SUMMARY
The food court at a shopping mall is a potential transfer point for pathogenic microbes, but to date, this environment has not been the subject of detailed molecular microbiological study. We used a combination of culture-based and culture-independent approaches to investigate the types and numbers of bacteria present on food court tables, and on a food court cleaning cloth. Bacteria were found at 102–105 c.f.u./m2 on food court tables and 1010 c.f.u./m2 on the cleaning cloth. Tag-pyrosequencing of amplified 16S rRNA genes revealed that the dominant bacterial types on the cleaning cloth were genera known to include pathogenic species (Stenotrophomonas, Aeromonas), and that these genera were also evident at lower levels on table surfaces, suggesting possible cross-contamination. The evidence suggests a public health threat is posed by bacteria in the food court, and that this may be due to cross-contamination between cleaning equipment and table surfaces.
Key words: Bacteriology, public health microbiology
Shopping malls are one of the major communal gathering places in urban societies, and are thus a prime location for the spread of microorganisms of public health concern. One area within the shopping mall that might be a key point for accumulation and transfer of microbes is the food court. This location has a high turnover of people, contains abundant microbial nutrients, and plays host to intimate contacts between patrons, food, and fomites such as tables – the latter would be expected to transfer microbes readily onto human skin [1, 2].
Several types of microbial transmission events could take place in the food court environment including: spread of highly contagious pathogens (such as common cold and influenza viruses) or opportunistic pathogens (e.g. Staphylococcus aureus) from one patron to another, spread of foodborne pathogens (e.g. Salmonella spp.) from food to patrons, and spread of environmental opportunistic pathogens (e.g. Pseudomonas aeruginosa) from the environment to patrons. In all of these cases, the role of proper cleaning practices in minimizing infection is paramount, especially in terms of minimizing the microbial load on fomites such as table surfaces.
Microbial contamination of public surfaces is well documented [3–7]. One study in a university [3] found the highest bacterial counts on telephone mouthpieces [up to 169 colony-forming units (c.f.u.)/cm2], lower counts on elevator buttons, computer keyboards, and tables (1–9 c.f.u./cm2), and detected two major species of concern, S. aureus and Stenotrophomonas (Sten.) maltophilia. An investigation of surfaces in trains, buses, and public areas of a hospital in London yielded a median count of 12 c.f.u./cm2, and S. aureus was detected at 8% of sites [6]. An analysis of computer keyboards in common areas in a university found that 60% of these were contaminated with methicillin-resistant S. aureus [8]. An extensive study across four cities in the USA [4] revealed mean bacterial counts on public surfaces ranging from 0·5 to 4 c.f.u./cm2, with faecal coliforms detected in 1·5% of samples. A handful of studies have used modern molecular ecological tools to investigate microbial communities on surfaces of public health significance [8, 9], but to date, there have been no detailed molecular microbiology studies focused on the shopping mall food court environment.
The aim of this study was to determine if the numbers and types of bacteria present on food court tables and cleaning equipment pose a potential public health risk. The specific objectives were to count aerobic heterotrophs on individual tables and on cleaning equipment, to compare the types of bacteria on the tables and the cleaning equipment, to detect potential pathogens, to detect faecal indicator organisms, and to compare a direct DNA extraction method with a culture-based method.
Study site and methods
A food court in a large shopping mall in Sydney, Australia was sampled on two dates, 5 months apart. On each date, swab samples were collected from five different tables, which were estimated to be cleaned 3–5 times per day. These samples were designated FC1–FC10. Each table was 70 cm × 100 cm (0·7 m2). Before sampling, nitrile gloves were donned, swabbed with 80% (v/v) ethanol, allowed to air dry, then the entire table surface was swabbed with a sterile moist wipe [a 10 cm × 10 cm section of a Wypall X50 (Kimberly Clarke, Australia)]. A negative control consisted of a sterile wipe handled for 1 min with ethanol-swabbed gloves, but not allowed to contact any other surfaces. Each wipe was transferred to a 50-ml tube and returned to the laboratory upon completion of sampling.
Glass beads (10 beads, each 5 mm diameter) and 30 ml buffer [20 mm phosphate-buffered saline containing 0·05% (v/v) Tween-80] were added to each tube and vortexed for 1 min. The wipe was drained against the side of the tube, excess liquid was squeezed out with sterile forceps, and the wipe was discarded. The remaining liquid was centrifuged (15 min at 5000 g), and the pellet was suspended in 1 ml buffer. A reusable cleaning cloth (sample CC; nylon-reinforced cellulose sponge, 3 mm thick) that had been used in the food court for several weeks was obtained on the first sampling date, and processed similarly to the table wipes, using a 10 cm × 10 cm section cut from the cloth.
Dilutions of the 1 ml suspensions derived from the table wipes or cleaning cloth were plated in triplicate onto R2A medium, and colonies counted after 2 weeks incubation at 25 °C. Two cell suspensions were subsequently made from colonies appearing on the R2A plates – the first of these was pooled from five plates (using one plate each from sample: FC1, FC2, FC3, FC4, FC5), while the second was pooled from triplicate plates from sample CC. In all these cases, plates containing about 100 colonies were used. DNA was extracted from cell suspensions via beadbeating [10]. The 16S rRNA genes were amplified by PCR using primers 27F and 519R, and the amplicons were sequenced by tag-pyrosequencing using the 27F primer (Research and Testing Laboratory, USA). The raw sequence data (about 10 000 sequences per sample) were subjected to a bioinformatics pipeline involving removal of chimeras and sequences containing large homopolymer tracts, then genus/species identities were inferred by automated BLAST search against the Ribosomal Database Project (RDP) [11]. The methods for PCR, sequencing and bioinformatic analysis have been described in detail elsewhere [12, 13].
Viable count results
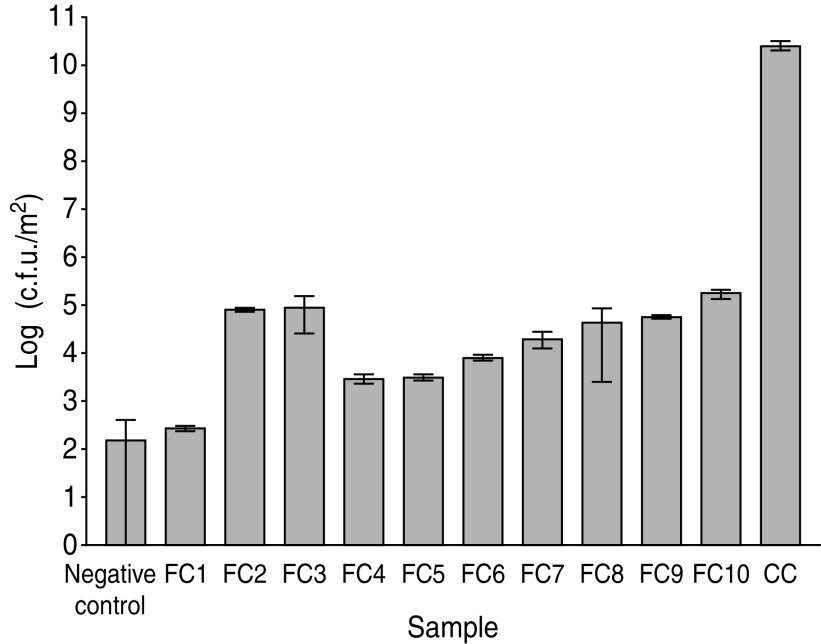
The viable counts of aerobic heterotrophs on table surfaces ranged from 2·7 × 102 to 1·7 × 105 c.f.u./m2 (Fig. 1). The average count was 4·2 × 104 c.f.u./m2 and the standard deviation was 5·6 × 104 c.f.u./m2. The negative control was not sterile, but gave a relatively low count (1·5 × 102 c.f.u./m2). One of the sampled tables yielded a count in the same range as the negative control, indicating this table was essentially uncontaminated, but the remainder of tables all yielded counts at least one order of magnitude greater. The viable count from the cleaning cloth (2·5 × 1010 c.f.u./m2) was much higher than any other sample, which may be due to the porous and moist environment in this material, which was unlike the table surfaces. Although there is no Australian or international standard for surface hygiene, a Canadian guideline [14] defines ‘clean’ as less than 5 × 104 c.f.u./m2.
Fig. 1.
Counts of aerobic heterotrophs on individual food court tables and a food court cleaning cloth. Columns represent the average count from three plates, and error bars show one standard deviation.
Bacterial community DNA sequencing results
Only very low yields of DNA were extracted from the table wipe samples, and these DNA samples either yielded no PCR products or very faint PCR products, which were insufficient for sequencing. It is possible that PCR inhibitors were co-extracted from the table surfaces or the wipes themselves, which contributed to this problem. These results limited the scope of sequencing analyses to the cells recovered directly from the cleaning cloth (CC, 5456 sequences, 395 ± 73 bp), colonies grown on R2A from the cleaning cloth (RCC, 8106 sequences, 384 ± 79 bp), and colonies grown on R2A from the table wipes (RTW, 6045 sequences, 413 ± 78 bp). Bacterial genera that contain pathogenic species were abundant in the food court samples, including: Stenotrophomonas, Acinetobacter, Aeromonas, Enterobacter, Citrobacter, Burkholderia, Pseudomonas, and Staphylococcus (Table 1). Complete details of sequencing results can be found in Supplementary Tables S2–S4.
Table 1.
Relative abundance and best Genbank matches of 16S rRNA gene sequences in different food court samples
| Cleaning cloth direct analysis (CC) | Cleaning cloth grown on R2A (RCC) | Table wipes grown on R2A (RTW) | |||
|---|---|---|---|---|---|
| Best match†‡ | % seqs | Best match | % seqs | Best match | % seqs |
| Stenotrophomonas* | 34·06 | Stenotrophomonas* | 46·92 | Staphylococcus* | 22·99 |
| Acinetobacter* | 16·10 | Chryseobacterium | 24·74 | Micrococcus | 14·18 |
| Aeromonas* | 14·06 | Delftia | 4·84 | Novosphingobium | 11·30 |
| Acidovorax | 8·00 | Pseudomonas* | 4·21 | Acinetobacter* | 6·58 |
| Janthinobacterium | 7·56 | Janthinobacterium | 3·22 | Paenibacillus | 5·38 |
| Chryseobacterium | 4·94 | Acinetobacter | 3·14 | Microbacterium | 4·76 |
| Pseudomonas* | 2·67 | Comamonas | 4·23 | Leifsonia | 4·58 |
| Delftia | 2·00 | Acidovorax | 1·77 | Massilia | 2·42 |
| Massilia | 1·41 | Enterobacter* | 1·60 | Bacillus | 2·17 |
| Aquitalea | 1·06 | Citrobacter* | 0·73 | Enterobacter* | 2·08 |
| Duganella | 0·86 | Escherichia* | 0·47 | Pantoea | 1·88 |
| Vogesella | 0·83 | Roseateles | 0·31 | Kocuria | 1·41 |
| Comamonas | 0·67 | Rhizobium | 0·30 | Kluyvera | 1·29 |
| Sphingobium | 0·48 | Duganella | 0·26 | Aerococcus | 1·13 |
| Sphingomonas | 0·42 | Herbaspirillum | 0·20 | Arthrobacter | 1·06 |
| Enterobacter* | 0·42 | Brevundimonas | 0·19 | Brevibacterium | 0·96 |
| Escherichia* | 0·37 | Pantoea | 0·28 | Geobacillus | 0·90 |
| Burkholderia* | 0·30 | Leptothrix | 0·10 | Corynebacterium* | 0·63 |
| Staphylococcus* | 0·28 | Achromobacter | 0·10 | Aeromonas* | 0·60 |
| Citrobacter* | 0·26 | Aeromonas* | 0·09 | Massilia | 0·58 |
Indicates bacterial genera that contain species of potential public health concern.
Only the 20 most abundant sequence types in each sample are shown in this table.
Automatic BLAST analysis with genus-level cut-off at 95% or higher sequence identity.
While it is not possible to obtain conclusive species-level identifications from the short pyrosequencing reads, we believe it is worthwhile to report the closest species matches for the food court sequences. The raw sequence data was reprocessed using the pyrosequencing pipeline at the RDP to yield a set of representative sequence types, and these were then individually analysed by BLASTn against GenBank. This analysis (Supplementary Table S1) revealed that all the sequence types assigned to Stenotrophomonas were most closely related to the opportunistic pathogen Sten. maltophilia, and all of the Aeromonas sequences were most closely related to the pathogenic species Aeromonas hydrophila. A small fraction of the Acinetobacter, Staphylococcus and Pseudomonas sequences were most closely related to pathogens (Acinetobacter baumannii, S. aureus, P. aeruginosa). The Escherichia sequences gave equal matches to E. coli and E. hermanii. The high numbers of sequences retrieved from diverse genera of Enterobacteriaceae (e.g. Escherichia, Citrobacter, Enterobacter, Pantoea) are suggestive of some level of faecal contamination on the food court tables – this may be due to the poor hygiene of the general public [15], or due to cross-contamination between the food court and bathrooms, perhaps mediated by use of the same cleaning cloth in both areas.
Comparison of the sequences from CC and RCC samples provides a measure of the bias of culturing vs. direct DNA extraction. As expected, the sequences from sample CC were more diverse overall than the sequences from sample RCC. The sequence composition of these two samples was similar, but not identical. In both cases, Stenotrophomonas was detected as the major type, and high levels of Acinetobacter were present. One notable difference was that Aeromonas sequences were two orders of magnitude less abundant in RCC compared to CC. This could be because the culture conditions used here were not appropriate for Aeromonas and/or because the aeromonads were overgrown by other bacteria on the general purpose R2A plates [16]. The comparison of samples CC and RCC highlights the usefulness of the culture-independent approach in detecting diverse types of pathogens.
Comparison of RCC and RTW samples revealed marked differences – the most obvious was that Gram-positive sequences were the most abundant types in the RTW sample, while few were detected in the RCC sample. The most likely explanation for this is that Gram-positives generally have higher desiccation resistance than Gram-negatives, and thus would be expected to be more abundant on a dry table surface compared to a moist cleaning cloth environment. Staphylococci in particular are capable of survival for months on dry, non-porous surfaces [17]. Normal flora from human skin (Staphylococcus warneri and Micrococcineae) were the dominant types in the RTW sample, but there was also evidence in this sample for cross-contamination with bacterial types detected in the cleaning cloth, such as Acinetobacter, Aeromonas, and sphingomonads.
Conclusions
The control of microbial contamination in any environment depends upon effective cleaning practices. In the food court studied, the tables were cleaned on a semi-continuous basis. However, such cleaning efforts would be confounded by the abundant bacterial community on the reusable cleaning cloth, which may be acting as both a reservoir and a vehicle for bacterial contamination. Of particular concern in this case is that the most dominant bacterial types detected on the cloth were most closely related to opportunistic pathogens such as Sten. maltophilia and Aeromonas hydrophila. While these organisms are not likely to be implicated in disease outbreaks in the same manner as foodborne infections such as Salmonella, they could cause sporadic cases of disease (such as wound infections), which would be difficult to trace back to the food court environment. While the experiments described here are a snapshot of a small number of tables and a single cleaning cloth in one shopping mall, the findings are of sufficient concern to warrant further investigation, to determine the extent to which these findings are encountered in other shopping malls.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268812002142.
click here to view supplementary material
Supplementary information supplied by authors.
ACKNOWLEDGEMENTS
We thank the cleaning staff of this shopping mall for their cooperation in obtaining samples. Melissa Coad and Kirsty McCully are acknowledged for assistance in sample collection and project management. This research was supported by funding from the United Voice Union, Australia.
DECLARATION OF INTEREST
None
REFERENCES
- 1.Rusin P, Maxwell S, Gerba C. Comparative surface-to-hand and fingertip-to-mouth transfer efficiency of gram-positive bacteria, gram-negative bacteria, and phage. Journal of Applied Microbiology 2002; 93: 585–592. [DOI] [PubMed] [Google Scholar]
- 2.Todd ECD, et al. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 6. Transmission and survival of pathogens in the food processing and preparation environment. Journal of Food Protection 2009; 72: 202–219. [DOI] [PubMed] [Google Scholar]
- 3.Brooke JS, et al. Investigation of bacterial pathogens on 70 frequently used environmental surfaces in a large urban US university. Journal of Environmental Health 2009; 71: 17–22. [PubMed] [Google Scholar]
- 4.Reynolds KA, et al. Occurrence of bacteria and biochemical markers on public surfaces. International Journal of Environmental Health Research 2005; 15: 225–234. [DOI] [PubMed] [Google Scholar]
- 5.Cosby CM, et al. Microbiological analysis of food contact surfaces in child care centers. Applied and Environmenal Microbiology 2008; 74: 6918–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otter JA, French GL. Bacterial contamination on touch surfaces in the public transport system and in public areas of a hospital in London. Letters in Applied Microbiology 2009; 49: 803–805. [DOI] [PubMed] [Google Scholar]
- 7.Mizumachi E, et al. Clonal distribution of enterotoxigenic Staphylococcus aureus on handles of handheld shopping baskets in supermarkets. Journal of Applied Microbiology 2011; 110: 562–567. [DOI] [PubMed] [Google Scholar]
- 8.Kassem II, Sigler V, Esseili MA. Public computer surfaces are reservoirs for methicillin-resistant staphylococci. ISME Journal 2007; 1: 265–258. [DOI] [PubMed] [Google Scholar]
- 9.Flores GE, et al. Microbial biogeography of public restroom surfaces. PLoS One 2011; 6: e28132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeates C, Gillings MR. Rapid purification of DNA from soil for molecular biodiversity analysis. Letters in Applied Microbiology 1998; 27: 49–53. [Google Scholar]
- 11.Cole JR, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Research 2009; 37: D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowd SE, et al. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiology 2008; 8: 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey MT, et al. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infection and Immunity 2010; 78: 1509–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.BC Centre for Disease Control Provincial Health Services Authority. Environmental hygiene monitoring: a guide for environmental health officers. Vancouver, Canada, 2010. [Google Scholar]
- 15.Judah G, et al. Dirty hands: bacteria of faecal origin on commuters' hands. Epidemiology and Infection 2010; 138: 409–414. [DOI] [PubMed] [Google Scholar]
- 16.Villari P, et al. A comparison of different culture media for the membrane filter quantification of Aeromonas in water. Letters in Applied Microbiology 1999; 29: 253–257. [DOI] [PubMed] [Google Scholar]
- 17.Neely AN, Maley MP. Survival of enterococci and staphylococci on hospital fabrics and plastic. Journal of Clinical Microbiology 2000;38: 724–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268812002142.
click here to view supplementary material
Supplementary information supplied by authors.