SUMMARY
A 3-year longitudinal study was conducted on a multi-site farrow-to-finish production system. For each of 18 cohorts at three finishing sites, 50 pigs were randomly selected. Faecal samples were collected every 2 weeks for 16 weeks. Salmonella was cultured from 453 (6·6%) of 6836 faecal samples. The pig-level incidence of Salmonella was 20·8% (187/899 pigs). Salmonella prevalence varied between cohorts and within pigs. The adjusted Salmonella prevalence decreased over the finishing period from 6·4% to 0·8%. Intermittent detection of Salmonella was found in more than 50% of pigs that were positive at more than one collection. The finding that the majority of pigs shed intermittently has implications for surveillance and research study design when determining Salmonella status. The variability in shedding over time, as well as between and within cohorts and pigs suggests that there may be time-variant risk factors for Salmonella shedding in swine.
Key words: Longitudinal, Salmonella, swine
INTRODUCTION
It has been well documented that Salmonella spp. are one of the major causes of foodborne diseases in the USA and worldwide [1–3]. In the USA alone, it is estimated that 1027 million non-typhoidal Salmonella human infections result in 19336 hospitalizations and 378 deaths annually [3], costing US$365 billion in direct medical expenditure per year [2]. Swine are a potential reservoir for human salmonellosis. The most common serotypes isolated in swine (S. Typhimurium, S. Heidelberg, S. Agona, S. Infantis) are common to those found in human cases [4, 5]. It has been suggested that reduction of Salmonella contamination of pork requires interventions at three levels: pre-harvest (farm), harvest (slaughter) and post-harvest (distribution systems and consumer handling) [6, 7]. In order to put in place on-farm control and intervention measures it is crucial to understand the dynamics of Salmonella infection in swine.
A large number of epidemiological studies have been conducted to determine prevalence and risk factors for Salmonella infection in swine. Most of these studies have used a cross-sectional study design. A limited number have assessed the faecal prevalence over time, with longitudinal studies showing high variability in Salmonella shedding at the farm, cohort and individual animal level [8–15]. Longitudinal studies at the pig level during the finishing phase have reported time variability of faecal shedding associated with cohort (or batch) of pigs [11–14]. Intermittent faecal shedding is also commonly reported in epidemiological studies of swine [12–14].
Therefore, longitudinal studies at the individual level based on bacteriological culture should be performed in order to investigate the dynamics of Salmonella infection in swine. The objective of this study was to describe the shedding pattern of Salmonella in faeces of naturally infected finishing pigs.
METHODS
A longitudinal study was conducted on a multi-site farrow-to-finish production system located in the Midwestern USA. The presence of Salmonella had been confirmed by culture of pooled faecal samples prior to initiation of the study. Selection criteria for the production system were willingness to cooperate in a long-term research project and to share health management and production records. The production system had three-site management, meaning that overall production was separated into three stages of production, breeding and farrowing, nursery (weaning until age ∼10 weeks) and finishing (10 weeks to slaughter, 24–26 weeks), with each stage housed at a separate site. The production system had all-in/all-out management in nursery and finishing sites. This system consisted of two farrowing sites (F1 and F2), two nursery sites (N1 and N2), and 12 finishing sites. The farrowing sites had a total inventory of 3700 sows (F1 = 1300, F2 = 2400), the average one-time inventory of the finishing sites was 25 000 (75 000 finishing pigs/year marketed). During the study period the system transitioned from two farrowing sites to one farrowing site of 3000 sows. The number of nursery and finisher sites remained unchanged. Three finishing sites (A, B, C) were conveniently selected, based on building design and willingness to participate in the study. At each finishing site one barn was selected for study inclusion. Site A had four barns in separate buildings. Pigs were allocated into 40 pens (20–25 pigs per pen). Sites B and C had identical building structures. Each site had four barns grouped in two buildings (two barns/rooms with one shared wall). Each barn housed about 1000 pigs. Pigs were housed in 12 pens; ten pens were initially stocked with pigs at placement (eight pens with a range of 100–125 pigs and two pens with a range of 40–50 pigs). The remaining two pens were used for sick pigs or pigs deemed to be at risk for illness. Sites A and B were finishing sites (age 10–26 weeks). Site C transitioned to a weaning-to-finishing site after the second cohort of pigs. For the weaning-to-finishing cohorts at site C, piglets were placed in the barn at age 3 weeks and remained until marketing. Finishing site A received pigs from nursery N1 in all cohorts; site B received pigs from nursery N2 in four cohorts and from N1 and N2 in the last two cohorts of pigs. The first cohort for site C was supplied from nursery N2, for all other cohorts piglets were placed directly from the farrowing sites due to the transition to a weaning-to-finishing site.
Sample collection
Nursery sampling
In order to evaluate the Salmonella status of the cohort of pigs prior to sampling during the finishing phase, ten pools were collected from the nursery rooms ∼1 week prior to movement to the finishing barn. A pool consisted of a minimum of 5 g fresh faecal material collected from five different locations on the same pen floor (25 g/pool). In the weaning-to-finishing site (site C), ten pools were collected from ten random pens when pigs were aged ∼9 weeks.
Environmental sampling
Finisher barns were cleaned and disinfected between batches of pigs. The disinfectants (Synergize, Preserve International, USA and VirkonS, Antec International, UK) were alternated following the standard operating procedure of the production system. In order to assess contamination, culture of environmental samples was performed after cleaning and disinfection and before placement of pigs in the barn. Drag swabs and environmental swab samples were obtained from cleaned and disinfected floors, walls, gates and feeders/drinkers following previously described methods [16]. Briefly, swabs were moistened with 10 ml of sterile buffered peptone water (BPW, Acumedia, Neogen Corporation, USA) before collection. To sample floors, one drag swab was used for four pens in site A. In sites B and C, one drag swab was used per 1–2 pens depending on pen size. To sample other environmental surfaces, a single 4 × 4 gauze moistened with BPW was used to sample each surface. Ten, five, three and two swabs were collected from floors, walls, gates and feeders/waterers, respectively in each barn prior to every cohort.
Individual faecal sampling
At the beginning of each cohort, 50 pigs (aged 10 ± 2 weeks) were randomly selected and individually identified with ear tags. Random number generation was conducted in Microsoft Excel 2007 (Microsoft, USA). In site A, a simple random sample was generated to select one pig from every pen (n = 40). Another ten pens were randomly selected to identify and select a second pig using a simple random sample (additional ten pigs for a total of n = 50/barn; 30 pens with one study pig, ten pens with two study pigs). A random proportional sampling scheme based on the number of pigs in each pen was conducted in sites B and C within each cohort. A range of 1–7 pigs per pen was selected. In sites B and C no pigs were selected for study inclusion from the pens identified for sick or at-risk pigs.
Individual faecal samples were collected from the rectum with gloved hand, and placed in sterile containers (Specimen cups, VWR International LLC, USA). Gloves for collecting the faecal samples were changed between pigs. After collection, samples were stored at ambient temperature for transport to the laboratory. Individual pig faecal samples were collected every 2 weeks for 16 weeks (eight total sampling periods per pig). A total of 400 individual samples (50 pigs × 8 sample periods) per cohort and 7200 faecal samples overall (400 samples × 18 cohorts) were planned for collection.
Bacteriological culture
Faecal samples
Bacteriological culture for Salmonella was performed by the Diagnostic Center for Population and Animal Health, Michigan State University. Faecal samples were transported to the laboratory the day of collection or stored for 48 h at 2·8 °C.
Faecal samples were cultured using standard enrichment methods from Davies et al. [17]. Briefly, for pooled samples from the nursery, 25 g of pooled faecal samples were diluted in 225 ml tetrathionate broth (TTB) (Becton Dickinson, USA) and incubated at 37 °C for 48 h. For individual pig faecal samples 10 g of the individual faecal samples were inoculated into 90 ml TTB and incubated at 37 °C for 48 h. After incubation, an aliquot (100 μl) of the faecal-TTB solution was inoculated into 9·9 ml Rappaport–Vassiliadis broth (RV, Becton Dickinson) and incubated at 42 °C for 24 h. The RV broth was plated onto xylose lysine tergoitol 4 agar (XLT4, Remel, Thermo Fisher Scientific, USA) selective agar plates and incubated at 37 °C, overnight. Suspect Salmonella colonies from microbiological culture were screened using Salmonella poly O antisera antiglutination (Becton Dickinson, USA).
Environmental samples
Environmental samples were cultured following the same protocol [17] using a volume of TTB sufficient to submerge the swabs (∼60 ml).
Data analysis
Bacteriological culture data were entered into an Excel 2007 spreadsheet using appropriate coding and subsequently verified for accuracy by checking each entry with the original hard copy result. The spreadsheets were transferred to a relational database (Access 2007, Microsoft Corporation). Data were retrieved from the database and imported into SAS 9.3 (SAS Institute Inc., USA) for data management and statistical analysis.
Descriptive statistics of demographic data (number of pigs sampled, gender), loss to follow-up and morbidity were presented in proportions.
Descriptive statistics of bacteriological culture were described for the nursery, barn environment, site, cohort, pig, and faecal sample (observation). Salmonella apparent prevalence (proportion of positive samples/tested) and respective 95% confidence intervals (CI) were estimated at each unit of observation: cohort (e.g. all collections combined within cohort), site (e.g. all cohorts combined), pig age (by collection period) and individual sample. Logistic models (Proc Glimmix, SAS 9.3, SAS Institute Inc.) accounting for clustering within pig, cohort and site, were applied to compare prevalence at each unit of investigation. Logistic models accounting for clustering within cohort and site were used to estimate adjusted prevalence for cohort and site, to compare the proportion of Salmonella-positive samples in sites, and to estimate the association between the proportion of Salmonella-positive samples by collection period and proportion of Salmonella-positive samples in nursery and environmental samples. Model-adjusted apparent prevalences are presented for cohorts, age and site. Pearson χ2 analysis with Bonferroni adjustment was used to compare the proportion of Salmonella-positive samples in the cohorts classified based on nursery and barn environmental Salmonella status (positive or negative). A significance level of 0·05 was used for all comparisons.
Patterns and duration of shedding were estimated for those pigs which met the following inclusion criteria: (1) survival until marketing (excluded n = 3 dead and n = 5 early shipment); (2) no more than one period from which a sample was not collected (excluded n = 1); (3) had no more than two negative cultures between two positive culture results (excluded n = 10).
In order to estimate the duration of shedding of individual pigs, we assumed that the shedding began 7 days prior the first detected positive culture and lasted until 7 days after the last isolation. The 7-day interval was selected taking into account data from experimental studies indicating that pigs start to shed Salmonella as early as 2–7 days post-exposure [18] and as late as 7–14 days [19, 20], after exposure to a Salmonella-contaminated environment or when commingled with pigs shedding Salmonella. This interval (7 days) is also the midpoint between two consecutive sampling periods.
RESULTS
Demographic results
A total of 900 pigs were selected for inclusion in the study. Forty-six percent (410/900) were barrows or castrated males and 54% (490/900) were females.
The total loss to follow-up for faecal sample collection was 5·1% (364/7200). Causes for loss to follow-up were: death, unable to collect a specimen (e.g. empty rectum, sick animal), or shipped to market prior to final collection. A total of 17 (17/900, 1·9%) pigs died during the study. The majority of the pigs were sampled eight (71·4%, 643/900) or seven (23·1%, 208/900) times.
At the observation level (individual pig × number of sample periods pig observed) diarrhoea was described in 2·4% (164/6836) of the observations. At the pig level, 15·1% (136/900) were observed to have diarrhoea at least once.
Nursery and barn environment
The total proportion of positive samples and respective 95% CI of nursery, barn environment samples and sites, stratified by cohort are summarized in Table 1. Pooled faecal samples from the source nursery were collected and cultured in 17/18 cohorts. Salmonella was detected in at least one nursery pool in 76·5% (13/17) of the cohorts. A total of 36·5% (62/170) of the pooled nursery samples were Salmonella positive. The proportion of positive nursery samples ranged from 0% to 100% in the cohorts (Table 1).
Table 1.
Proportion of samples positive for Salmonella spp. by site and cohort (samples represent individual faecal samples, pooled faecal samples from the source nursery and barn environmental swabs) and respective 95% confidence intervals (CI)
| Site/cohort | Environment* | Nursery† | Cohort | ||||
|---|---|---|---|---|---|---|---|
| Total positive faecal samples tested, n/N (%) | Adjusted Salmonella apparent prevalence‡ | ||||||
| % | (95% CI) | % | (95% CI) | % | (95% CI) | ||
| Site A | |||||||
| 1 | 5·0 | (0·1–24·9) | 60·0 | (26·2–87·9) | 30/388 (7·7) | 7·6 | (4·5–12·5) |
| 2 | 25·0 | (8·7–49·1) | 80·0 | (44·4–97·5) | 42/396 (10·6) | 10·4 | (6·7–15·7) |
| 3 | 0·0 | n.a. | n.s. | n.s. | 7/390 (1·8) | 1·9 | (0·7–5·4) |
| 4 | 85·0 | (62·1–96·8) | 60·0 | (26·2–87·8) | 46/382 (12·0) | 12·1 | (7·9–18·3) |
| 5 | 5·0 | (0·1–24·9) | 20·0 | (2·5–55·6) | 16/386 (4·1) | 4·7 | (2·5–8·6) |
| 6 | 5·0 | (0·1–24·9) | 10·0 | (2·5–44·5) | 6/396 (1·5) | 1·9 | (0·7–5·0) |
| Site B | |||||||
| 1 | 10·0 | (1·2–31·7) | 10·0 | (0·25–44·5) | 12/383 (3·1) | 3·3 | (1·5–7·1) |
| 2 | 10·0 | (1·2–31·7) | 100·0 | (69·2–100) | 156/354 (44·1) | 43·1 | (35·7–50·9) |
| 3 | 0·0 | n.a. | 20·0 | (2·5–55·6) | 4/379 (1·1) | 1·5 | (0·50–4·5) |
| 4 | 30·0 | (11·9–54·3) | 100·0 | 69·2–100) | 57/339 (16·8) | 16·4 | (11·3–23·0) |
| 5 | 5·0 | (0·1–24·9) | 0·0 | n.a. | 2/362 (0·6) | 1·1 | (0·3–4·0) |
| 6 | 0·0 | n.a. | 0·0 | n.a. | 16/386 (4·1) | 4·2 | (2·1–8·3) |
| Site C | |||||||
| 1 | 0·0 | n.a. | 0·0 | n.a. | 1/387 (0·3) | 0·6 | (0·1–3·0) |
| 2 | 0·0 | n.a. | 80·0 | (44·4–97·5) | 24/393 (6·1) | 5·7 | (3·2–10·1) |
| 3 | 0·0 | n.a. | 0·0 | n.a. | 6/390 (1·5) | 1·6 | (0·6–4·6) |
| 4 | 15·0 | (3·2–37·9) | 60·0 | (26·2–87·8) | 18/371 (4·9) | 4·5 | (2·4–8·8) |
| 5 | 5·0 | (0·1–24·9) | 10·0 | (2·5–55·6) | 0/376 (0·0) | 0·0 | n.a. |
| 6 | 0·0 | n.a. | 10·0 | (2·5–44·5) | 10/378 (2·6) | 2·5 | (1·0–6·1) |
n.s., Not sampled; n.a., not applicable.
Total of 20 environmental samples per cohort.
Total of 10 pooled samples per cohort.
Adjusted apparent prevalence accounting for clustering.
Environmental samples were collected for all cohorts. Salmonella was detected in at least one environmental swab in 61·1% (11/18) of the cohorts. The total number of positive swabs was 40 (40/360, 11·1%). The proportion of positive barn environmental swabs ranged from 0% to 85% in the cohorts (Table 1).
Site, cohort and age apparent prevalence
Salmonella was isolated from at least one sample type (nursery, environmental or individual faecal) sample at all three sites. In 17/18 cohorts at least one individual faecal sample was positive. Salmonella was cultured from 6·6% (453/6836, 95% CI 6·0–7·2) of individual faecal samples. The proportion of positive faecal samples within a cohort (eight collection periods combined per cohort) ranged from 0% to 44·1%. Within site, the adjusted Salmonella apparent prevalence per cohort (six cohorts/site) ranged from 1·9% (95% CI 0·7–5·4) to 12·1% (95% CI 7·9–18·3) in site A, 1·1% (95% CI 0·3–4·0) to 43·1% (95% CI 35·7–50·9) in site B and 0% to 5·7% (95% CI 3·2–10·1) in site C (Table 1).
For the 17 cohorts with both nursery and environmental swab collections, there were nine cohorts with at least one positive sample in both sample types that also had at least one individual faecal sample positive. Three cohorts were Salmonella positive in the nursery but Salmonella negative for environmental swabs. One cohort was negative in the nursery and had at least one environmental swab positive. Three cohorts were negative for both sample types. One cohort had at least one positive sample for both nursery and environmental samples but was negative for individual faecal samples. The proportion of Salmonella-positive samples was significantly greater in those cohorts in which both the nursery and the barn environment were Salmonella positive (P < 0·05) (Table 2). No significant difference was found in cohorts negative for both types of samples and nursery positive and environment negative or nursery negative and environment positive (P > 0·05) (Table 2). There was a positive association between the proportion of positive samples at each collection and the proportion of positive pooled nursery samples (P < 0·0001). There was also a trend of a positive association between the proportion of positive barn environmental swabs (P = 0·07) and the proportion of positive faecal samples at each collection.
Table 2.
Distribution of cohorts and proportion of samples positive for Salmonella spp. by the Salmonella status of nursery and environmental swabs*
| Nursery and environment status | No. of positive cohorts | No. of negative cohorts | Positive faecal samples tested (n/N) | Proportion of positive faecal samples (%)† | 95% CI |
|---|---|---|---|---|---|
| Nursery+ environment+ | 9 | 1 | 383/3771 | 10·2a | 9·2–11·2 |
| Nursery+ environment– | 3 | 0 | 38/1150 | 3·3bd | 2·4–4·5 |
| Nursery− environment+ | 1 | 0 | 2/362 | 0·6ce | 0–2·0 |
| Nursery− environment– | 3 | 0 | 23/1163 | 2de | 1·3–3·0 |
CI, Confidence interval.
Seventeen cohorts are represented, one cohort was excluded as no nursery samples were collected.
Different superscript letters indicate a significant difference (P < 0·05) of proportion of positive faecal samples.
Although there was a numerical difference of the overall proportion of positive samples in the sites [site A: 6·3% (147/2338); site B: 11·2% (247/2203); site C (2·6% (59/2295)], no significant difference in the adjusted Salmonella prevalence (P > 0·05) between sites was found (site A: 5·1%, 95% CI 1·7–13·8; site B: 5·5%, 95% CI 1·9–15·1; site C: 1·8%, 95% CI 0·6–5·5).
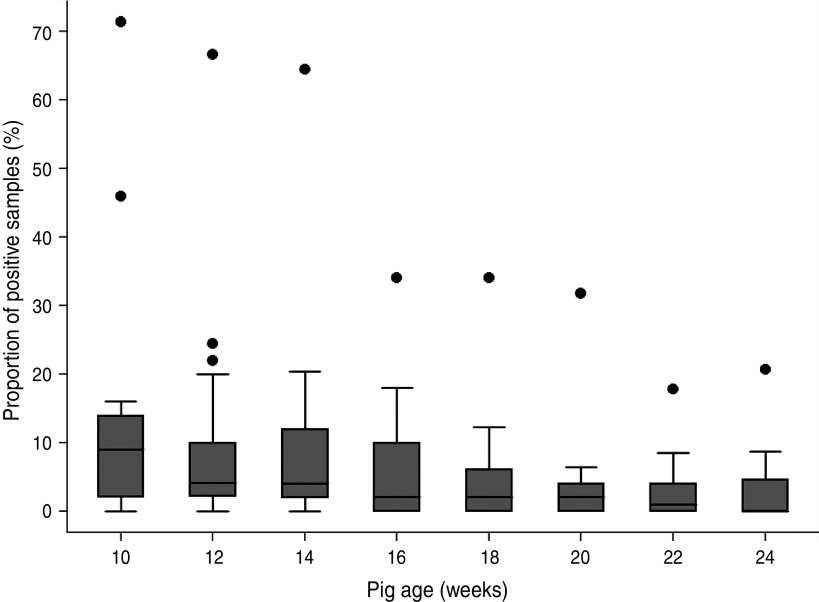
For all 18 cohorts, the proportions of positive samples per cohort were plotted by age (Fig. 1). The overall median was 2·0%; 25%, 75% and 95% quartiles were 0%, 7·4% and 25·5%. The adjusted Salmonella apparent prevalences were 6·4% (95% CI 3·2–12·6), 4·3% (95% CI 2·1–8·8) and 3·7% (95% CI 1·8–7·8) at ages 10, 12 and 14 weeks, respectively, and were significantly different (P < 0·0001) from the end of finishing period (at 24 weeks: 0·8%, 95% CI 0·3–2·0). There were no significant differences (P > 0·05) in adjusted Salmonella apparent prevalence in the last four collections (18 weeks: 1·2%, 95% CI 0·6–2·7; 20 weeks: 0·9%, 95% CI 0·6–1·5; 22 weeks: 0·6%, 95% CI 0·3–1·5; 24 weeks: 0·3%, 95% CI 0·3–2·0).
Fig. 1.
Box plot representing the distribution of Salmonella-positive faecal samples within each cohort by pig age.
Pig apparent prevalence and duration of shedding
Most pigs were detected as Salmonella positive for the first time at the first collection period (age 10 weeks, 61·5%, 115/187). This was followed by collections 2 and 3 (ages 12 and 14 weeks, 14·4%, 27/187), collection 4 (age 16 weeks, 4·3%, 8/187), collection 6 (age 20 weeks, 3·7%, 7/187) and collections 5, 7 and 8 (ages 18, 22 and 24 weeks, 0·5%, 1/187).
Overall incidence of Salmonella was 20·8% (187/899 pigs, 95% CI 18·2–23·6). Of the positive pigs, 87 were culture positive once (46·5%) and 27 (14·4%), 31 (16·6%), 17 (9·1%), 10 (5·4%), six (3·2%), and seven (3·7%) pigs were positive, two, three, four, five, six, and seven times, respectively. Only two pigs were Salmonella positive in all eight collection periods. The duration of shedding was clustered within site and cohort. The majority of the pigs with two or more positive samples belonged to site B (61/100 pigs), with sites A and C having 30 and nine pigs detected as culture positive for Salmonella at two or more collection periods, respectively. In site B, two cohorts had the majority (53/61) of pigs with two or more positive samplings (40 and 13 pigs, in cohorts 2 and 4, respectively). In site A, two cohorts had the majority of pigs with two or more positive samples (19/30; nine and 10 pigs in cohorts 2 and 4, respectively).
There were 95 pigs detected positive on more than two sampling occasions that had consecutive sampling collections. Of these, 46·3% (44/95) had consecutive positive culture samplings, 23·2% (22/95) had one culture-negative faecal sample between positive culture samples and 30·5% (29/95) were culture negative on two or more occasions between the first and last culture-positive sample collection period for each pig.
A total of 168 pigs met the inclusion criteria for estimation of shedding period. The median time of shedding was 14 days (s.d. = 32·5, range 14–112 days). Eighty-five (50·6%) pigs shed for 14 days, 15 (8·9%) pigs shed for 28 days, 18 (10·7%) pigs shed for 42 days, 11 (6·6%) pigs shed for 56 days and 39 (23·2%) pigs shed for between 70 and 112 days.
DISCUSSION
Estimates of Salmonella prevalence in finishing pigs in the USA range from 3·4% to 48% [10, 11, 15, 21–26]. The observed proportion of Salmonella-positive samples and cohort prevalence were within the range of these reports. The overall incidence of positive pigs was 20·8%, which is, to the best of our knowledge, the first estimate of incidence in naturally infected swine in one large swine production system in the USA.
Several longitudinal studies have been conducted at the farm [8, 9, 15, 27, 28] and cohort/pig group [10, 29, 30] level. A limited number of studies have repeatedly sampled individual pigs [11–14]. Similar to these previous studies, we report variability of prevalence by cohort and within pig. This may suggest there are risk factors at the cohort and pig level that might be associated with Salmonella prevalence. This variability reinforces that point estimates of prevalence might misclassify farm and pig status and that prospective studies are needed to assess time-dependent risk factors for Salmonella in swine with consideration for risk factors that may be distributed at different levels of organization (farm, cohort, pig) [31].
The majority of the pigs were detected as Salmonella positive at the beginning of the finishing period (age 10 weeks). Although individual sampling during the nursery phase was not performed in this study, Salmonella was isolated in nursery pool samples from a majority of the cohorts and there was a positive association between the nursery pool prevalence and the proportion of positive individual samples by collection period. This suggests that pigs were exposed to Salmonella in the nursery and may have been shedding at arrival to the finishing barn. Salmonella shedding during the nursery phase has been reported [13], in some cases representing the peak prevalence during the nursery period [12]. Several authors have reported increased prevalence when pigs were moved to finishing units [13, 30], which may be a result of multiple potential factors: stress caused by transportation, comingling with new pigs, changes in feed type and exposure to residual contamination [13, 30].
Contaminated facilities are a source of Salmonella [10, 11, 32, 33] and may in part explain the high prevalence of Salmonella at the first collection period. In agreement with other authors we observed that cleaning and disinfection did not eliminate Salmonella in the barn environment. The elimination of Salmonella from the barn environment is difficult and residual contamination might be responsible for new infections [11, 14, 32, 33]. The trend of a positive association between the proportion of positive barn environmental swabs and the proportion of positive individual samples suggests that the contaminated environment may have contributed to Salmonella infections in the finishing phase.
There was a significant difference in Salmonella prevalence between age groups and shedding decreased as pig age increased. Other authors have reported a decrease in prevalence during the finishing period [12, 13, 30]. It is unclear whether this association represents the natural history of Salmonella in swine, with young animals being more susceptible and ultimately clearing the infection over time, or if other factors are involved. Further research to understand whether control of Salmonella in young pigs ultimately would decrease the risk of shedding at the time of harvest is warranted.
More than 50% of the Salmonella-positive pigs were detected at two sampling periods. Other studies that have followed pigs over time have reported a lower percentage of pigs that were detected on more than two occasions. Beloeil et al. [14] reported that a majority of pigs shed only once in weekly samplings. In other studies the comparison is not as direct, since in this study the sampling period was more frequent than other reports [11, 12]. In this study, pigs identified as Salmonella positive on more than two occasions were clustered within site and cohort. This is in agreement with Kranker et al. [12] who reported characteristic patterns (shorter or longer periods of shedding) by cohort. This might suggest that there are cohort-level effects that are related to duration of shedding or transmission dynamics.
The median and range of shedding duration in this study is similar to that described by Kranker et al. [12] who reported a mean duration of shedding of 18 or 26 days (range 7–101 days). Although our estimates are limited by an imperfect diagnostic test, the sampling frequency and the assumption of no new infections, these data present critical information regarding the duration of shedding in naturally infected swine. Further research to understand risk factors for duration of Salmonella shedding in swine are warranted.
There was intermittent detection of shedding in more than 50% of the pigs with multiple culture-positive collections. Salmonella carriers can shed intermittently and for long periods [11, 12, 34]. It is difficult to separate intermittent shedding of Salmonella from intermittent detection or new infections. Despite being an imperfect diagnostic test, faecal culture is considered the ‘gold standard’ for Salmonella isolation. Estimates of the relative sensitivity of faecal culture range from 6·5% to 95%, depending on culture method and parallel estimation of the sensitivity [17, 22, 35–38]. Although a relative short sampling interval (2 weeks) was conducted in this study, new infections could occur between sampling occasions. Therefore, the intermittent shedding could be intermittent detection of an ongoing infection or a new infection after clearance of a previous infection.
These data represent one production company in one region of the USA. Although this may limit external validity, we believe that this limitation is minimal. This farm is typical of many US swine production systems in size and production practices. Furthermore, there are many similarities between the results in this study compared to studies both in the USA and other countries. A further limitation for interpretation is in regard to the univariate analyses reported in this paper. Statistical inferences regarding these associations should be interpreted with caution, as univariate analyses may bias the results reported. Further risk-factor analyses using multivariate analysis were not the aim of this paper and are currently in process. Despite this limitation, the findings presented in this paper are consistent with what has been previously reported in the literature [11–14, 30, 32, 33].
These descriptive data regarding the incidence, duration and pattern of shedding in swine provide critical data for understanding risk factors for Salmonella in finishing swine. The variability and clustering of Salmonella shedding by site, cohort and pig not only suggest a need to evaluate time-variant risk factors, but also guide the design of future epidemiological studies for identification of potential risk factors at different levels of clustering (site, cohort, pig). Future research of the epidemiology of Salmonella in swine should focus on longitudinal study designs focused on multilevel and time-variant risk factors. This study also reinforces that point-source estimates of prevalence might misclassify herd or pig Salmonella status.
ACKNOWLEDGEMENTS
This work was supported by USDA-NRI, Epidemiologic Approaches to Food Safety Grant 2007–01775. The authors thank the participating pork producers and their staff for collaborating in the investigation, and staff and students at Michigan State University for their technical support.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Greig JD, Ravel A. Analysis of foodborne outbreak data reported internationally for source attribution. International Journal of Food Microbiology 2009; 130: 77–87. [DOI] [PubMed] [Google Scholar]
- 2.Center for Disease Control and Prevention. Vital signs: incidence and trends of infection with pathogens transmitted commonly through food: foodborne diseases active surveillance network, 10 U. S. Sites, 1996–2010. Morbidity and Mortality Weekly Report 2011; 60: 749–755. [PubMed] [Google Scholar]
- 3.Scallan E, et al. Foodborne illness acquired in the United States – major pathogens. Emerging Infectious Diseases 2011; 17: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foley SL, Lynne AM, Nayak R. Salmonella challenges: prevalence in swine and poultry and potential pathogenicity of such isolates. Journal of Animal Science 2008; 86: E149–162. [DOI] [PubMed] [Google Scholar]
- 5.Center for Disease Control and Prevention. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food – 10 states, 2009. Morbidity and Mortality Weekly Report 2010; 59: 418–422. [PubMed] [Google Scholar]
- 6.Boyen F, et al. Non-typhoidal Salmonella infections in pigs: a closer look at epidemiology, pathogenesis and control. Veterinary Microbiology 2008; 130: 1–19. [DOI] [PubMed] [Google Scholar]
- 7.Lo Fo Wong DMA, et al. Epidemiology and control measures for Salmonella in pigs and pork. Livestock Production Science 2002; 76: 215–222. [Google Scholar]
- 8.Rajic A, et al. Longitudinal study of Salmonella species in 90 Alberta swine finishing farms. Veterinary Microbiology 2005; 105: 47–56. [DOI] [PubMed] [Google Scholar]
- 9.Farzan A, et al. A longitudinal study of the Salmonella status on Ontario swine farms within the time period 2001–2006. Foodborne Pathogens and Disease 2008; 5: 579–588. [DOI] [PubMed] [Google Scholar]
- 10.Dorr PM, et al. Longitudinal study of Salmonella dispersion and the role of environmental contamination in commercial swine production systems. Applied and Environmental Microbiology 2009; 75: 1478–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funk JA, Davies PR, Nichols MA. Longitudinal study of Salmonella enterica in growing pigs reared in multiple-site swine production systems. Veterinary Microbiology 2001; 83: 45–60. [DOI] [PubMed] [Google Scholar]
- 12.Kranker S, et al. Longitudinal study of Salmonella enterica serotype Typhimurium infection in three Danish farrow-to-finish swine herds. Journal of Clinical Microbiology 2003; 41: 2282–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nollet N, et al. Distribution of Salmonella strains in farrow-to-finish pig herds: a longitudinal study. Journal of Food Protection 2005; 68: 2012–2021. [DOI] [PubMed] [Google Scholar]
- 14.Beloeil PA, et al. Longitudinal serological responses to Salmonella enterica of growing pigs in a subclinically infected herd. Preventive Veterinary Medicine 2003; 60: 207–226. [DOI] [PubMed] [Google Scholar]
- 15.Rostagno MH, Hurd HS, McKean JD. Variation of bacteriologic and serologic Salmonella enterica prevalence between cohorts within finishing swine production farms. Food Research International 2012; 45: 867–870. [Google Scholar]
- 16.Kingston DJ. A comparison of culturing drag swabs and litter for identification of infections with Salmonella spp. in commercial chicken flocks. Avian Diseases 1981; 25: 513–516. [PubMed] [Google Scholar]
- 17.Davies PR, et al. Comparison of methods for isolating Salmonella bacteria from faeces of naturally infected pigs. Journal of Applied Microbiology 2000; 89: 169–177. [DOI] [PubMed] [Google Scholar]
- 18.Fedorka-Cray PJ, et al. Transmission of Salmonella Typhimurium to swine. Veterinary Microbiology 1994; 41: 333–344. [DOI] [PubMed] [Google Scholar]
- 19.van Winsen RL, et al. Monitoring of transmission of Salmonella enterica serovars in pigs using bacteriological and serological detection methods. Veterinary Microbiology 2001; 80: 267–274. [DOI] [PubMed] [Google Scholar]
- 20.Osterberg J, Lewerin SS, Wallgren P. Direct and indirect transmission of four Salmonella enterica serotypes in pigs. Acta Veterinaria Scandinavica 2010; 52: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies PR, et al. Prevalence of Salmonella in finishing swine raised in different production systems in North Carolina, USA. Epidemiology and Infection 1997; 119: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurd HS, et al. Estimation of the Salmonella enterica prevalence in finishing swine. Epidemiology and Infection 2004; 132: 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahnson PB, et al. Herd-level risk factors for Salmonella enterica subsp. enterica in U. S. market pigs. Preventive Veterinary Medicine 2006; 76: 249–262. [DOI] [PubMed] [Google Scholar]
- 24.Gebreyes WA, Thakur S, Morrow WEM. Comparison of prevalence, antimicrobial resistance, and occurrence of multidrug-resistant Salmonella in antimicrobial-free and conventional pig production. Journal of Food Protection 2006; 69: 743–748. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, et al. Sub-iliac lymph nodes at slaughter lack ability to predict Salmonella enterica prevalence for swine farms. Foodborne Pathogens and Disease 2010; 7: 795–800. [DOI] [PubMed] [Google Scholar]
- 26.USDA-APHIS. Salmonella on U. S. swine sites – prevalence and antimicrobial susceptibility. (http://www.aphis.usda.gov/animal_health/nahms/swine/downloads/swine2006/Swine2006_is_salmonella.pdf). Accessed 23 February 2011.
- 27.van der Wolf PJ, et al. A longitudinal study of Salmonella enterica infections in high-and low-seroprevalence finishing swine herds in The Netherlands. Veterinary Quarterly 2001; 23: 116–121. [DOI] [PubMed] [Google Scholar]
- 28.Rajic A, et al. Salmonella infections in ninety Alberta swine finishing farms: serological prevalence, correlation between culture and serology, and risk factors for infection. Foodborne Pathogens and Disease 2007; 4: 169–177. [DOI] [PubMed] [Google Scholar]
- 29.Merialdi G, et al. Longitudinal study of Salmonella infection in Italian farrow-to-finish swine herds. Zoonoses and Public Health 2008; 55: 222–226. [DOI] [PubMed] [Google Scholar]
- 30.Vigo GB, et al. Salmonella enterica subclinical infection: bacteriological, serological, pulsed-field gel electrophoresis, and antimicrobial resistance profiles-longitudinal study in a three-site farrow-to-finish farm. Foodborne Pathogens and Disease 2009; 6: 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dohoo IR, Martin W, Stryhn H. Veterinary Epidemiologic Research, 2nd edn. Charlottetown, Prince Edward Island, Canada: VER Inc., 2010, pp. 173–174. [Google Scholar]
- 32.Mannion C, et al. Efficacy of cleaning and disinfection on pig farms in Ireland. Veterinary Record 2007; 161: 371–375. [DOI] [PubMed] [Google Scholar]
- 33.Zewde BM, et al. Comparison of swiffer wipes and conventional drag swab methods for the recovery of Salmonella in swine production systems. Journal of Food Protection 2009; 72: 142–146. [DOI] [PubMed] [Google Scholar]
- 34.Scherer K, et al. Time course of infection with Salmonella Typhimurium and its influence on faecal shedding, distribution in inner organs, and antibody response in fattening pigs. Journal of Food Protection 2008; 71: 699–705. [DOI] [PubMed] [Google Scholar]
- 35.Funk JA, Davies PR, Nichols MA. The effect of faecal sample weight on detection of Salmonella enterica in swine feces. Journal of Veterinary Diagnostic Investigation 2000; 12: 412–418. [DOI] [PubMed] [Google Scholar]
- 36.Rostagno MH, et al. Culture methods differ on the isolation of Salmonella enterica serotypes from naturally contaminated swine faecal samples. Journal of Veterinary Diagnostic Investigation 2005; 17: 80–83. [DOI] [PubMed] [Google Scholar]
- 37.Love BC, Rostagno MH. Comparison of five culture methods for Salmonella isolation from swine faecal samples of known infection status. Journal of Veterinary Diagnostic Investigation 2008; 20: 620–624. [DOI] [PubMed] [Google Scholar]
- 38.Funk J. Pre-harvest food safety diagnostics for Salmonella serovars. Part 1: Microbiological culture. Journal of Swine Health and Production 2003; 11: 87–90. [Google Scholar]