SUMMARY
The objective of this study was to characterize Listeria monocytogenes isolated from farmed Atlantic salmon (Salmo salar) and the processing environment in three different Norwegian factories, and compare these to clinical isolates by multiple-locus variable-number tandem repeat analysis (MLVA). The 65 L. monocytogenes isolates obtained gave 15 distinct MLVA profiles. There was great heterogeneity in the distribution of MLVA profiles in factories and within each factory. Nine of the 15 MLVA profiles found in the fish-associated isolates were found to match human profiles. The MLVA profile 07-07-09-10-06 was the most common strain in Norwegian listeriosis patients. L. monocytogenes with this profile has previously been associated with at least two known listeriosis outbreaks in Norway, neither determined to be due to fish consumption. However, since this profile was also found in fish and in the processing environment, fish should be considered as a possible food vehicle during sporadic cases and outbreaks of listeriosis.
Key words: Clinical isolates, genotyping, Listeria monocytogenes, MLVA profiles, seafood
INTRODUCTION
Listeria monocytogenes has been recognized as an important pathogen during the last decades, causing the disease known as listeriosis in animals and humans [1–3]. Several epidemiological investigations have shown that listeriosis may result from the consumption of contaminated food [4]. Listeriosis is typically associated with high-risk groups, such as individuals with an impaired immune system, including pregnant women, newborns, the elderly, injecting drug users and HIV patients [5, 6]. Infections by L. monocytogenes in humans and animals are mainly associated with septicaemia, meningitis, encephalitis and abortion [7].
In connection with outbreaks caused by L. monocytogenes it is necessary to use a method that can identify strains and facilitate the search for the source of infections. Several molecular methods have been applied for typing L. monocytogenes, such as serotyping, restriction enzyme analysis (REA), ribotyping, pulsed-field gel electrophoresis (PFGE) and amplified fragment length polymorphism (AFLP) [8]. MLVA typing has been an emerging method in recent years and is considered as a robust, reliable and discriminatory tool for genotyping bacterial isolates aiding outbreak investigations [9, 10]. This method has been used for typing several pathogenic organisms such as Salmonella enterica Typhimurium [11–13], Bacillus anthracis [14], Clostridium perfringens [15] and Escherichia coli [16, 17].
Murphy et al. [18] were the first to apply the MLVA method to type L. monocytogenes. These authors examined 25 isolates of L. monocytogenes from sliced vacuum-smoked salmon and 20 non-salmon retail products from Ireland. They found the MLVA technique was able to discriminate between isolates from smoked salmon and non-salmon samples, indicating different sources of contamination of the products.
In a comparative study of 79 Norwegian and 61 Swedish L. monocytogenes isolates, Lindstedt et al. [8] found 28 MLVA and 24 PFGE profiles in the Norwegian isolates, and 42 MLVA and 43 PFGE profiles in the Swedish isolates. Thus there appears to be a difference in the discriminatory power between sample sets. However, the speed of the method and labour demand support using the MLVA method compared to PFGE as was shown by Sperry et al. [19].
In the present study, the MLVA method was applied for genotyping L. monocytogenes, to examine the distribution of strains in different fish-processing factories in Norway, and to compare the isolates from these factories with isolates from listeriosis patient.
MATERIALS AND METHODS
Bacterial strains and DNA extraction
Three fish-processing factories designated A, B, and C, located in different areas of Norway, were examined for the presence of L. monocytogenes during five visits over a 6-month period in 2007. A total of 65 L. monocytogenes isolates were found and subsequently genotyped by MLVA. Of these, 23 isolates were obtained from fish samples and 42 isolates were from environmental samples. All isolates were kept in Microbank vials at −80 °C and resuscitated on blood agar (TSS) (trypticase soy agar with 5% sheep blood; bioMérieux, France). Genomic DNA in samples was extracted using the BioRobot EZ1 (Qiagen, Germany) instrument. The EZ1 DNA Tissue kit (Qiagen) was used for DNA extraction and purification based on cell lysis and immunomagnetic separation by paramagnetic monodisperse latex according to the manufacturer's instructions.
Variable number tandem repeat (VNTR) loci and primer sequence
Five VNTR loci were identified on the genome of the two fully sequenced L. monocytogenes strains (EGDe serotype 1/2a, Genbank accession no.: NC_003210; and F2365 serotype 4b, Genbank accession no.: NC_002973) [8]. The five primer sets for PCR amplification, VNTR loci (LMV1, LMV2, LMV6, LMV7, LMV9) were chosen as described by Lindstedt et al. [8]. The forward primer of each set was labelled at its 5′ end with a fluorescent dye including hexachlorofluorescein (HEX), carboxyfluorescein (6-FAM), and tetrachlorofluorescein (TET) [20]. The length of repeat units for the five loci ranged from 6 to 15 base pairs (bp).
Multiplex PCR reactions
The PCR reactions were performed in a Gene Amp PCR System 9700 thermocycler using the Qiagen Multiplex PCR kit from Applied Biosystems (USA). The primers were multiplexed in two separate reactions (R1 and R2) in a total of 50 μl according to the manufacturer's instructions. The R1 reaction contained 1 μl (10 μm) each of LMV2 and LMV6 primers. The R2 reaction contained 1 μl (10 μm) each of primers LMV1, LMV7, and LMV9. The reaction mixture was prepared for amplification of template DNA in one sample. A volume of 2 μl extracted template DNA was added to the individual PCR tubes containing 48 μl reaction mix to a total of 50 μl. The mixture was then mixed gently. The PCR tubes were placed in individual thermocyclers for both reactions. The PCR program started with an initial heat step at 95 °C for 15 min to activate the DNA polymerase. PCR reactions were run under the following conditions: 30 cycles at 94 °C for 30 s for denaturation, R1 at 60 °C and R2 at 63 °C for 90 s for annealing, and at 72 °C for 90 s for extension, with a final extension step at 72 °C for 10 min [8].
Sample pooling and capillary electrophoresis
The two PCR multiplexed solutions were mixed prior to the electrophoretic analysis which provides a fluorescent signal from each amplicon. The products from R1 and R2 solutions were pooled into a single tube as follows: 10 μl R2 was added to 50 μl R1 in a single tube and mixed gently. From this pooled solution, 1·5 μl was added to 12 μl of Formamid (Applied Biosystems) and 2 μl of GeneFlo-625 TAMRA (CHIMERx, USA) internal size standard. The run sample was denatured for 3 min at 94 °C and cooled to room temperature before being loaded onto the capillary sequencer. Amplicons were separated by capillary electrophoresis on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). The capillary electrophoresis was run at 60 °C for 35 min per sample using a 47 cm capillary filled with performance-optimized polymer 4 (POP4), 10 × Genetic Analyzer buffer with EDTA, with an injection voltage of 15 kV for 5 s and a running voltage of 15 kV. Data were collected and the sizes of PCR amplicons were determined by the software. Each peak was identified according to colour and size and was binned into alleles so that each new multiple of repeat was assigned to a distinct allele number.
Data analysis
A typical electropherogram is shown in Figure 1. Each peak represents a PCR product and was equivalent to a band obtained by agarose gel electrophoresis. Loci LMV2 and LMV6 labelled with the dye 6-FAM were depicted as blue peaks, loci LMV1 and LMV9 labelled with the dye HEX were depicted by black, and locus LMV7 labelled with the dye TET was depicted as green. Thus, blue, black and green peaks represented different VNTR loci. The DNA size standard labelled with the dye TAMRA was depicted in red which presented the internal size standard and each standard size was 25 bp in length. Sizes were automatically recorded using GeneScan software. The MLVA pattern of each sample varied depending on the number of tandem repeats at each VNTR locus in the isolate under study and gave its own distinct fingerprint. Variation in size according to multiples of the repeat was interpreted as separate alleles.
Fig. 1.
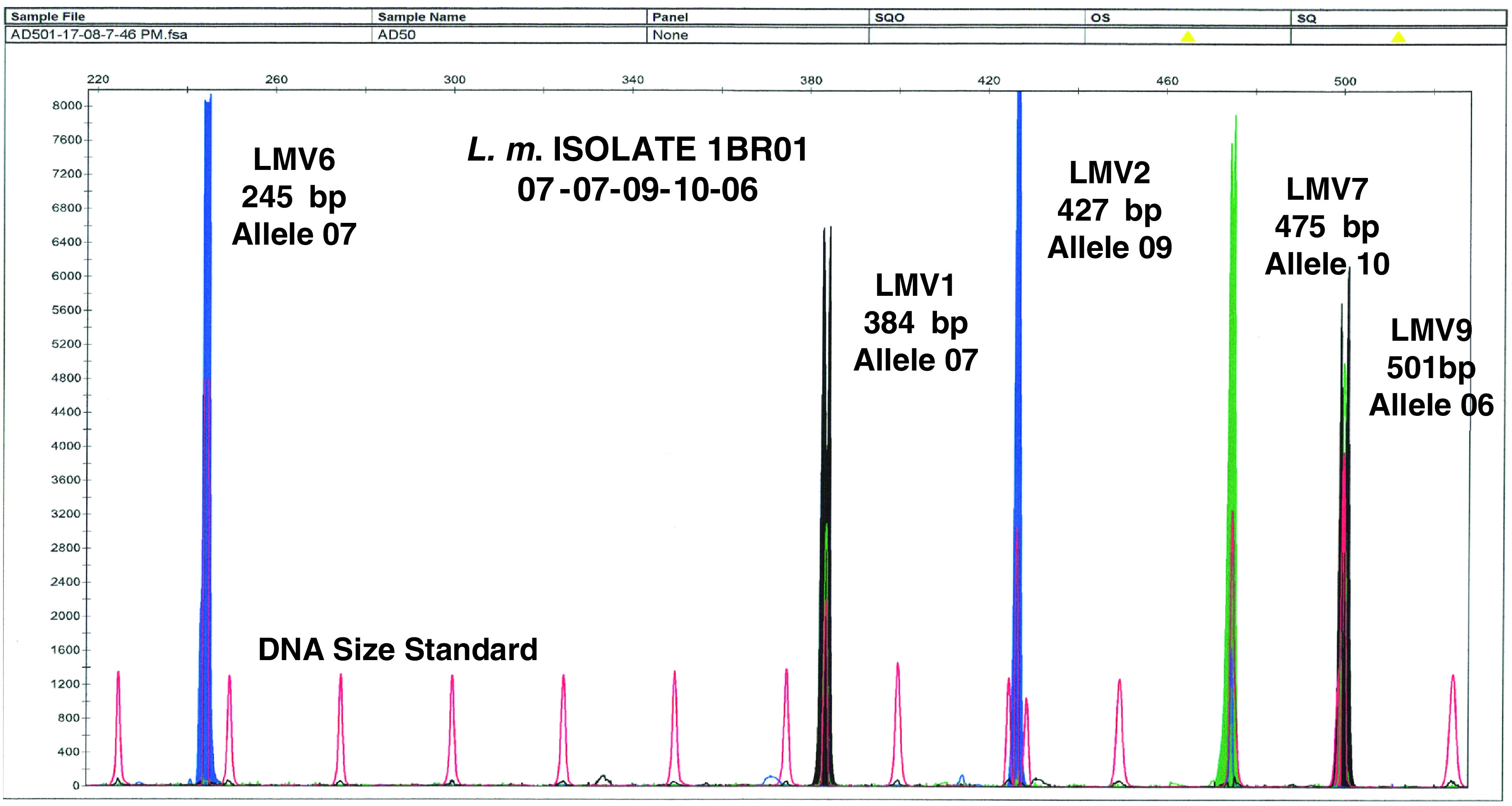
Multicolour capillary electrophoresis-based variable number tandem repeat (VNTR) of L. monocytogenes isolates. Blue, black and green peaks represent different VNTR loci and red peaks represent the internal size standard.
The MLVA profile was presented in the order of the allele string as follows: LMV6-LMV1-LMV2-LMV7-LMV9. This allele string of five numbers identified the bacterial isolate [12]. As an example, isolate 1BR01 obtained in factory A was defined by the string 07-07-09-10-06, indicating that it had allele number 07 (245 bp) at locus LMV6, allele number 07 (384 bp) at locus LMV1, allele number 09 (427 bp) at locus LMV2, allele number 10 (475 bp) at locus LMV7, and allele number 06 (501 bp) at locus LMV9. The allele numbers were then entered into a BioNumerics version 4.5 (Applied Maths, Belgium) database as character values, and a dendrogram was constructed using categorical coefficients and the Ward algorithm. The minimum spanning tree (MST) was also constructed. Based on the dendrogram, pattern similarities were easily visualized and matching profiles were also rapidly identified. Furthermore, under the same running conditions, the MLVA profiles can rapidly be shared electronically between laboratories by email [20].
RESULTS
Characteristics of selected VNTR loci
The results from MLVA typing showed that all loci varied greatly in their degree of polymorphism from the LMV2 locus with seven different alleles to the LMV9 locus with two alleles. The LMV2 locus was highly polymorphic whereas the LMV9 locus was less so. Generally, all loci were presented in the 65 L. monocytogenes isolates with no missing amplicons in the four loci LMV2, LMV6, LMV7, and LMV9. For the LMV1 locus some amplicons were missing. Simpson diversity index (DI) was calculated based on the formula:
This factor was used to evaluate the discriminatory power of subtyping methods as described previously [21]. In this study, the genetic diversity estimated for five variable tandem repeat loci varied greatly, ranging from 0·034 to 0·79 (Table 1). The highest DI (0·79) was found at the LMV2 locus with the distribution of seven alleles, whereas the lowest DI (0·034) was found at the LMV9 locus with the distribution of two alleles. The remaining showed variable values ranging from 0·58 to 0·72, of which the DI value 0·58 was found at the LMV7 locus, 0·65 at the LMV6 locus, and 0·72 at the LMV1 locus with one missing PCR amplicon as stated above. The studies of Lindstedt et al. [12, 13] have shown that the high polymorphism in allele distribution at each locus in a set of five loci results in a high degree of strain discriminative genotyping.
Table 1.
Characteristics of selected variable number tandem repeat loci
| Locus | Repeat length | Copy number | Size range of amplicon (bp) (min–max) | No. of alleles* | Percentage of missing amplicons | Polymorphism index (DI)† |
|---|---|---|---|---|---|---|
| LMV1 | 6 | 18·3 | 366–409 | 06 | 1·5 (1/65) | 0·72 |
| LMV2 | 9 | 20·9 | 382–573 | 07 | 0 | 0·79 |
| LMV6 | 15 | 4·4 | 214–276 | 05 | 0 | 0·65 |
| LMV7 | 9 | 14·6 | 448–538 | 06 | 0 | 0·58 |
| LMV9 | 9 | 7·8 | 501–530 | 02 | 0 | 0·034 |
Not including the unamplifiable allele at the LMV1 locus.
Simpson diversity index (DI) = 1 – ∑(allele frequency)2.
Distribution of L. monocytogenes strains in the fish-processing factories
A total of 15 distinct MLVA profiles identified the 65 L. monocytogenes isolates from the fish-processing factories. In general, the profile heterogeneity between factories and within each factory was large. Seven different profiles were found in isolates from factory A, seven other profiles in factory B, and two other profiles in factory C. There was only one profile (08-08-16-19-06) found in both factories A and B. The MLVA profile distribution from the fish-processing factories is shown in Table 2.
Table 2.
Distribution of multiple-locus variable-number tandem repeat analysis (MLVA) profiles of the 65 L. monocytogenes isolates from factories A, B and C
| MLVA profile | No. of isolates (%) in each factory | Total number of isolates (%) of MLVA profiles | ||
|---|---|---|---|---|
| A | B | C | ||
| 05-08-14-10-06 | 11 (27·5%) | 11 (16·9%) | ||
| 06-00-14-10-06 | 1 (16·7%) | 1 (1·5%) | ||
| 06-08-04-19-06 | 3 (15·8%) | 3 (4·6%) | ||
| 06-08-14-18-06 | 1 (5·3%) | 1 (1·5%) | ||
| 06-08-15-10-06 | 7 (17·5%) | 7 (10·8%) | ||
| 06-08-16-10-06 | 2 (5%) | 2 (3·1%) | ||
| 06-09-04-10-06 | 5 (83·3%) | 5 (7·7%) | ||
| 06-09-26-16-06 | 1 (2·5%) | 1 (1·5%) | ||
| 06-10-04-16-06 | 10 (52·6%) | 10 (15·4%) | ||
| 06-10-16-21-06 | 1 (5·3%) | 1 (1·5%) | ||
| 06-11-15-18-06 | 1 (5·3%) | 1 (1·5%) | ||
| 07-07-09-10-06 | 15 (37·5%) | 15 (23·1%) | ||
| 07-10-15-18-06 | 3 (7·5%) | 3 (4·6%) | ||
| 08-08-16-19-06 | 1 (2·5%) | 2 (10·5%) | 3 (4·6%) | |
| 09-04-18-06-09 | 1 (5·3%) | 1 (1·5%) | ||
| Total | 40 | 19 | 06 | 65 (100%) |
The MLVA typing revealed high variety of profiles in factory A, where a total of 40 isolates were characterized by seven different MLVA profiles. The most common profiles were 07-07-09-10-06 (37·5%), 05-08-14-10-06 (27·5%) and 06-08-15-10-06 (17·5%). In factory B, 19 isolates were identified by seven distinct MLVA profiles. The profile with the highest frequency was 06-10-04-16-06. This profile accounted for 52·6%, indicating that it was the predominant strain in factory B. This profile was found frequently at various environmental sites such as run-off water, cooling water, conveyor belts and floors. In factory C, six isolates revealed two distinct MLVA profiles. The profile 06-09-04-10-06 was the most common, and accounted for 83·3%, whereas 06-00-14-10-06 only accounted for 16·7%. The strains defined by 06-09-04-10-06 were found on raw fish, in swabs of fish at the head-cutting line, and in various environmental sites such as gutting machine, conveyor belt and drain at gutting area. There was only one isolate defined by the profile 06-00-14-10-06 that existed in the drain at the fine cutting line.
Comparison between strains from fish-processing factories and listeriosis patients
A database of 71 clinical isolates previously characterized by MLVA in the Norwegian Institute of Public Health, Oslo, was compared with the MLVA profiles of the 65 L. monocytogenes strains from the fish-processing factories. Of the 15 distinct MLVA profiles in the current study, nine were found to match profiles obtained from listeriosis patients (Fig. 2). These were 09-04-18-06-09, 08-08-16-19-06, 05-08-14-10-06, 06-10-04-16-06, 06-10-16-21-06, 06-08-14-18-06, 06-11-15-18-06, 06-00-14-10-06, and 07-07-09-10-06. Human strains were isolated from blood, spinal fluid, human placenta, and some of unknown source in patients suffering from listeriosis in Norway and Sweden. The profile 07-07-09-10-06 was found frequently from blood culture and spinal fluid, and has been involved in two known listeriosis outbreaks in Norway. One of these outbreaks was connected to consumption of cold meat cuts sliced on a contaminated slicer. The second outbreak could be linked to contaminated ecological Camembert cheese, where the concentration of L. monocytogenes was found to be as high as 3·6 × 108 per serving [22]. In this outbreak 17 patients were infected and three died, giving a mortality rate of 18%.
Fig. 2.
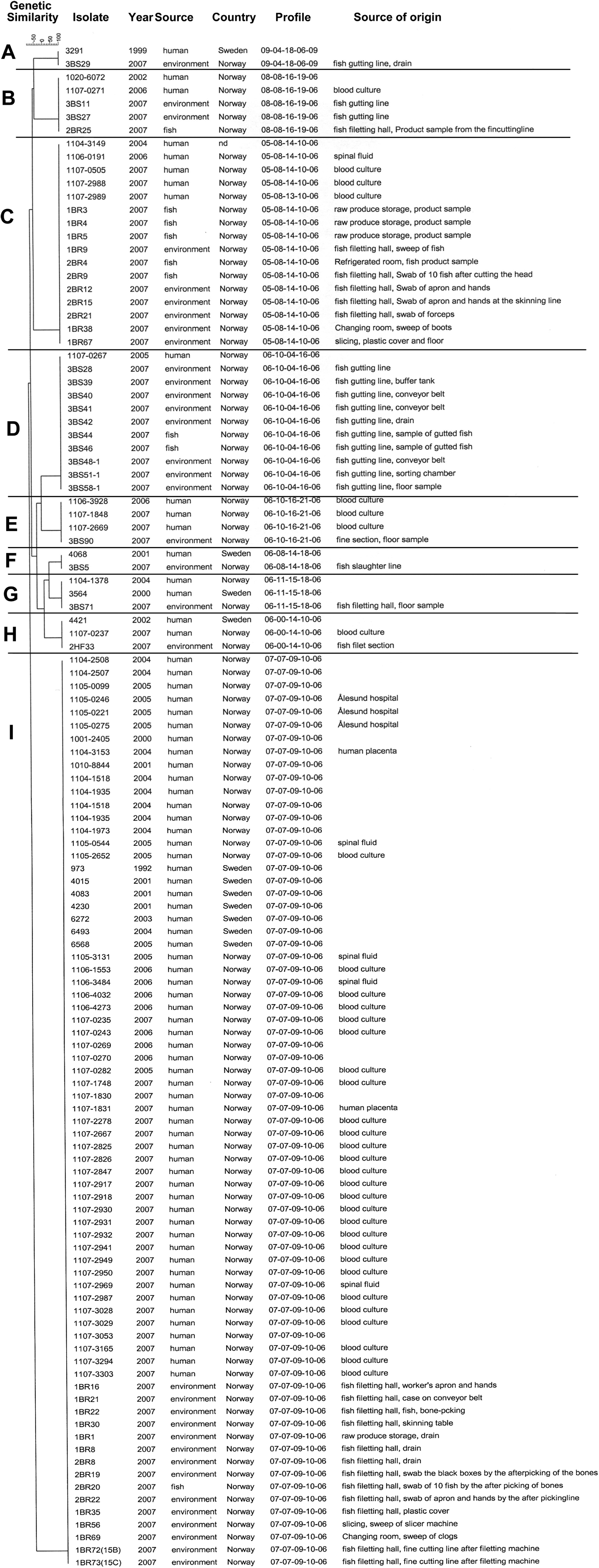
Dendrogram of L. monocytogenes strains from fish-processing factories and listeriosis patients. Some human isolates are identified by the clinical source. The nine overlapping multiple-locus variable-number tandem repeat analysis (MLVA) profiles comprising both fish-processing factories and human isolates are indicated by A–I.
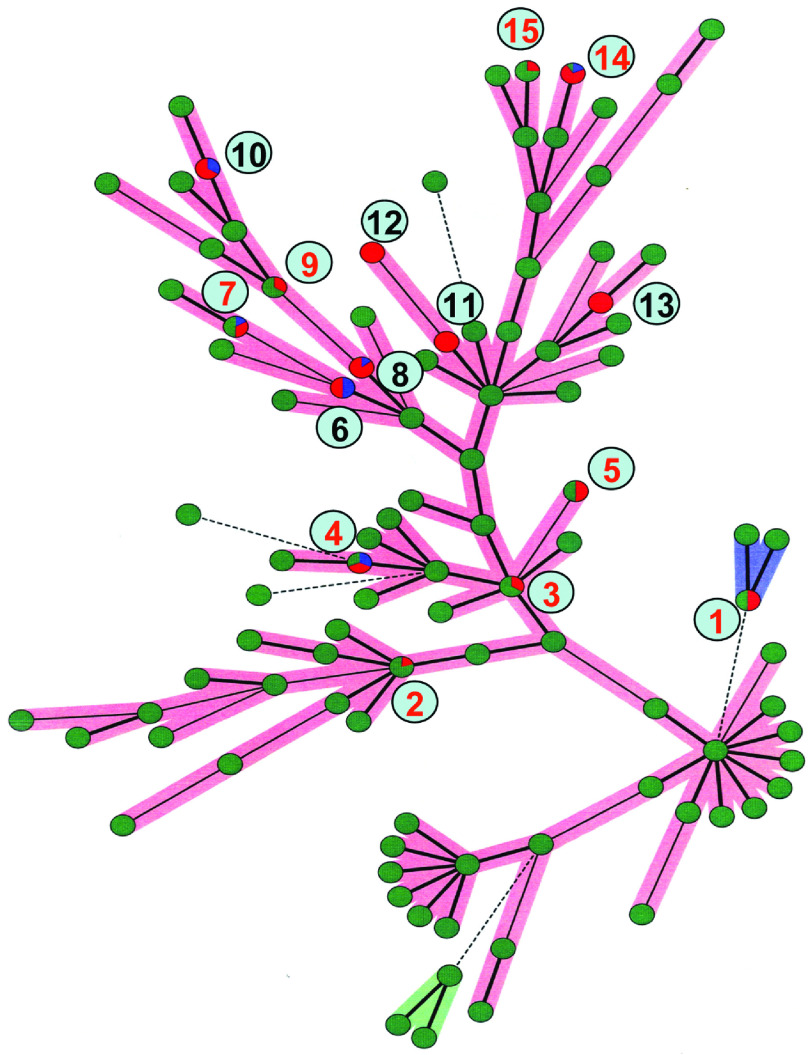
Minimum spanning tree (MST)
The clonal relationships between the 65 isolates from the fish-processing factories and human strains were constructed with MLVA profiles by the MST method [12]. Figure 3 presents the phylogenetic tree built with MLVA profiles of all isolates. A total of 15 distinct MLVA profiles were included. In our study we used five specific loci and MLVA profiles matching all five loci were regarded as clonally related. The results showed that nine MLVA profiles from fish-processing factories were found to match human profiles.
Fig. 3.
The minimum spanning tree diagram showing the clonal relationships between isolates. Difference in the relationship between profiles is indicated by thick lines, thin lines, and dotted lines. The thickness and dotting of lines indicate the distance between the circles, thus a thicker line denotes a closer distance than a thin line, and a thin line denots a closer distance than a dotted line. Single-coloured green dots represent profiles obtained from human isolates only, red dots represent isolates from fish-processing environment only and blue circles represents fish isolates only. The multicolour dots show shared multiple-locus variable-number tandem repeat analysis (MLVA) profiles. The diagram includes 15 distinct MLVA profiles, numbered from 1 to 15 in the figure. Nine MLVA profiles from fish samples or from the fish-processing environment matched human profiles, and are numbered in red (nos. 1, 2, 3, 4, 5, 7, 9, 14, 15). For the remaining profiles (nos. 6, 8, 10, 11, 12, 13) there was no overlap between human profiles and profiles from fish samples or from the fish-processing environment.
DISCUSSION
Evaluation of the MLVA method
The MLVA method is one of several molecular techniques used for genotyping bacterial pathogens to investigate the epidemiology of outbreaks, as well as to trace the contamination routes in food-processing factories. In the MLVA approach applied in the present study, DNA extraction by a BioRobot EZ1 instrument using the technique of immunomagnetic separation by monodisperse latex beads, results in better purification of DNA compared to DNA extraction procedures not involving such a step. Capillary electrophoresis is the preferred method due to the availability of multiple fluorescent labels that results in accuracy and reproducibility compared to the conventional agarose gel electrophoresis approach [23]. In particular, the primers labelled with multicolour fluorescent dyes and the internal size standard result in easy recognition of separated peaks based on size and colour at each locus from the resulting electropherogram. Moreover, the use of multicolour dyes, rather than a single dye as previously used, helps to overcome the size overlap of two VNTR loci.
Several studies have compared and correlated the various typing methods used for L. monocytogenes. Since PFGE is known as the gold standard method, most studies have compared alternative techniques with PFGE. Jadhav et al. [24] reviewed the literature on relevant methods for subtyping L. monocytogenes. After comparing MLVA, MLST (multi-locus sequence-based typing), ribotyping and PFGE these authors, found the discriminatory power of MLVA (using three tandem repeat regions) was higher than the other three methods. When considering the discriminatory index, the following order was observed in terms of decreasing discriminatory power: MLVA > PFGE > MLST > ribotyping. Furthermore, the results obtained from MLVA typing can be shared quickly with other laboratories based on the dendrogram or MST [12]. It is also important to consider that while PFGE takes up to a week for completion, the MLVA analysis can be run within a day.
L. monocytogenes strains from fish-processing factories and listeriosis patients
The profile 07-07-09-10-06 was found to be predominant in factory A. This profile was also frequently found in samples from blood, spinal fluid and placenta from Norwegian and Swedish patients. The profile has been involved in at least two known listeriosis outbreaks in Norway. Lindstedt et al. [8] found profile 07-07-09-10-06 to be present both in the environment and in food. In the present study, factory A had L. monocytogenes 07-07-09-10-06 in fish samples, processing line samples and in various environmental sites such as drains, floors, equipment, machines (slicing machine), and aprons and gloves of workers. In the present study, L. monocytogenes 07-07-09-10-06 was found to be widespread in the processing environment and accounted for the highest prevalence of the detected MLVA profiles. This suggests that the strains belonging to this MLVA profile are persistent in the factory processing environment. Previous studies have demonstrated that certain clones of L. monocytogenes may establish and persist for years in each individual food-processing plant [25]. The profile 07-07-09-10-06 was found in clinical sites of listeriosis patients and in a fish-processing factory. However, some studies reported that most environmental strains of L. monocytogenes from the smoked-fish industry were of a lineage different from that of human clinical isolates, and that the clinical strains were more virulent than isolates from food [25]. The two other common profiles in factory A were 05-08-14-10-06 and 06-08-15-10-06. Only profile 05-08-14-10-06 was found in isolates from listeriosis patients in Norway. The profile 08-08-16-19-06 was also found in human isolates but this strain accounted for the lowest level in this factory. In our study, these strains were found widely in raw fish, fish samples taken during production and in a variety of processing environmental sites such as table, boxes, forceps, boots, floor, fillet machine, aprons and gloves of personnel, indicating that raw material may be the initial source of contamination in the factory. The dominant strain in factory B was identified as profile 06-10-04-16-06. Except for one profile, the six remaining profiles including 09-04-18-06-09, 08-08-16-19-06, 06-10-04-16-06, 06-10-16-21-06, 06-08-14-18-06, and 06-11-15-18-06 were found to match human isolates. In particular, profile 09-04-18-06-09 was found in isolates from Sweden and profile 06-11-15-18-06 was also found in the environment. In the factory, all these strains were widespread in the processing environment, especially in various drains, floors, and equipment. Unlike factories A and B, the distribution of strains in factory C was relatively homogenous as only two MLVA profiles were found. The most common profile was 06-09-04-10-06, this profile was, however, not found in human isolates.
Some cases of listeriosis with fish as the most likely source have been reported [26–28]. Listeriosis with Atlantic salmon as the food source has not been identified in Norway. In a study by Peiris et al. [29] on gravad and cold-smoked salmon in Sweden, the authors conclude that the isolation of strains identical or closely related to clinical isolates, as assessed by serotyping and PFGE, suggests that these products are a possible source for listeriosis. Based on the available information from the literature and on the findings of the present study, fish should be considered as one of several possible sources of L. monocytogenes when investigating sporadic cases of foodborne listeriosis or outbreaks.
CONCLUSION
The 65 L. monocytogenes isolates from the three fish-processing factories were characterized into 15 distinct MLVA profiles. This typing method showed that all strains were highly heterogenic between factories and within each factory. Seven profiles were found in factories A and B, two profiles in factory C. One profile was found to be present in both factories A and B. The profile 07-07-09-10-06 was predominant in factory A, profile 06-10-04-16-06 was predominant in factory B, whereas 06-09-04-10-06 was predominant in factory C. Of the 15 profiles, nine were found to match MLVA profiles from human isolates. Of these 07-07-09-10-06 was regularly found in Norwegian and Swedish patients. Furthermore, this specific profile has been reported to be involved in at least two known listeriosis outbreaks in Norway. The results from the present study show that some MLVA profiles are found both in fish and in fish-processing factories and in human isolates, showing that an epidemiological link can not be excluded. On the basis of the results we have presented, it cannot be concluded that people have been infected by L. monocytogenes through the consumption of salmon products. However, lightly preserved fish and fish products should be considered as one of several possible sources of L. monocytogenes when investigating the epidemiology of foodborne listeriosis.
ACKNOWLEDGEMENTS
The skilled technical assistance of Tone Galluzzi and Kjersti Borlaug during collection of samples and preparation of bacterial isolates is greatly appreciated. Traute Vardund is acknowledged for skilful preparation of DNA samples and for help in running the MLVA analysis.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Wesley IV. Listeriosis in animals. In: Ryser ET, Martin EH, eds. Listeria, Listeriosis and Food Safety. Boca Raton: CRC Press, 2007, pp. 55–84. [Google Scholar]
- 2.Painter J, Slutsker L. Listeriosis in humans. In: Ryser ET, Martin EH, eds. Listeria, Listeriosis and Food Safety. Boca Raton: CRC Press, 2007, pp. 85–109. [Google Scholar]
- 3.Jemmi T, Stephan R. Listeria monocytogenes: foodborne pathogen and hygiene indicator. Revue Scientific et Technique de l'Office International des Epizooties 2006; 25: 571–580. [PubMed] [Google Scholar]
- 4.Adams MR, Moss MO. Food Microbiology, 5th edn. Cambridge: RSC Publishing, 2008, pp. 224–231. [Google Scholar]
- 5.Farber JM, Peterkin PI. Listeria monocytogenes, a foodborne pathogen. Microbiological Reviews 1991; 55: 476–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orsi RH, Bakker HC, Wiedmann M. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. International Journal of Medical Microbiology 2011; 301: 79–96. [DOI] [PubMed] [Google Scholar]
- 7.Paoli GC, Bhunia AK, Bayles DO. Listeria monocytogenes. In: Fratamico PM, Bhunia AK, Smith JL, eds. Foodborne Pathogens. Norfolk, UK: Caister Academic Press, 2005, pp. 295–325. [Google Scholar]
- 8.Lindstedt BA, et al. Multiple-locus variable number tandem repeats analysis of Listeria monocytogenes using multicolor capillary electrophoresis and comparison with pulsed-field gel electrophoresis typing. Journal of Microbiological Methods 2008; 72: 141–148. [DOI] [PubMed] [Google Scholar]
- 9.Lindstedt BA. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 2005; 26: 2567–2582. [DOI] [PubMed] [Google Scholar]
- 10.Van Belkum A. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA). Immunology and Medical Microbiology 2007; 49: 22–27. [DOI] [PubMed] [Google Scholar]
- 11.Larsson JT, et al. Development of a new nomenclature for Salmonella Typhimurium multilocus variable number of tandem repeats analysis (MLVA). Eurosurveillance 2009; 14: 1–5. [PubMed] [Google Scholar]
- 12.Lindstedt BA, et al. Multiple-locus variable number tandem repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. Journal of Microbiological Methods 2004; 59: 163–172. [DOI] [PubMed] [Google Scholar]
- 13.Lindstedt BA, et al. Harmonization of the multiple-locus variable-number tandem repeat analysis method between Denmark and Norway for typing Salmonella Typhimurium isolates and closer examination of the VNTR loci. Journal of Applied Microbiology 2007; 102: 728–735. [DOI] [PubMed] [Google Scholar]
- 14.Keim P, et al. Multiple-locus variable number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. Journal of Bacteriology 2000; 182: 2928–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawires YS, Songer JG. Multiple-locus variable tandem repeat analysis for strain typing of Clostridium perfringens. Anaerobe 2005; 11: 262–272. [DOI] [PubMed] [Google Scholar]
- 16.Miko A, et al. Evaluation of multiple-locus variable number of tandem-repeats analysis (MLVA) as a method for identification of clonal groups among enteropathogenic, enterohaemorrhagic and avirulent Escherichia coli O26 strains. FEMS Microbiology Letters 2010; 303: 137–146. [DOI] [PubMed] [Google Scholar]
- 17.Solecki O, et al. E coli O157 from sheep in northeast Scotland: prevalence, concentration shed, and molecular characterization by multilocus variable tandem repeat analysis. Foodborne Pathogens and Disease 2009; 6: 849–854. [DOI] [PubMed] [Google Scholar]
- 18.Murphy M, et al. Development and application of multiple-locus variable number of tandem repeat analysis (MLVA) to subtype a collection of Listeria monocytogenes. International Journal of Food Microbiology 2007; 115:187–194. [DOI] [PubMed] [Google Scholar]
- 19.Sperry KEV, et al. Multiple-locus variable-number tandem-repeat analysis as a tool for subtyping Listeria monocytogenes strains. Journal of Clinical Microbiology 2008; 46: 1435–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yazdankhah SP, Lindstedt BA. Variable number tandem repeat typing of bacteria. In: Bergman NH, ed. Methods in Molecular Biology, Comparative Genomics, vol. 2, New Jersey, USA: Totowa, 2007, pp. 395–405. [DOI] [PubMed] [Google Scholar]
- 21.Cho S, et al. Multiple-locus variable number tandem repeat analysis of Salmonella enteritidis isolates from human and non human resources using a single multiplex PCR. FEMS Microbiology Letters 2007; 275: 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnsen BO, et al. A large outbreak of Listeria monocytogenes infection with short incubation period in a tertiary care hospital. Journal of Infection 2010; 61: 465–470. [DOI] [PubMed] [Google Scholar]
- 23.Slack A, et al. An improved multiple-locus variable number of tandem repeats analysis for Leptospira interrogants serovar Australis: a comparison with fluorescent amplified fragment length polymorphism analysis and its use to redefine the molecular epidemiology of this serovar in Queensland, Australia. Journal of Medical Microbiology 2006; 55: 1549–1557. [DOI] [PubMed] [Google Scholar]
- 24.Jadhav S, Bhave M, Palombo EA. Methods used for the detection and subtyping of Listeria monocytogenes. Journal of Microiological Methods 2012; 88: 327–341. [DOI] [PubMed] [Google Scholar]
- 25.Møretrø T, Langsrud S. Listeria monocytogenes: Biofilm formation and persistence in food-processing environments. Biofilms 2004; 1: 107–121. [Google Scholar]
- 26.Facinelli B, et al. Ignorance about Listeria. British Medical Journal 1989; 299: 73. [Google Scholar]
- 27.Ericsson H, et al. An outbreak of listeriosis suspected to have been caused by rainbow trout. Journal of Clinical Microbiology 1997; 35: 2904–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miettinen H, Wirtanen G. Prevalence and location of Listeria monocytogenes in farmed rainbow trout. International Journal of Food Microbiology 2005; 104: 135–143. [DOI] [PubMed] [Google Scholar]
- 29.Peiris IP, et al. Gravad (Gravlax) and cold-smoked salmon, still a potential source of listeriosis. Journal of Foodservice 2009; 20: 15–20. [Google Scholar]