SUMMARY
A large outbreak of cholera reported during April–July 2009 in the Kendrapada district of Odisha, India was investigated. Forty-one rectal swabs and 41 water samples, collected from diarrhoeal patients and from different villages were bacteriologically analysed for the isolation of bacterial enteriopathogens, antibiogram profile and detection of various toxic genes. The bacteriological analysis of rectal swabs and environmental water samples revealed the presence of V. cholerae O1 Ogawa biotype El Tor. The V. cholerae strains were resistant to ciprofloxacin, co-trimoxazole, chloramphenicol, streptomycin, ampicillin, furazolidone and nalidixic acid. The multiplex polymerase chain reaction (PCR) assay on V. cholerae strains revealed the presence of ctxA and tcpA genes. The mismatch amplification of mutation assay (MAMA) PCR on clinical and environmental isolates of V. cholerae revealed that the strains were El Tor biotype, which harboured the ctxB gene of the classical strain. The random amplified polymorphic DNA PCR analysis and pulsed-field gel electrophoresis results indicated that the V. cholerae isolates belonged to the same clone. This investigation gives a warning that the El Tor variant of V. cholerae has spread to the coastal district causing a large outbreak that requires close monitoring and surveillance on diarrhoeal outbreaks in Odisha.
Key words: El Tor variant Vibrio cholerae, Odisha, spread
INTRODUCTION
The disease cholera, caused by Vibrio cholerae, has been reported throughout the world and is of primary importance in developing countries. This disease is endemic, epidemic and pandemic in nature in modern times. V. cholerae has many serogroups and serotypes. Of the serogroups mainly V. cholerae O1 and O139 are the outbreak/epidemic strains. V. cholerae O1 has two biotypes (classical and El Tor), which are traditionally distinguished by phenotypic tests and genotypic differences. However, three variants of the El Tor biotype have been described recently: Matlab (in Bangladesh) variants in 2002 which could not be biotyped because they possessed a mixture of the characteristics of both classical and El Tor [1]; Mozambique variant in 2004–2005, which has a typical El Tor genome but a tandem repeat of the classical ctx prophage in the small chromosome [2]; and El Tor variant (a typical El Tor biotype and an El Tor ctx prophage that produces ctx of the classical type); predominant in Bangladesh since 2001 [3]. The El Tor variant of V. cholerae has also been described in other regions of Asia and Africa [4]. The V. cholerae strain, responsible for the expanding cholera epidemic in Haiti during 2010 was due to El Tor variant O1 strains that are also predominant in South Asia, including Bangladesh [5, 6]. Similarly, outbreaks of cholera caused by a strain of the V. cholerae O1 El Tor variant have been reported in Punjab and Haryana during July–September 2007 [7]. Large outbreaks of cholera have also been reported from South India [8]. Epidemics of severe cholera caused by El Tor variants of V. cholerae O1 Ogawa in Odisha were reported from six tribal blocks of three districts of Odisha during July–September 2007 [9]. In this study we investigated the causative agent of the diarrhoeal outbreak, the antibiogram profile and detection of various toxic genes of V. cholerae and their clonality, which were reported during April–July 2009 in the Rajnagar block of Kendrapada district, on the eastern coast of Odisha, India.
METHODS
Case definition
A case of severe diarrhoea was defined as the occurrence of ⩾3 watery stools in a 24-h period associated with a clinical history of vomiting, abdominal cramping and severe dehydration in residents of Rajnager block between April and July 2009.
Data collection
An investigating team from the Microbiology Department, Regional Medical Research Centre (RMRC), Bhubaneswar and local health workers from the Rajnagar Community Health Centre (CHC) searched actively to identify severe diarrhoeal cases in the diarrhoea-affected villages. They collected a clinical history of diarrhoeal patients, and risk factors for the acquisition and spread of the infection in that block. All severe diarrhoeal cases were recorded and plotted in an epidemic curve to check the progress of the outbreak. Simultaneously, severe diarrhoeal cases were checked by month for the last 3 years from the hospital records to confirm the diarrhoeal outbreak. The occurrence of severe diarrhoeal cases was plotted in the block map by month to observe the spread of the disease. Prior to onset of the diarrhoeal outbreak in Rajnagar block a large festival had been held on 14 April 2009, with about 30 000 people attending between 08:00 and 18:00 hours with some returning later in the evening. Suspecting the festival as the source of infection detailed information was collected regarding the festival; e.g. types of food, source of drinking water available at the festival. The first reported diarrhoeal patient was identified from the hospital record and traced back to her village. This person had returned to her village from the local festival and after 24 h developed severe diarrhoea associated with vomiting, abdominal cramping and severe dehydration. A house-to-house survey was conducted in the villages using a questionnaire to collect information on the villagers' knowledge, attitude and practices regarding diarrhoea as well as socio-demographic information on the community in the diarrhoea-affected villages.
Sample collection
Rectal swabs from severe diarrhoeal patients were collected in Cary–Blair transport medium from Rajnagar CHC and also from the households of diarrhoea-affected villages of Rajnagar block, Kendrapada district. The rectal swabs were collected from patients prior to the administration of antibiotics. The samples were transported to the RMRC, Bhubaneswar within 24 h of collection for bacteriological analysis. After isolation of V. cholerae from the rectal swabs as the aetiological agent of the outbreak, water samples were collected non-randomly from water bodies, e.g. ponds, rivers, small roadside water reservoirs as well household water from various different diarrhoea-affected villages in order to determine the source of infection.
Processing of samples
Rectal swabs were inoculated onto MacConkey agar, Hektoen enteric agar (HEA), thiosulphate-citrate-bile-salt sucrose agar (TCBS) and enrichment was performed in Selenite-F broth and alkaline peptone water (APW) [10]. Water samples were processed through membrane filtration, sedimentation and enriched in double-strength APW medium and then subcultured on TCBS plates to check for growth of V. cholerae strains [9]. V. cholerae was identified by standard biochemical tests and serotyping was performed using antisera obtained from Becton Dickinson (BD, USA).
Control measures
All the drinking water sources from different villages in the Rajnagar area were chlorinated. A public awareness campaign was initiated based at sub-centres located in various villages. The sub-centre staff were instructed to refer all cases of diarrhoea to Rajnagar CHC. Intravenous fluids, oral rehydration solution and essential drugs were stocked at the sub-centres. Pharmacists and nurses at the sub-centres were instructed to provide intravenous fluid while referring cases to the PHC.
Antimicrobial susceptibility testing
The sensitivity and the resistance pattern of V. cholerae O1 strains were tested with antibiotic-impregnated commercial disks (Himedia, India) using ampicillin (10 μg), chloramphenicol (30 μg), co-trimoxazole (25 μg), ciprofloxacin (5 μg), furazolidone (100 μg), gentamicin (10 μg), neomycin (30 μg), nalidixic acid (30 μg), norfloxacin (10 μg), streptomycin (10 μg), tetracycline (30 μg), azithromycin (15 μg) and polymyxin B (50 μg). V. cholerae O1 strains were subcultured in tryptic soy broth (BD, USA) and plated on Mueller–Hinton agar (BD, USA). Plates were incubated for 24 h at 37°C. Characterization of strains as susceptible or resistant was determined based on the size of the inhibition zones around each antibiotic disk according to the manufacturer's instructions following the Kirby–Bauer technique [11].
Polymerase chain reaction (PCR) assay
A multiplex PCR-based assay was employed to determine the presence of the A subunit of the cholera toxin gene (ctxA) and to biotype the V. cholerae strains by targeting tcpA (encoding the major structural subunit of the toxin co-regulated pilus), which is specific for El Tor and classical strains, according to the method described by Khuntia et al. [12].
Mismatch amplification mutation assay (MAMA)-PCR for differentiation of the cholera toxin B subunit of classical and El Tor biotypes of V. cholerae O1
The MAMA-PCR was designed to detect the nucleotide sequence difference at position 203 of the ctxB gene for the identification of cholera toxin of classical and El Tor biotypes. For this, a conserved forward primer (FW-com) and two allele-specific primers, Rv-cla and Rv-elt, were designed that are able to amplify ctxB of classical and El Tor biotypes, respectively [13]. Using the above technique the environmental and clinical isolates of all V. cholerae O1 strains were subjected to simplex PCR assay to detect the V. cholerae O1 El Tor variant that harbours the classical ctxB gene and V. cholerae O1 that harbours the El Tor ctxB gene separately. The random amplified polymorphic DNA (RAPD) PCR assay was performed on selected V. cholerae strains with 1281 and 1283 primers to check the clonality as in a previous study [14].
Pulsed-field gel electrophoresis (PFGE) analysis
Genomic DNA of V. cholerae O1 was prepared in agarose plugs to perform PFGE. As described by Sinha et al. [15], PFGE was performed using the counter-clamped homogenous electric field method on the CHEF Mapper system (Bio-Rad, USA) with 1% PFGE grade agarose in 0·5× TBE buffer. Run condition was generated by an auto-algorithm mode of CHEF Mapper, the PFGE system was comparable to a size range of 20–300 kb markers for V. cholerae strains. The gel was stained in 10 μg/ml ethidium bromide solution for 30 min, de-stained in water for 15 min and photographed under UV light in an Alpha Imager (Alpha InfoTech Corporation, USA).
RESULTS
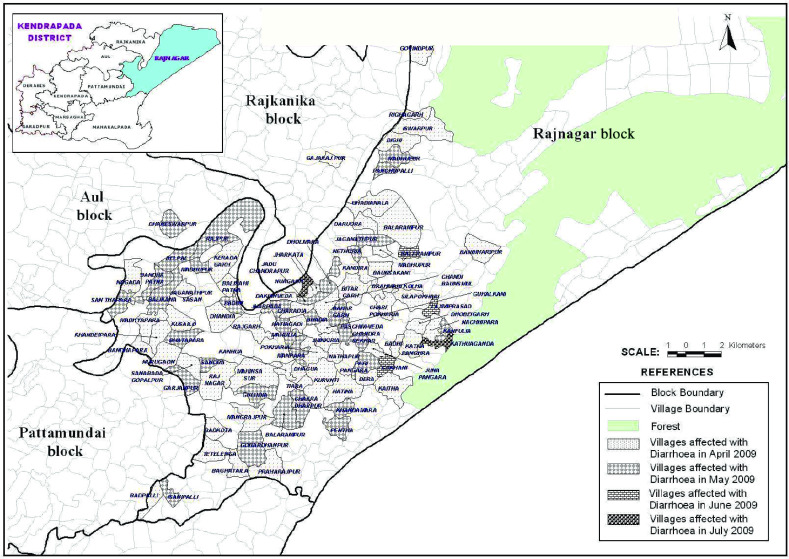
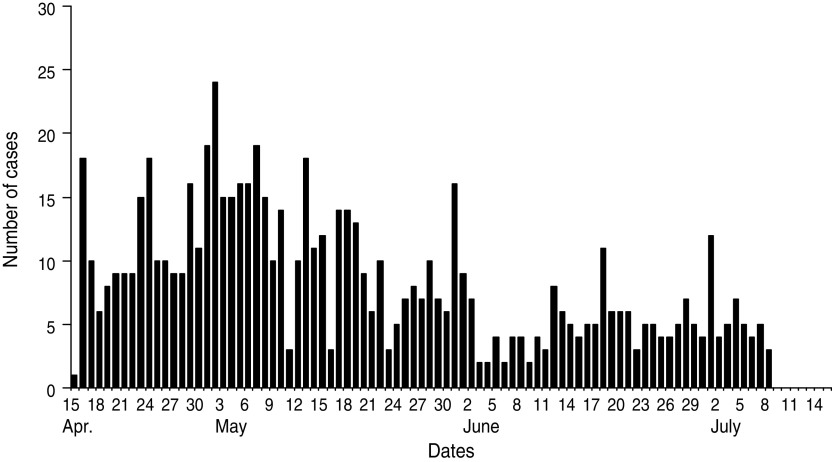
The diarrhoeal cases were reported to Rajnagar CHC on 15 April 2009 at midnight. The diarrhoeal patients that had attended the festival originated from different villages. According to data available from Rajnagar CHC, of the total 18 gram panchayats (GP; rural local governments), 224 villages and 162 984 inhabitants there were 16 GP, 108 villages and 96 878 inhabitants that were affected by severe cases of diarrhoea, respectively (Table 1). Of these, 715 severe diarrhoeal cases and seven deaths were reported with an attack rate of 0·7% and a case-fatality rate of 0·98%. Diarrhoea-affected villages and cases were analysed by month. This analysis indicated that the highest number of diarrhoea-affected villages were reported during April (n = 63) followed by May (n = 41), June (n = 3) and July (n = 1) (Figs 1, 2). Figure 3 depicts all cases of diarrhoea according to date. Interestingly, the epidemic curve produced many peaks, which indicates that the clustering of cases was reported in different time periods from different villages. After appropriate control measures initiated by the government of Odisha, the number of cases of diarrhoea declined gradually and after 8 July 2009 no further cases were reported.
Table 1.
Severe diarrhoea in Rajnagar block, Kendrapada district, April–July 2009
| Number | |
|---|---|
| Total gram panchayats | 18 |
| Diarrhoea-affected gram panchayats | 16 |
| Total villages | 224 |
| Diarrhoea-affected villages | 108 |
| Total population | 162 984 |
| Diarrhoea-affected population | 96 878 |
Fig. 1.
[colour online]. Diarrhoea-affected Rajnagar block, Kendrapada district (April–July 2009).
Fig. 2.
[colour online]. Diarrhoea-affected villages in Rajnagar block by month (April–July 2009).
Fig. 3.
Number of cases of severe diarrhoea by date, Rajnagar block, April–July 2009.
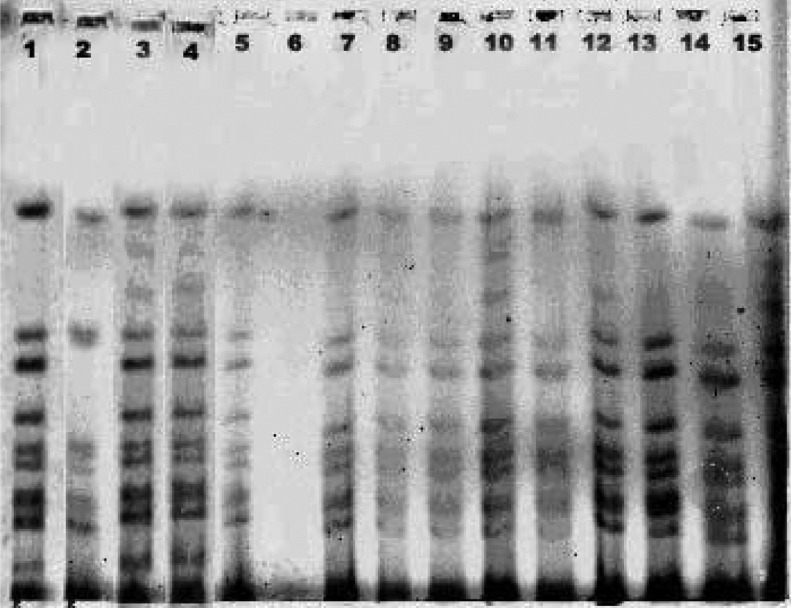
The rectal swabs collected from diarrhoeal patients from Rajnagar CHC and also from the diarrhoea-affected villages were analysed bacteriologically. The water samples from the ponds, tube wells and roadside water reservoirs from 12 affected villages were analysed for the presence of V. cholerae. Of 41 rectal swabs, 27 were culture positive and of these, 18 (67%) were V. cholerae O1 Ogawa biotype El Tor, followed by seven (26%) E. coli and two (7%) Aeromonas spp. (Table 2). Similarly, 2/41 water samples collected from different ponds, roadside small water reservoirs, rivers, and tube well water were analysed and the pond water tested positive for V. cholerae O1 Ogawa (Table 3). The antibiogram profile of V. cholerae isolates isolated from diarrhoeal patients and the pond water indicated that the V. cholerae strains were sensitive to tetracycline, norfloxacin, gentamicin, azithromycin, neomycin, erythromycin and resistant to ciprofloxacin, co-trimoxazole, chloramphenicol, streptomycin, ampicillin, nalidixic acid and furazolidone. The molecular analysis of all V. cholerae strains isolated from diarrhoeal patients and pond water samples revealed that the El Tor variants of V. cholerae O1 Ogawa with the ctxB gene of the classical strain was the causative organism of this outbreak, which was confirmed by MAMA-PCR. The RAPD-PCR analysis by 1281 and 1283 primers on representative V. cholerae isolates indicated that they belonged to same clone. Similarly, the PFGE analysis on a number of representative strains along with the 2007 tribal cholera epidemic strain revealed them to be clonal in nature (Fig. 4).
Table 2.
Bacteriological analysis of enteropathogens isolated from diarrhoeal patients
| No. (%) | |
|---|---|
| Total samples | 41 (100) |
| Culture negative | 14 (34) |
| E. coli | 7 (26) |
| V. cholerae O1 Ogawa | 18 (67) |
| Salmonella spp. | 0 |
| Shigella spp. | 0 |
| Aeromonas spp. | 2 (7) |
Table 3.
Analysis of water samples collected from Rajnagar block
| Sampleno. | Category | Totalsamples | No. positivefor V. choleraeO1 Ogawa (%) |
|---|---|---|---|
| 1 | Pond | 29 | 2 (7) |
| 2 | Road side small reservoir | 4 | 0 |
| 3 | River | 1 | 0 |
| 4 | Tube well | 7 | 0 |
| Total | 41 | 2 (5) |
Fig. 4.
PFGE analysis of Vibrio cholerae O1 Ogawa isolates during 2000, 2007 and 2009. 1, 569B-V.cholerae O1 classical; 2, VC 20-V. cholerae O1 El Tor; 3, 1669N-El Tor variant V. cholerae with ctxB gene of classical strain; 4, DM8 (2007); 5, JP47 (2000); 6, water; 7, RJ15 (2009); 8, RJ7 (2009); 9, KP3 (2007); 10, RJ9 (2009); 11, RJ10 (2009); 12, RJ16 (2009); 13, DM7 (2007); 14, KP14 (2007), 15, DM1 (2007).
DISCUSSION
Cholera, a waterborne disease, is transmitted due to contamination of water sources. The present cholera outbreak might have originated from the festival during April 2009 and continued for 3 months up to July 2009 affecting many villages. The water samples collected from the pond water were positive for V. cholerae O1 Ogawa and therefore contact with pond water is a possible source of infection. Analysis of samples of severe diarrhoeal patients from Rajnagar CHC according to month and year (2006–2009) indicated that this was a definite outbreak of diarrhoea which occurred during April–July 2009. The early reporting of the causative organism, chlorination of different water sources and intervention of appropriate control measures stopped the spread of the disease to other adjacent blocks.
Cholera is one of the major health problems in Odisha leading to great morbidity and mortality. Since the last two decades several studies on diarrhoeal disorders have revealed the causative agents of cholera as V. cholerae O1 Ogawa/Inaba biotype El Tor, V. cholerae O139 and V. choleare O1 El Tor variant [9, 14]. During the super-cyclone in 1999 there was a cholera outbreak in Odisha in nine districts including Kendrapada district due to V. cholerae O1 Ogawa El Tor biotype and V. cholerae O139 serogroup [14]. The present large cholera outbreak in the Rajnagar area was due to an El Tor variant of V. cholerae O1 strains that carried the ctxB gene of the classical strain, which was detected for the first time in that area.
Since 1995 interesting fluctuations in the pattern of resistance to various antibiotics have been witnessed in V. cholerae O1 and O139 serogroups in Odisha. The antibiotic resistance profile of V. cholerae O1 and O139, isolated during the 1999 super-cyclone cholera outbreak was: ampicilin, co-trimoxazole, furazolidone, neomycin, nalidixicacid, streptomycin (for O1), and ampicilin, furazolidone, neomycin, streptomycin (for O139) [14]. One hundred percent nalidixic acid-resistant V. cholerae O139 and 100% ampicilin-susceptible V. cholerae O1 have been reported from 2000 onwards. Although emergence of fluoroquinolone-resistant V. cholerae were reported before 2007, 64·8% ciprofloxacin- and 63·2% norfloxacin-resistant V. cholerae were isolated during the 2007 cholera outbreak where tetracycline was the best alternative drug of choice [9]. In the present study, with the exception of norfloxacin, a high percentage of resistance to ciprofloxacin, ampicillin, nalidixic acid, furazolidone, streptomycin and chloramphenicol has been observed which is in accord with reports during the 2007 cholera epidemic in the tribal areas of Odisha [9]. Similar cholera outbreaks have been reported during 2004 from Southern India [16]. Multiple antibiotic resistance in V. cholerae has emerged as a major problem worldwide [17]. Since 1996 there has been a progressive increasing trend of antibiotic resistance towards common fluroquinolones in India, i.e. ciprofloxacin and norfloxacin [9, 18–20]. The V. cholerae strains isolated from this outbreak were sensitive to tetracycline which is different from reports in other regions of India [21]. Multidrug resistance prevents additional changes to disease management. Some antibiotics are deemed unusable for certain population groups, i.e. tetracycline is not recommended for use in children and quinolone is not recommended for pregnant women and children [22]. These rapid shifts in antimicrobial resistance are in contrast to the previous decade when antibiotic resistance in V. cholerae O1 was not a usual phenomenon and multiple antiobiotic-resistant strains were rare. However, this pattern of quick shifts in antibiogram profile indicates an enhanced mobility in genetic elements, which confers resistance to antibiotics. Recent studies by Beaber & Waldor [23] have shown that SXT constin, a mobile genetic element that is a conjugative self-transmissible plasmid, is responsible for antibiotic resistance. Recent studies suggest SXT constin as an important element for horizontal dissemination of antibiotic-resistant genes in bacteria. In SXT constin, the antibiotic resistance genes are clustered in certain frameworks. These genes confer resistance to chloramphenicol, sulfamethaxazole, streptomycin and trimethoprim [24]. Therefore, it can be assumed that V. cholerae strains might harbour SXT constin as reported by Goel et al. [16].
Cholera pathogenesis is a complex process, which involves the synergetic action of several genes. Cholera toxin, constituting AB subunits is considered as the most important epidemic marker in various toxins produced by V. cholerae [25]. Several studies have indicated that there is continual change in the sequence of the ctxB gene. Biotype-specific ctxф is found in V. cholerae strains. El Tor biotype strains harbour ctxElTф while classical strains have ctxclassф. However, the genomic structure of ctxф, in which the cholera toxin genes are maintained, differs between the classical and El Tor biotypes [2, 26]. Interestingly, the new V. cholerae O1 variants carrying the ctxB gene of the classical strains were first isolated in Bangladesh in patients with severe diarrhoea during 2002. In this study all the El Tor V. cholerae isolates possessed the ctxB gene of classical biotype and belonged to the same clone of V. cholerae strains that caused the 2007 cholera epidemic in Odisha (Fig. 4). Similar results with El Tor variant V. cholerae strains causing outbreaks and epidemics of cholera in different countries in Asia, USA, and Africa have been reported [4–6, 9, 18]. Studies from India, Bangladesh and USA show that the classical cholera toxin-producing El Tor strains are now replacing the seventh pandemic El Tor strains [5, 6, 8, 9]. The El Tor variant V. cholerae with classical ctxB trait, spread to the Rajnagar area and caused a large cholera outbreak subsequent to the 2007 cholera epidemic in the southern tribal districts in Odisha.
The present study indicates that after the large cholera epidemic of 2007 in the tribal areas of Odisha, the epidemic spread to the coastal areas with a high percentage of antibiotic resistance. The present cholera outbreak is an early warning that future outbreaks/epidemics of diarrhoeal disorders in Odisha may occur due to this El Tor variant of V. cholerae O1 strain. This requires implementation of continuous surveillance and monitoring of diarrhoeal outbreaks/epidemics in different regions of Odisha and India.
ACKNOWLEDGEMENTS
The authors are grateful to the medical officer and staff of Rajnagar CHC, Kendrapada district for their cooperation and help during the field visits and sample collection. Thanks are also due to the Indian Council of Medical Research (ICMR), New Delhi for funding.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Safa A, et al. Genetic characteristics of Matlab variants of Vibrio cholerae O1 that are hybrids between classical and El Tor biotypes. Journal of Medical Microbiology 2006; 55: 1563–1569. [DOI] [PubMed] [Google Scholar]
- 2.Ansaruzzaman M, et al. Cholera in Mozambique, variant of Vibrio cholerae. Emerging Infectious Diseases 2004; 10: 2057–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair GB, et al. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. Journal of Clinical Microbiology 2006; 44: 4211–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Safa A, et al. Vibrio cholerae O1 hybrid El Tor strains, Asia and Africa. Emerging Infectious Diseases 2008; 14: 987–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nair GB, et al. Cholera due to altered El Tor strains Vibrio cholerae O1 in Bangladesh. Journal of Clinical Microbiology 2006; 38: 4211–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin CS, et al. The origin of the Haitian cholera outbreak strain. New England Journal of Medicine 2011; 364: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taneja N, et al. Outbreaks caused by new variant of V. cholerae O1 El Tor, India. Emerging Infectious Diseases 2009; 15: 352–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar P, et al. A large cholera outbreak due to a new cholera toxin variant of the Vibrio cholerae O1 El Tor biotype in Orissa, Eastern India. Journal of Medical Microbiology 2009; 58: 234–238. [DOI] [PubMed] [Google Scholar]
- 9.Pal BB, et al. Epidemics of severe cholera caused by El Tor Vibrio cholerae O1 Ogawa possessing the ctxB gene of the classical biotype in Orissa, India. International Journal of Infectious Diseases 2010; 14: 384–389. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization. Manual for Laboratory Investigations of Acute Enteric infections, CDD/83.3. Geneva: World Heath Organization, 1987. [Google Scholar]
- 11.Bauer AW, et al. Antibiotic susceptibility testing by a standardized single disc method. American Journal of Clinical Pathology 1996; 45: 493–496. [PubMed] [Google Scholar]
- 12.Khuntia HK, Pal BB, Chhotray GP. Quadruplex PCR assay for simultaneous detection of biotype, serotype, toxigenic potential and central regulating factors of Vibrio cholerae. Journal of Clinical Microbiology 2008; 46: 2399–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morita M, et al. Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiology Immunology 2008; 52: 314–317. [DOI] [PubMed] [Google Scholar]
- 14.Chhotray GP, et al. Incidence and molecular analysis of V. cholerae associated with cholera outbreak subsequently to the super cyclone in Orissa, India. Epidemiology and Infection 2002; 128: 131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinha S, et al. Molecular comparison of toxigenic clinical and non-toxigenic environmental strains of Vibrio cholerae O1 Ogawa isolated during an outbreak of cholera in South India. Indian Journal of Medical Research 2001; 114: 883–889. [PubMed] [Google Scholar]
- 16.Goel AK, et al. Molecular characterization of Vibrio cholerae outbreak strains with altered El Tor biotype from southern India. World Journal of Microbiology and Biotechnology 2010; 26: 281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faruque AS, et al. Emergence of multidrug-resistant strains of Vibrio cholerae O1 in Bangladesh and reversal of their susceptibility to tetracycline after two years. Journal of Health Population and Nutrition 2007; 25: 241–243. [PMC free article] [PubMed] [Google Scholar]
- 18.Garg P, et al. Emergence of fluoroquinolone-resistant strains of Vibrio cholerae O1 biotype El Tor among hospitalized patients with cholera in Calcutta, India. Antimicrobial Agents and Chemotherapy 2006; 45: 1605–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishna BV, Patil AB, Chandrasekhar MR. Fluoroquinolone-resistant Vibrio cholerae isolated during a cholera outbreak in India. Transactions of Royal Society of Tropical Medicine and Hygiene 2006; 100: 224–226. [DOI] [PubMed] [Google Scholar]
- 20.Goel AK, Jiang SC. Genetic determinants of virulence, antibiogram and altered biotype among the Vibrio cholerae O1 isolates from different cholera outbreaks in India. Infection Genetics and Evolution 2010; 10: 815–819. [DOI] [PubMed] [Google Scholar]
- 21.Jesudason MV. Change in serotype and appearance of tetracycline resistance in V. cholerae O1 in Vellore, South India. Indian Journal of Medical Microbiology 2006; 24: 152–153. [DOI] [PubMed] [Google Scholar]
- 22.Sabeena F, et al. In vitro susceptibility of Vibrio cholerae O1 biotype El Tor strains associated with an outbreak of cholera in Kerala, Southern India. Journal of Antimicrobial and Chemotherapy 2001; 47: 361–362. [DOI] [PubMed] [Google Scholar]
- 23.Beaber JW, Waldor MK. Identification of operators and promoters that control SXT conjugative transfer. Journal of Bacteriology 2004; 186: 5945–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beaber JW, Hochhut B, Waldor MK. Genomics and functional analysis of SXT, an integrating antibiotic resistance gene transfer elements derived from Vibrio cholerae. Journal of Bacteriology 2002; 184: 4259–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaper JB, Morris JG Jr., Levine MM. Cholera. Clinical Microbiology Review 1995; 8: 48–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 1996; 272: 1910–1914. [DOI] [PubMed] [Google Scholar]