SUMMARY
This longitudinal field study investigated the hypothesis that persistently high antibody levels indicate a high risk of Salmonella Dublin shedding in animals in 14 endemically infected dairy herds. A hierarchical multivariable logistic regression was used to analyse 6614 paired faecal cultures and four types of temporal antibody profiles from cattle aged ≥180 days. Age and repeated measurements on animals nested within herds were taken into account. Overall, the prevalence of faecal shedders was low (0·3% and 2·8% in the lowest and highest risk groups, respectively). An important predictor of faecal shedding was young age. There was a significant, but modest increase in risk in cattle with persistently high or recently increased antibody levels, but no difference between these two groups. Contrary to previous recommendations, the detection of carriers by the use of repeated antibody testing is not likely to be a plausible control option in most Salmonella Dublin-infected dairy herds.
Key words: Bacterial infections, control, epidemiology, Salmonella, Veterinary epidemiology and bacteriology
INTRODUCTION
Salmonella enterica subsp. enterica serovar Dublin (S. Dublin) is a gastrointestinal bacterial infection prevalent in many cattle herds worldwide. It causes increased morbidity, mortality and production losses [1–3]. Even though it is host-adapted, it occasionally causes human infections that tend to be severe due to the invasive nature of this infection [4].
Controlling S. Dublin in cattle herds requires intervention to minimize the exposure to bacteria in the environment or shed by other animals in the herd [5, 6]. A test-and-cull strategy to remove persistently infected cattle has long been considered an important control element [7–10]. However, this recommendation is mainly based on limited, potentially biased study material or experimentally induced infections [7, 11, 12]. If a test strategy involving repeated antibody testing of all cattle or groups of cattle in infected herds is implemented as part of a control programme, and previous recommendations concerning the interpretation of the obtained antibody profiles for each individual animal are used [8, 11, 13], it may lead to a long list of heifers and cows suspected as S. Dublin carriers, particularly under high prevalence conditions [14]. It is often not economically feasible for the farmer to cull so many animals. Furthermore, there are indications in previous studies that not all of the suspected carrier animals actually pose a risk to the herd [15, 16]. Hence, there is a need to quantify the risk posed by cattle with different temporal antibody profiles (TAPs) to facilitate prioritization of risk management or culling decisions in the control of S. Dublin.
The objective of this study was to investigate the hypothesis that cattle with persistently high antibody levels are at higher risk of shedding S. Dublin through faeces than cattle with recent increases, fluctuating or moderately high antibody levels, or low antibody levels. This study focused on S. Dublin for two reasons: (1) S. Dublin is the most commonly isolated serotype in Danish cattle, this is also true for several other countries; (2) detection of persistently infected carriers by use of serology is to the best of the author's knowledge only used for control of S. Dublin.
MATERIALS AND METHODS
Selection of herds and sampling
In 2000, a total of 14 dairy herds in the southern part of the Jutland peninsula of Denmark were selected to participate in a field study based on having bulk-tank milk S. Dublin ELISA results >50 ODC% (background-corrected optical density values [17]). At that time around 25% of the ∼9000 Danish dairy herds had bulk-tank milk S. Dublin ELISA values >50 ODC%. Herd size of the 14 selected herds was between 15 and 121 lactating cows (and between 69 and 262 animals in total) across all herd visits. Eleven of the herds consisted mostly of the Danish Holstein breed and other three herds consisted mostly of the Jersey breed. Management, housing system and feeding practices were not recorded, but were likely to be similar to other S. Dublin-infected herds in Denmark at the time.
S. Dublin was isolated from faecal samples at least once from these herds during the study period, from the beginning of 2000 to the beginning of 2002, and there were indications of the herds being endemically infected throughout the study period (i.e. continued serological responses in all age groups of cattle, or faecal or environmental samples being culture positive). All except one of the 14 herds were visited five times with about 3 months between each visit; the last herd was visited four times. At each visit, blood samples were collected from all calves, young stock and dry cows on the premises, and milk samples were collected from all lactating cows at the morning milking for serological analysis. Faecal samples were collected rectally from all accessible animals and placed into marked containers with the aim of obtaining at least 50 g from each animal. The samples were transported directly to the Danish Cattle Health Laboratory (DCHL) in Ladelund, and stored at <5 °C until required for analysis, which took place within a few days after arrival. At the laboratory, the faecal samples were pooled five at a time using 5 g per sample which was mixed to produce a 25 g pool before analysis.
Laboratory analyses
Pooled faecal samples were examined at DCHL for the presence of Salmonella bacteria using standard procedures described previously [18, 19]. If the pool was found positive for Salmonella the individual samples were cultured using 25 g of faecal material to try to locate those animals that were positive in the pool. It has been estimated that using the pooling procedure may lower the sensitivity of the culture method to approximately half of the sensitivity of the method using individual samples at the first step [20]. Serotyping and confirmation of positive isolates took place at the Danish Veterinary Institute in Copenhagen (now the National Food Institute, Technical University of Denmark). Whereas the analytical sensitivity of the test in the laboratory is reasonably high, i.e. ∼80% in samples with 10 c.f.u./g faeces [21], the diagnostic sensitivity for detection of infected cattle in naturally infected herds has been estimated to be very low, ∼6–14% in subclinically infected cattle [22].
The S. Dublin ELISA used in this study was performed at DCHL with a small modification from a previously described ELISA method [23]. An O antigen-based Salmonella serogroup D lipopolysaccharide preparation produced at the Danish Veterinary Institute in Copenhagen was used in the assay. By this method the ELISA mainly targets S. Dublin in cattle. However, cross-reactions with other serovars that share O antigens with S. Dublin may occur [24]. The laboratory procedure has been described in detail by Nielsen & Ersbøll [18]. An ODC% value, which is a background-corrected proportion of the test sample's optic density (OD) to the positive reference sample, was calculated as follows:
where 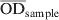 is the average value of two test wells,
is the average value of two test wells, 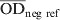 and
and 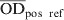 are the average values of four reference wells in the ELISA plates. The ODC% values were used to categorize cattle into antibody profile groups as described below.
are the average values of four reference wells in the ELISA plates. The ODC% values were used to categorize cattle into antibody profile groups as described below.
Definition of antibody profile groups in individual cattle
ELISA results from animals aged <90 days were discarded before categorization of cattle, because the diagnostic sensitivity and specificity of the test are known to be compromised by impaired capability of antibody production in calves aged <11–12 weeks [25] and maternally derived antibodies from colostrum [20]. The final dataset contained 3097 animals that were tested at least twice in the study herds. A total of 335 (9·8%) animals were excluded due to lack of sufficient samples. The ELISA results from the animals that were aged ≥180 days at each herd visit were used to group the animals into four TAPs on each of the last four sampling dates in the herds. The categorization explanations, criteria and distribution of animals and faecally positive animals in each TAP category are given in Table 1. Using these definitions, an animal that was only sampled once could not be included in the dataset. The age on the visiting date was recorded. The age distribution in the TAP categories is also given in Table 1. Thus, with about 3 months between each sampling date, the definition of the TAP categories was based on up to 1 year's samples from the animals.
Table 1.
Temporal antibody profile (TAP) criteria and distributions of animals, observations, faecal culture-positive observations and age in 14 dairy herds that were endemically infected with S. Dublin in Denmark between 2000 and 2002
| Group and short explanation | Criteria | No. of animals* | n (%) observations | n (%) faecal culture-positive observations† | Average age, years (95% CI) |
|---|---|---|---|---|---|
| TAP1: Persistently high antibody levels | The current and the previous two samples ≥80 ODC% | 126 | 182 (2·8%) | 3 (1·7%) | 3·8 (3·6–4·1) |
| TAP2: Recent increase in antibody levels | The current sample ≥50 ODC% and the previous one or two samples <25 ODC% | 214 | 214 (3·2%) | 6 (2·8%) | 2·7 (2·4–2·9) |
| TAP3: Fluctuating or moderately high antibodies | One of the previous samples ≥25 ODC%, but not three samples ≥80 ODC% | 1696 | 4022 (61%) | 31 (0·8%) | 3·4 (3·3–3·4) |
| TAP4: Recent low antibody levels | The current and the previous one or two samples <25 ODC% | 1061 | 2196 (33%) | 6 (0·3%) | 2·9 (2·8–3·0) |
CI, Confidence interval; ODC%, background-corrected optical density value.
The TAPs were based on the two or three most recent samples from each animal aged ≥180 days. The TAP groups were mutually exclusive.
Number of animals represented in the number of observations.
Percentage of the sample event observations in which S. Dublin bacteria were isolated.
Statistical analyses
SAS® version 9.2 (SAS Institute Inc., USA) was used for the data management, descriptive and statistical analyses. A hierarchical multivariable logistic regression analysis was used to statistically compare the effects of TAP categories and age of the animals, and to predict the probability of faecal excretion of S. Dublin. The model took into account repeated measurements at the animal level nested within the herd using generalized estimating equations (GEE) using a repeated statement in proc genmod in SAS. A significance level of 5% was used to evaluate the statistical evidence of the effect of the predictors. The interaction between age and TAP categories was similarly tested at the 5% significance level.
RESULTS
There were a total of 6614 observations representing 3097 animals aged ≥180 days in the dataset for analysis. This left 1750 observations that did not fit into any of the TAP categories and were excluded from the dataset for analysis because of too young age or because there were too few samples of the animals.
S. Dublin bacteria (or in three cases non-typable Salmonella) were isolated in 46 (0·7%) of the 6614 observations and 14 (0·8%) of the 1750 observations that were not included in the analysis. Eleven of these 14 isolates were found in cattle with ELISA results ≥50 ODC%, and the last three had ODC% values of 24, 29 and 37, respectively. Eight of the 14 animals were aged <1 year.
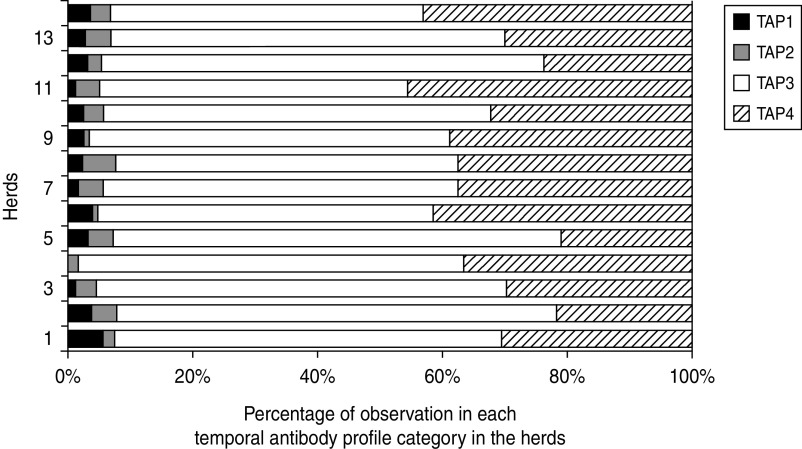
Table 1 shows the distribution of animals, observations, faecal culture-positive observations and age within each of the TAPs. Despite the fact that the percentage of faecal culture-positive animals was highest in the TAP1 and TAP2 categories, the absolute number of cattle shedding S. Dublin was to be found highest in animals with fluctuating or moderately high antibody levels. Figure 1 shows the distribution of observations in the four TAP categories in the study herds.
Fig. 1.
Distribution of S. Dublin temporal antibody profiles (TAP) in 14 Danish dairy herds. TAP1, Persistently high antibody levels; TAP2, recently increased antibody levels; TAP3, fluctuating or moderately high antibody levels; TAP4, recent low antibody levels.
The parameter estimates, odds ratios and P values from the final multilevel, multivariable logistic regression model are given in Table 2. Increasing age was clearly associated with decreasing probability of faecal shedding (Fig. 2). Furthermore, differences in faecal shedding probabilities were found to differ significantly between TAP1 and TAP4, but no statistical difference was found between TAP1 and TAP2 or TAP3. TAP2 on the other hand had a significantly higher probability of faecal shedding of S. Dublin than both TAP3 and TAP4.
Table 2.
Final hierarchical multivariable logistic regression model of predictors for S. Dublin isolation from faecal cultures in 14 endemically infected dairy herds
| Predictors | β | s.e. | OR (95% CI) | P |
|---|---|---|---|---|
| Intercept | −4·680 | 0·477 | ||
| Age, years | −0·544 | 0·136 | 0·6 (0·4–0·8) | <0·0001 |
| Temporal antibody profile (TAP) | 0·002 | |||
| TAP1: Persistently high antibodiesa,b | 2·103 | 1·056 | 8·2 (1–65) | |
| TAP2: Recent increasea | 2·183 | 0·585 | 8·9 (3–28) | |
| TAP3: Fluctuating or moderately high antibodiesb | 1·258 | 0·440 | 3·5 (1–8) | |
| TAP4: Recent low antibodiesc | 0 | — |
OR, Odds ratio; CI, confidence interval.
a,b,c Variable levels with different superscript letters were significantly different at the 5% significance level.
Fig. 2.
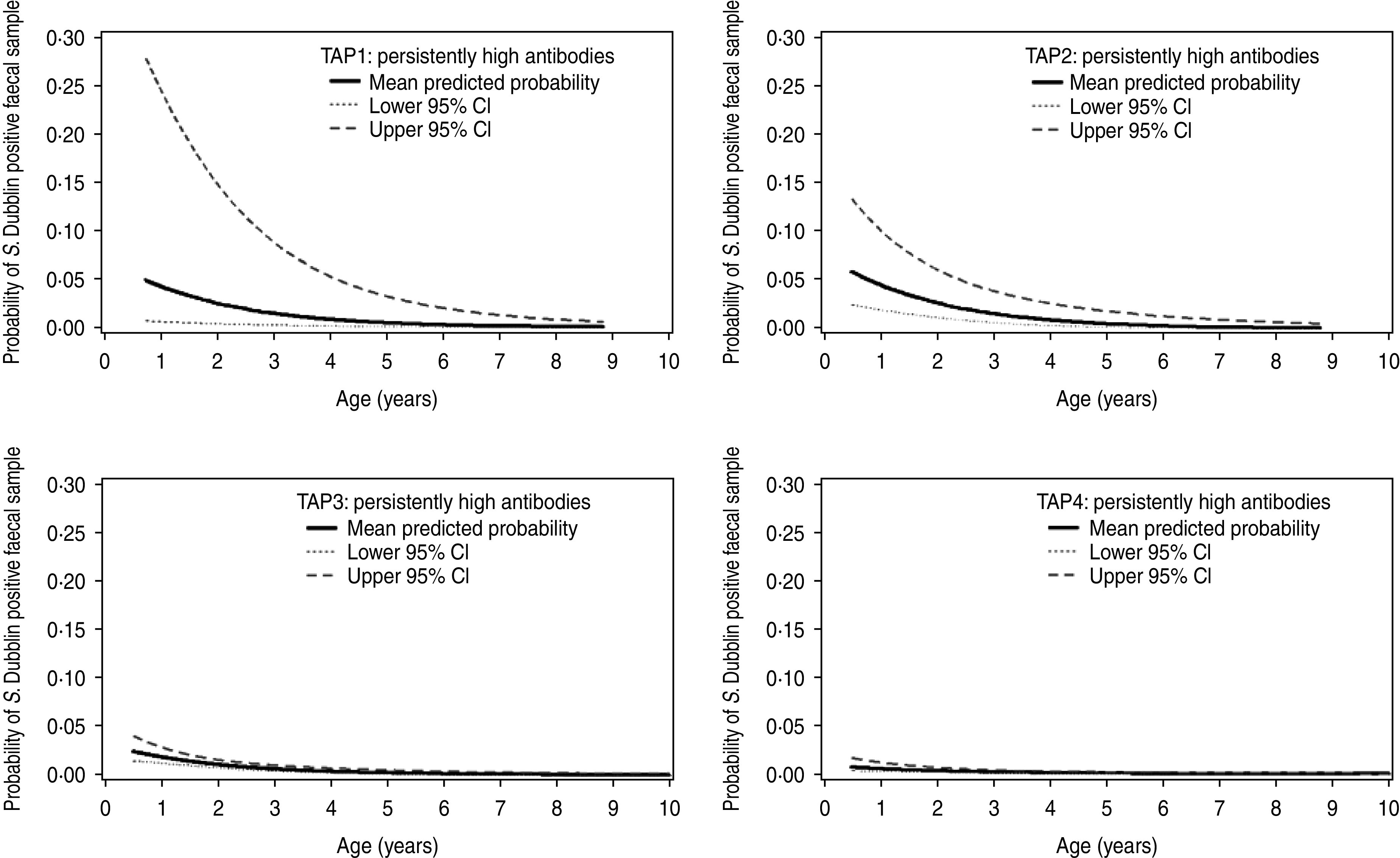
Model-predicted probabilities of faecal shedding of S. Dublin and 95% confidence intervals (CI) vs. age in cattle aged ≥180 days with four different temporal antibody profiles (TAP) in 14 Danish endemically infected dairy herds.
DISCUSSION
In this study, a large field data collection from 14 endemically infected dairy herds was used to investigate the hypothesis that cattle with persistently high antibody levels are at high risk of shedding S. Dublin and therefore are candidates to be culled or at least managed so that they do not spread the infection to herd-mates. Despite that fact that there were seropositive animals in many of the age groups at most of the herd visits, indicative of the herds being endemically infected, the general probability of shedding was very low (on average 0·7%) in all groups of cattle aged ≥180 days; only 46/6614 samples were found culture positive for S. Dublin. Apart from S. Dublin, only non-typable strains were isolated in three samples of faecal cultures from 3/14 herds. These were thought very likely to be S. Dublin by the Danish Veterinary Institute in Copenhagen (Dorte Lau Baggesen, personal communication), and were therefore included as such. Based on this study material there was no evidence that animals with persistently high antibodies over a period of at least 6 months were at higher risk of shedding S. Dublin bacteria in their faeces than other seropositive cattle. The only cattle that had a significantly lower probability of a positive faecal culture than the rest of the TAP categories were those with persistently low antibody levels (TAP4) (Table 2). In general the proportion of observations in the TAP1 and TAP2 categories were low compared to TAP3 and TAP4 in these 14 dairy herds (Fig. 1). However, some of the TAP3 observations were based on two consecutively high antibody measurements of ≥80 ODC%, which could not yet be categorized as persistently high until one more sample was available. TAP1 and TAP2 could not be assigned to the same animal more than twice, whereas the other two categories could be assigned to the same animal up to three times. However, that was not the explanation for the large differences in proportions of observations in the TAP categories. The highest number of TAP1 observations in one herd was 25 (3% out of 897 observations in herd 13) and the highest proportion of TAP1 observations within a herd was 6% (19 observations out of 334 in herd 1). These numbers indicate how many suspected carriers were present in the herds at any given time point and which animals should be considered for culling, if recommendations from previous studies are followed in a control scenario [8, 26].
TAP2 had a significantly higher probability of faecal shedding of S. Dublin than both TAP3 and TAP4. This group was also generally younger than the other groups, but since age was accounted for in the model, this was not the only explanation. A recent increase in antibody levels in the TAP2 category was indicative of recent exposure, which also increases the risk that the animal is still infected and may be excreting bacteria [27, 28].
The animals in the TAP3 category had a significantly higher probability of faecal excretion than TAP4, and as the largest group TAP3 was the group excreting bacteria most frequently in absolute numbers. The group consisted of cattle with fluctuating or continuously moderately high antibody measurements. The most likely explanation for such TAPs is that the animals have been exposed repeatedly over time from herd-mates or a contaminated environment. This would probably mainly include exposure to much smaller doses of bacteria than under experimental infection trials, which in turn may lead to lower or slower immune responses together with few clinical signs [28, 29].
The results from the modelling of faecal excretion provided uncertain parameter estimates due to the low number of positive faecal cultures. This results in very wide confidence intervals for the odds ratios, which should therefore be interpreted with caution. This requires some reflection on how to quantify the risk associated with individual animals in endemically infected herds. It can be argued that 14 dairy herds is not a sufficiently large sample of herds, and that some endemically infected herds might have higher prevalence levels of faecal shedders than the herds in the present study. However, previous studies have suggested that this is not the case [7, 30]. Rather than selecting animals with persistently high antibodies to follow and observe in order to discover if they shed bacteria, as was done in the study of carriers by House et al. [7], the present study was based on repeated paired samples on all cattle present on the farm over a 1-year period, and therefore has the potential to provide much less biased results in the evaluation of differences between TAP categories. However, such a sampling frame is very time-consuming and expensive, so adding more herds is not an obvious choice under economic limitations. The cheaper of the two laboratory procedures is the ELISA. Hence, a much cheaper sampling frame would be to start by testing all animals with ELISA three times at intervals of 2–3 months, and then test all or a stratified random sample of these with faecal culture.
Furthermore, the faecal culture test used in this study is known to have poor diagnostic sensitivity (∼6–14%) for detection of infected cattle [22], mainly because these animals may not necessarily be shedding bacteria. The test probably has better sensitivity (∼80%) for detection of infectious animals (i.e. faecal shedders) [21]. Access to methods with improved sensitivity for detection of bacterial shedding would be useful for research studies of potentially persistently infected S. Dublin carriers.
Regardless of the limitations in sample size of faecal culture-positive animals, there was a very clear association between age and the probability of faecal shedding in this study (Fig. 2). The younger the animals the more likely they were to excrete S. Dublin. The highest estimated probability occurred in calves aged ≥180 days. Here it averaged 5–6% in TAP1 and TAP2, ∼2·5% in TAP3 and 1·5% in TAP4, whereas cattle aged >3 years on average were faecal culture positive for S. Dublin less than 2% of the time regardless of the TAP.
The implication of this study is that S. Dublin carriage detection based on repeated antibody measurements should be regarded as a very uncertain method for use as a control element in persistently infected dairy herds. The age associations indicated a more likely benefit of directing the focus towards methods to prevent the spread of bacteria between calves and young stock, including consistent sectioning and careful cleaning of the environment and housing equipment on a regular basis as suggested in previous studies [6, 31, 32].
ACKNOWLEDGEMENTS
The author thanks the farmers for their participation and assistance with the sampling in the project. Thanks are also due to the technicians from the Danish Dairy Board, who collected all of the samples and from the Danish Cattle Health Laboratory for handling, storing and analysing the samples. The work was funded by the Danish Dairy Board and was performed as part of the ‘Kongeå project’.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Nielsen TD, et al. Association between bulk-tank milk Salmonella antibody level and high calf mortality in Danish dairy herds. Journal of Dairy Science 2010; 93: 304–310. [DOI] [PubMed] [Google Scholar]
- 2.Richardson A, Watson WA. A contribution to the epidemiology of Salmonella Dublin infection in cattle. British Veterinary Journal 1971; 127: 173–182. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen TD, et al. Evaluation of milk yield losses associated with Salmonella antibodies in bulk-tank milk in bovine dairy herds. Journal of Dairy Science 2012; 95: 4873–4885. [DOI] [PubMed] [Google Scholar]
- 4.Jones TF, et al. Salmonellosis outcomes differ substantially by serotype. Journal of Infectious Diseases 2008; 198: 109–114. [DOI] [PubMed] [Google Scholar]
- 5.Boqvist S, Vågsholm I. Risk factors for hazard of release from Salmonella control restriction on Swedish cattle farms from 1993 to 2002. Preventive Veterinary Medicine 2005; 71: 35–44. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen LR, Nielsen SS. A structured approach to control of Salmonella Dublin in 10 Danish dairy herds based on risk scoring and test-and-manage procedures. Food Research International 2011; 45: 1158–1165. [Google Scholar]
- 7.House JK, et al. Enzyme-linked immunosorbent assay for serologic detection of Salmonella dublin carriers on a large dairy. American Journal of Veterinary Research 1993; 54: 1391–1399. [PubMed] [Google Scholar]
- 8.Smith BP, et al. Identification of Salmonella dublin carrier cattle. Proceedings of the International Symposium on Salmonella and Salmonellosis. Zoopôle, Ploufragan,France, 1992, pp. 225–230. [Google Scholar]
- 9.Richardson A. The Transmission of Salmonella dublin to calves from adult carrier cows. Veterinary Record 1973; 92: 112–115. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen BB, Vestergaard E-M. Use of ELISA in the eradication of Salmonella Dublin infection. Proceedings of the International Symposium on Salmonella and Salmonellosis. Zoopôle, Ploufragan, France: 1992, pp. 220–224. [Google Scholar]
- 11.Spier SJ, et al. Use of ELISA for detection of immunoglobulins G and M that recognize Salmonella dublin lipopolysaccharide for prediction of carrier status in cattle. American Journal of Veterinary Research 1990; 51: 1900–1904. [PubMed] [Google Scholar]
- 12.Spier SJ, et al. Persistent experimental Salmonella dublin intramammary infection in dairy cows. Journal of Veterinary Internal Medicine 1991; 5: 341–350. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen LR, et al. Salmonella Dublin infection in dairy cattle: risk factors for becoming a carrier. Preventive Veterinary Medicine 2004; 65: 47–62. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen LR, Dohoo I. Culling decisions of dairy farmers during a 3-year Salmonella control study. Preventive Veterinary Medicine 2011; 100: 29–37. [DOI] [PubMed] [Google Scholar]
- 15.Hoorfar J, Wedderkopp A, Lind P. Comparison between persisting anti-lipopolysaccharide antibodies and culture at postmortem in salmonella-infected cattle herds. Veterinary Microbiology 1996; 50: 81–94. [DOI] [PubMed] [Google Scholar]
- 16.Lomborg S, et al. Effects of experimental immunosuppression in cattle with persistently high antibody levels to Salmonella Dublin lipopolysaccharide O-antigens. BMC Veterinary Research 2007; 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen LR, Ersbøll AK. Factors associated with variation in bulk-tank-milk Salmonella Dublin ELISA ODC% in dairy herds. Preventive Veterinary Medicine 2005; 68: 165–179. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen LR, Ersbøll AK. Age stratified validation of an indirect Salmonella Dublin serum ELISA for individual diagnosis in cattle. Journal of Veterinary Diagnostic Investigation 2004; 16: 205–211. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen LR, et al. Prevalence and risk factors for Salmonella in veal calves at Danish cattle abattoirs. Epidemiology and Infection 2011; 139: 1075–1080. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen LR. Salmonella Dublin in dairy cattle: Use of diagnostic tests for investigation of risk factors and infection dynamics (PhD thesis). The Royal Veterinary and Agricultural University, 2003, pp. 1–219.
- 21.Baggesen DL, et al. Growth inhibitory factors in bovine faeces impairs detection of Salmonella Dublin by conventional culture procedure. Journal of Applied Microbiology 2007; 103: 650–656. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen LR, Toft N, Ersbøll AK. Evaluation of an indirect serum ELISA and a bacteriological faecal culture test for diagnosis of Salmonella serotype Dublin in cattle using latent class models. Journal of Applied Microbiology 2004; 96: 311–319. [DOI] [PubMed] [Google Scholar]
- 23.Hoorfar J, et al. Serodiagnosis of Salmonella dublin infection in Danish dairy herds using O-antigen based enzyme-linked immunosorbent assay (published Erratum appears in Can. J. Vet. Res. 1995, 59, p. 25). Canadian Journal of Veterinary Research 1994; 58: 268–274. [PMC free article] [PubMed] [Google Scholar]
- 24.Konrad H, et al. Production of Salmonella serogroup D (O9)-specific enzyme-linked immunosorbent assay antigen. American Journal of Veterinary Research 1994; 55: 1647–1651. [PubMed] [Google Scholar]
- 25.Da Roden L, et al. Effect of calf age and Salmonella bacterin type on ability to produce immunoglobulins directed against Salmonella whole cells or lipopolysaccharide. American Journal of Veterinary Research 1992; 53: 1895–1899. [PubMed] [Google Scholar]
- 26.Smith BP, et al. Detection of Salmonella dublin mammary gland infection in carrier cows, using an ELISA for antibody in milk or serum. American Journal of Veterinary Research 1989; 50: 1352–1360. [PubMed] [Google Scholar]
- 27.Nielsen LR, van den Borne B, van Schaik G. Salmonella Dublin infection in young dairy calves: Transmission parameters estimated from field data and an SIR model. Preventive Veterinary Medicine 2007; 79: 46–58. [DOI] [PubMed] [Google Scholar]
- 28.Robertsson JA. Humoral antibody responses to experimental and spontaneous Salmonella infections in cattle measured by ELISA. Zentralblatt fur Veterinarmedizin, B 1984; 31: 367–380. [DOI] [PubMed] [Google Scholar]
- 29.Wray C, Sojka WJ. Salmonella dublin infection of calves: use of small doses to simulate natural infection on the farm. Journal of Hygiene 1981; 87: 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veling J, et al. Herd-level diagnosis for Salmonella enterica subsp. enterica serovar Dublin infection in bovine dairy herds. Preventive Veterinary Medicine 2002; 53: 31–42. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen LR, Kudahl AB, Østergaard S. Age-structured dynamic, stochastic and mechanistic simulation model of Salmonella Dublin infection within dairy herds. Preventive Veterinary Medicine 2012; 105: 59–74. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen TD, et al. Effect of management on prevention of Salmonella Dublin exposure of calves during a one-year control programme in 84 Danish dairy herds. Preventive Veterinary Medicine 2012; 105: 101–109. [DOI] [PubMed] [Google Scholar]