had the option of participating in additional sessions with animals during the week. On three different afternoons, the pupils could participate in contact sessions with the animals, in which they could be in the pens with the animals and pet them. These sessions mainly involved close contact with young kids and lambs, which had been born 3–4 weeks previously. The pupils also had the option of participating in routine farm work three mornings over the week. This included feeding, milking and mucking out (removing faeces and urine-soaked bedding from an animal enclosure and replacing it with clean dry bedding). Thus, the pupils had the option of participating in six additional sessions with the animals. Using this information, a dose–response relationship was detected between illness and number of additional contact sessions with animals (Table 3).
Table 2.
Exposure to risk factors, outbreak cases due to Cryptosporidium infection in schoolchildren visiting a holiday farm in Norway, 5–23 March 2012 (n = 145)
| Contact with animals | Exposed | Not exposed | RR* (95% CI) | P value | ||||
|---|---|---|---|---|---|---|---|---|
| Ill | Total | Ill | Total | |||||
| Goat/kids | 40 | 145 | 28% | 0 | 0 | – | – | – |
| Lambs | 13 | 27 | 48% | 27 | 118 | 23% | 2·1 (1·3–3·5) | 0·01 |
| Drinking milk directly from goat | 25 | 72 | 35% | 14 | 69 | 20% | 1·7 (1·0–3·0) | 0·05 |
| Attending >50% of animal contact sessions† | 18 | 34 | 53% | 22 | 111 | 20% | 2·67 (1·6–4·4) | <0·01 |
RR, Risk ratio; CI, confidence interval.
Risk ratios are crude.
Contact sessions with the animals includes petting and playing with the animals and routine farm work.
Table 3.
Dose–response relationship between illness and contact with animals in schoolchildren visiting a holiday farm in Norway, 5–23 March 2012
| No. of animal contact sessions | Cases | Total | Attack rate (%) | Risk ratio | 95% CI |
|---|---|---|---|---|---|
| 1 | 2 | 24 | 8 | Reference | – |
| 2 | 8 | 36 | 22 | 2·7 | 0·6–11·5 |
| 3 | 7 | 28 | 25 | 3·0 | 0·7–13·1 |
| 4 | 11 | 23 | 48 | 5·7 | 1·4–23·1 |
| 5 | 3 | 5 | 60 | 7·2 | 1·6–32·5 |
| 6 | 2 | 3 | 67 | 8·0 | 1·7–37·7 |
CI, Confidence interval.
Microbiological investigation
Nine faecal samples received early in the investigation were analysed for Salmonella, Shigella, Yersinia, Campylobacter, norovirus, sapovirus, Giardia cysts and Cryptosporidium oocysts; all were positive for Cryptosporidium and negative for other enteropathogens. Further samples were only screened for Cryptosporidium and Giardia. In total, Cryptosporidium oocysts were detected in 13/41 stool samples from pupils, and in 2/8 samples from farm staff. Real-time PCR was positive for C. parvum in all 15 positive samples. From four pupils in the cohort, samples were subtyped at the GP60 gene. All were allele type IIa A19G1R1. The sequence has been submitted to GenBank (accession no. KC679056). The remaining samples were not subtyped due to resource constraints. Of the 26 animal faecal samples examined, samples from four goat kids and two lambs were positive for Cryptosporidium oocysts. Most of these samples contained low oocyst concentrations, but the faecal sample from one lamb contained a very high concentration of Cryptosporidium oocysts (>1 × 106 oocysts/g faeces). Molecular analysis demonstrated that the animals were infected with two different species of Cryptosporidium, i.e. C. parvum and C. xiaoi; positive PCR results were obtained from four animals, all of which were infected with C. parvum, and two of these animals, one lamb and one goat kid, were also infected with C. xiaoi. At both the SSU rRNA gene and the actin gene, sequences obtained were identical to sequences already available in GenBank (e.g. accession no. GQ983355 for C. parvum sequence, and accession nos. JX258864 and GQ337964 for C. xiaoi sequences). For the GP60 gene, the subtype of C. parvum was identical to that found in the human patients, IIa A19G1R1 (accession no. KC679056).
Environmental investigation
No irregularities were found by the local food safety authority during inspection of the kitchen or in the overall organization of the holiday farm. The kitchen staff had good routines regarding preparation and storage of food, cleaning and personal hygiene. None of the kitchen staff reported sickness in the days or weeks before the outbreak. Neither of the two positive farm staff worked in the kitchen.
Farm staff working with the animals normally did not work in the kitchen; pupils helping in the kitchen washed their hands under supervision before starting work and wore an apron.
The visitors were all informed about the importance of good hand hygiene upon arrival at the holiday farm, and were told to count to 20 while washing their hands after being in contact with animals and before eating. Washbasins were found in each bedroom, as well as in the washrooms and shower rooms. The holiday farm also had a dispenser with alcohol-based disinfectant in the dining room, but it should be noted that alcohol-based disinfectants are ineffective against Cryptosporidium. In the questionnaire, 75% of children claimed to wash their hands for 20 s, and 11% stated that they washed their hands for more than half a minute.
The holiday farm has a private water supply with a UV disinfection system. The well was inspected for possible surface water leakage and the UV system was controlled; no irregularities were detected and the UV system emitted sufficient energy to inactivate Cryptosporidium. The wastewater system is separate. Past records of water quality tests taken regularly since installation of the UV disinfection system in 2009, have all shown good drinking-water quality. The water samples were negative for parasites.
DISCUSSION
This was the second registered outbreak of cryptosporidiosis in visitors at this holiday farm, and therefore it was considered particularly important to identify the source of oocysts and the transmission vehicle. This investigation indicates that the outbreak was probably due to direct or indirect contact with infected animals. Dose–response analysis showed that risk of illness increased with time spent with the animals. During the outbreak period, goat kids and lambs were aged 3–4 weeks, and at their most playful and appealing. Cases were distributed over a long period, reflecting a continuously present source of infection as well as the relatively long incubation period associated with cryptosporidiosis. Farm staff had observed that some of the kids and lambs had loose stools, but had assumed that this was due to the transition from drinking only milk to the start of eating grass. None of the food items consumed at the holiday farm were significantly associated with illness in the epidemiological study, and food samples were not available for sampling. However, it cannot be excluded that food items could have acted as vehicles for the oocysts, with contamination from hands to food items, and be responsible for some of the infections.
During the previous outbreak in 2009 [10], faecal samples from 20 animals were analysed (five calves, five lambs, 10 goat kids). Only one sample (from a lamb) was positive, with only a single oocyst detected, and therefore molecular characterization was not attempted. The vehicle for transmission was not definitively identified, but several possible routes of transmission were recognized, including an infected food handler, water, and contact with animals. It might be speculated that contact with animals was the source of infection in the earlier outbreak, but that due to lack of awareness, the agent of infection was identified relatively late and when animal samples were analysed, oocyst shedding had decreased to below the levels of detection. In that outbreak no attempt was made to correlate animal contact with infection. Following the outbreak in 2009, a UV disinfection system for drinking water was installed and improvements in hand hygiene routines were recommended.
C. parvum oocysts from both animal and patient samples in the recent (2012) outbreak were subtyped as IIa A19G1R1, the same subtype associated with the 2009 outbreak. C. parvum subtyping is rarely performed in Norway, so the prevalence and distribution of this subtype is unknown. However, a limited survey (six flocks) of Cryptosporidium infections in sheep and lambs in Norway [6] which included genotyping, did not identify C. parvum infections at all; rather, they were infected with non-zoonotic species (C. xiaoi) or species associated with only sporadic human infection (C. ubiquitum). In other European countries, this subtype of C. parvum has only been reported sporadically from humans, calves and hedgehogs [25–28]. With two outbreaks 3 years apart with the same rare subtype, it is likely that the source of the infection has been present on the farm since the first outbreak in 2009, or even before, and has become established in the animals there. Surveying young animals for this subtype of Cryptosporidium in the spring is recommended, particularly to determine whether it may also infect other animals such as calves. Both this outbreak and the previous outbreak in 2009 occurred in March, when oocyst shedding in farm animals may be considered to be higher due to newly born naive animals being most susceptible to becoming infected, either from oocysts in the environment or from low-level, probably asymptomatic, shedding in adult animals. C. parvum infections in farm animals are particularly associated with pre-weaned calves, and calves are considered the most usual source of zoonotic transmission [29]. In Norway no previous outbreaks of cryptosporidiosis have been associated with sheep or goats. However, outbreaks of cryptosporidiosis elsewhere have previously been associated with lambs [30] and during 2013 another cryptosporidiosis outbreak in Norway (with another subtype of C. parvum) was associated with goat kids and lambs, while a review of the literature suggested that Cryptosporidium infections in sheep/goats were of greater zoonotic threat than Giardia infections [31], although it was emphasized that there is considerable variation in prevalence and genotypic variation.
Close contact with animals is often a highlight for children visiting holiday farms, and the farm involved in this outbreak would like to continue to offer their guests close-up experiences with animals. However, since animals can be sources of zoonotic pathogens, infection risks should be minimized by ensuring good hand-washing routines, as also pointed out by the Centers for Disease Control and Prevention (CDC) in their recommendations for Cryptosporidium prevention for camps [32].
Although hand-washing routines were already in place, with staff at the farm informing visitors about the importance of good hand hygiene, especially after contact with animals, after toilet use, and before eating, direct observation of hand-washing routines is necessary to ensure compliance, especially in schoolchildren. In the wake of the current outbreak, recommendations to the holiday farm primarily concern more easily accessible washbasins, and more clearly visible posters emphasizing the importance of hand-washing after all contact with animals and before eating. Changing of clothes and shoes before entering the dining room and hand-washing under surveillance of a teacher are other recommendations particularly applicable to schoolchildren. The holiday farm was also instructed not to offer non-pasteurized milk.
However, there are some limitations with this study that should be taken into account. It is possible that some children included as cases in the descriptive study may have been misclassified due to the broad case definition and could potentially have experienced gastrointestinal symptoms caused by a different pathogen. None of the cases that visited the farm during the first week of the outbreak submitted a faecal sample. The first faecal samples were submitted from the school visiting the farm during the second week of the outbreak, prior to the initiation of the outbreak investigation. These samples were tested for multiple pathogens. After Cryptosporidium infection was identified in these patients, samples from subsequent cases were only tested for Cryptosporidium and Giardia. The case definition for the analytical study was therefore further restricted to include only children with symptom duration of >1 day in order to limit misclassification. However, misclassification of controls as cases would only have reduced the power to detect associations between exposure and illness.
Recall bias is a concern in the study and can be attributed to the amount of time between being at the farm and being interviewed. In this case, the children were asked to complete the questionnaire at school after they had returned from the trip, and while they were supervised by their teachers, it is possible that some children were influenced by their peers in the way they completed the questionnaire. In addition, accurate recall may not be reliable in children who may not fully understand the nature and importance of the investigation. This may only bias the results if there is a reason to believe that this would affect cases and controls differently, which we believe would be minimal in this case, especially regarding the exposures related to behaviour and activities, as the children were not informed about any suspected source beforehand.
That the same school was involved in a previous outbreak at the same farm may also have introduced bias. Although this may have heightened awareness in the school staff and improved the timeliness of the investigation, experiences from the previous outbreak may have influenced perceptions of the cause of the outbreak among the teachers. However, as there were no conclusions about the source in the previous investigation, this is unlikely to have affected the results.
In this report, we describe the largest documented outbreak of cryptosporidiosis in Norway to date. The source of this outbreak was probably oocysts in the faeces of infected lambs and/or goat kids. Zoonotic transmission from ovines and caprines has not previously been documented in Norway, and this outbreak indicates that they should be considered a potential source of zoonotic parasites, as well as other pathogens. The origin of this genotype of C. parvum, that has not previously been documented in Norway and is rarely documented elsewhere, is unknown, but it would be interesting to determine the extent of its establishment and in which animal species.
Even when apparently satisfactory hand-washing routines have been established, the persistence of oocysts in adhesion to surfaces means that hygienic routines must be followed scrupulously to minimize the risk of transmission of infection.
ACKNOWLEDGEMENTS
We acknowledge the following colleagues for their contribution to the outbreak investigation: Frank Thrana (municipality doctor in Tønsberg), Christian Nelke (municipality doctor in Nore and Uvdal), and Kjersti Soli (Norwegian Food Safety, district of Kongsberg). The staff at the parasitology laboratory at NVH (Teresa Hagen, Ahmed Abdelghani, Kristoffer Tysnes) and at the microbiology laboratory at Vestfold Hospital (Anita Bratfoss Andreassen) are gratefully acknowledged for their technical support.
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Baldursson S, Karanis P. Waterborne transmission of protozoan parasites: review of worldwide outbreaks – an update 2004–2010. Water Research 2011; 45: 6603–6614. [DOI] [PubMed] [Google Scholar]
- 2.Robertson LJ, Chalmers RM. Foodborne cryptosporidiosis: is there really more in Nordic countries? Trends in Parasitology 2013; 29: 3–9. [DOI] [PubMed] [Google Scholar]
- 3.Evans MR, Gardner D. Cryptosporidiosis outbreak associated with an educational farm holiday. Communicable Disease Report. CDR Review 1996; 6: R50–R51. [PubMed] [Google Scholar]
- 4.Sayers GM, et al. Cryptosporidiosis in children who visited an open farm. Communicable Disease Report. CDR Review 1996; 6: R140–R144. [PubMed] [Google Scholar]
- 5.Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Experimental Parasitology 2010; 124: 80–89. [DOI] [PubMed] [Google Scholar]
- 6.Robertson LJ, Gjerde BK, Furuseth HE. The zoonotic potential of Giardia and Cryptosporidium in Norwegian sheep: a longitudinal investigation of 6 flocks of lambs. Veterinary Parasitology 2010; 171: 140–145. [DOI] [PubMed] [Google Scholar]
- 7.Nygard K, et al. Are domestic Cryptosporidium and Giardia infections in Norway underdiagnosed? [in Norwegian]. Tidsskrift for Den Norske Laegeforening 2003; 123: 3406–3409. [PubMed] [Google Scholar]
- 8.Robertson L, et al. A small outbreak of human cryptosporidiosis associated with calves at a dairy farm in Norway. Scandinavian Journal of Infectious Diseases 2006; 38: 810–813. [DOI] [PubMed] [Google Scholar]
- 9.Hajdu A, et al. Investigation of Swedish cases reveals an outbreak of cryptosporidiosis at a Norwegian hotel with possible links to in-house water systems. BMC Infectious Diseases 2008; 8: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rimseliene G, et al. An outbreak of gastroenteritis among schoolchildren staying in a wildlife reserve: thorough investigation reveals Norway's largest cryptosporidiosis outbreak. Scandinavian Journal of Public Health 2011; 39: 287–295. [DOI] [PubMed] [Google Scholar]
- 11.Robertson LJ, et al. Cryptosporidium parvum infections in Bergen, Norway, during an extensive outbreak of waterborne giardiasis in autumn and winter 2004. Applied and Environmental Microbiology 2006; 72: 2218–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smittskyddsinstitutet. Cryptosporidium i Østersund. (http://www.slv.se/upload/dokument/livsmedelsforetag/dricksvatten/SMI%20Cryptosporidium-i-Ostersund-2011-15-4.pdf), 2011. Accessed 16 May 2013.
- 13.Hamnes IS, Gjerde B, Robertson L. Prevalence of Giardia and Cryptosporidium in dairy calves in three areas of Norway. Veterinary Parasitology 2006; 140: 204–216. [DOI] [PubMed] [Google Scholar]
- 14.Isenberg HD (ed.). Clinical Microbiology Procedures Handbook, 2nd edn, Washington, DC: ASM Press, 2004, volume 1, pp. 3.8.1.1–3.8.3.1. [Google Scholar]
- 15.Hoehne M, Schreier E. Detection of Norovirus genogroup I and II by multiplex real-time RT-PCR using a 3′-minor groove binder-DNA probe. BMC Infectious Diseases 2006; 6: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jothikumar N, et al. Rapid and sensitive detection of noroviruses by using TaqMan-based one-step reverse transcription-PCR assays and application to naturally contaminated shellfish samples. Applied and Environmental Microbiology 2005; 71: 1870–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logan C, O'Leary JJ, O'Sullivan N. Real-time reverse transcription PCR detection of norovirus, sapovirus and astrovirus as causative agents of acute viral gastroenteritis. Journal of Virological Methods 2007; 146: 36–44. [DOI] [PubMed] [Google Scholar]
- 18.Hadfield SJ, et al. Detection and differentiation of Cryptosporidium spp. in human clinical samples by use of real-time PCR. Journal of Clinical Microbiology 2011; 49: 918–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fayer R, Xiao LH (eds). Cryptosporidium and Cryptosporidiosis. Boca Raton, USA: CRC Press Inc., 2007. [Google Scholar]
- 20.Sulaiman IM, Lal AA, Xiao L. A population genetic study of the Cryptosporidium parvum human genotype parasites. Journal of Eukaryotic Microbiology 2001; (Suppl.): 24S–27S. [DOI] [PubMed] [Google Scholar]
- 21.Robertson LJ, et al. Application of genotyping during an extensive outbreak of waterborne giardiasis in Bergen, Norway, during autumn and winter 2004. Applied and Environmental Microbiology 2006; 72: 2212–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International Organization for Standardization: Water quality – isolation and identification of Cryptosporidium oocysts and Giardia cysts from water. ISO 15553. Geneva, 2006. [Google Scholar]
- 23.Anon. Method 1622: Cryptosporidium in water by filtration/IMS/FA DRAFT. United States Environmental Protection Agency, Office of Water, Washington, DC, 20460, EPA-821-R97-023, 1997. [Google Scholar]
- 24.Anon. Method 1623.1: Cryptosporidium and Giardia in water by filtration/IMS/FA. United States Environmental Protection Agency, Office of Water (MS-140), Washington, DC, 20460; EPA-821-R-12-001, January 2012. [Google Scholar]
- 25.Brook EJ, et al. Molecular epidemiology of Cryptosporidium subtypes in cattle in England. Veterinary Journal 2009; 179: 378–382. [DOI] [PubMed] [Google Scholar]
- 26.Dyachenko V, et al. Occurrence and molecular characterization of Cryptosporidium spp. genotypes in European hedgehogs (Erinaceus europaeus L.) in Germany. Parasitology 2010; 137: 205–216. [DOI] [PubMed] [Google Scholar]
- 27.Soba B, et al. Molecular characterisation of Cryptosporidium isolates from humans in Slovenia. Clinical Microbiology and Infection 2006; 12: 918–921. [DOI] [PubMed] [Google Scholar]
- 28.Wielinga PR, et al. Molecular epidemiology of Cryptosporidium in humans and cattle in The Netherlands. International Journal for Parasitology 2008; 38: 809–817. [DOI] [PubMed] [Google Scholar]
- 29.Xiao L, Feng Y. Zoonotic cryptosporidiosis. FEMS Immunology and Medical Microbiology 2008; 52: 309–323. [DOI] [PubMed] [Google Scholar]
- 30.Pritchard GC, et al. Cryptosporidium species in lambs submitted for diagnostic postmortem examination in England and Wales. Veterinary Record 2008; 163: 688–689. [PubMed] [Google Scholar]
- 31.Robertson LJ. Giardia and Cryptosporidium infections in sheep and goats: a review of the potential for transmission to humans via environmental contamination. Epidemiology and Infection 2009; 137: 913–921. [DOI] [PubMed] [Google Scholar]
- 32.Centers for Disease Control and prevention (CDC). Prevention and control of Crypto at camps (http://www.cdc.gov/parasites/crypto/camps.html). Accessed 12 August 2013.