Abstract
Myeloproliferative neoplasms (MPNs) are associated with significant alterations in the bone marrow microenvironment that include decreased expression of key niche factors and myelofibrosis. Here, we explored the contribution of TGF-β to these alterations by abrogating TGF-β signaling in bone marrow mesenchymal stromal cells. Loss of TGF-β signaling in Osx-Cre–targeted MSCs prevented the development of myelofibrosis in both MPLW515L and Jak2V617F models of MPNs. In contrast, despite the absence of myelofibrosis, loss of TGF-β signaling in mesenchymal stromal cells did not rescue the defective hematopoietic niche induced by MPLW515L, as evidenced by decreased bone marrow cellularity, hematopoietic stem/progenitor cell number, and Cxcl12 and Kitlg expression, and the presence of splenic extramedullary hematopoiesis. Induction of myelofibrosis by MPLW515L was intact in Osx-Cre Smad4fl/fl recipients, demonstrating that SMAD4-independent TGF-β signaling mediates the myelofibrosis phenotype. Indeed, treatment with a c-Jun N-terminal kinase (JNK) inhibitor prevented the development of myelofibrosis induced by MPLW515L. Together, these data show that JNK-dependent TGF-β signaling in mesenchymal stromal cells is responsible for the development of myelofibrosis but not hematopoietic niche disruption in MPNs, suggesting that the signals that regulate niche gene expression in bone marrow mesenchymal stromal cells are distinct from those that induce a fibrogenic program.
Keywords: Hematology, Oncology
Keywords: Bone marrow, Fibrosis, Leukemias
Introduction
The bone marrow provides a specialized microenvironment that supports hematopoiesis. Hematopoietic niches in the bone marrow are dependent, in part, on the constitutive production of certain cytokines and chemokines by a heterogeneous population of stromal cells. For example, expression of Kit ligand (Kitlg) and CXCL12 by perivascular mesenchymal stromal cells (MSCs) plays an essential role in hematopoietic stem cell (HSC) maintenance (1–3). There are emerging data suggesting that malignant hematopoietic cells induce alterations in the bone marrow microenvironment that contribute to hematopoietic phenotypes (4). A better understanding of the interactions of malignant hematopoietic cells with bone marrow MSCs may identify novel targets for therapeutic intervention.
Myeloproliferative neoplasms (MPNs) are chronic myeloid malignancies characterized by overproduction of myeloid lineage cells (5). Activating mutations in genes impacting the JAK/STAT signaling pathway are the major causes of these cancers, including mutations in JAK2, CALR, and MPL (6). Alterations in the bone marrow microenvironment are a prominent feature of MPNs. Most striking is the development of myelofibrosis, which is characterized by extensive collagen deposition in the bone marrow and is associated with a poor prognosis (7, 8). Recent data show that RNA expression of key niche factors, including Kitlg and Cxcl12, is reduced in bone marrow MSCs from mouse models of MPNs and from patients with myelofibrosis (9–11). This is relevant, since studies by our group and others have shown that deleting these niche factors from stromal cells results in a loss of hematopoietic stem/progenitor cells (HSPCs) in the bone marrow and development of extramedullary hematopoiesis (1–3). There is also evidence that expression of CXCR4, the major receptor for CXCL12, is reduced on HSPCs in patients with MPNs, which would further reduce CXCL12 signaling (12). On the other hand, Migliaccio et al. reported that CXCL12 protein expression, as measured by immunostaining, is increased in the bone marrow of patients with MPNs and in the Gata1low mouse model of MPN (13). Thus, the impact of the MPN on bone marrow stromal niche factor expression (particularly CXCL12) remains unclear. Finally, there is evidence for altered bone metabolism in MPNs, with osteosclerosis reported in patients with myelofibrosis (14). Collectively, these alterations in the bone marrow microenvironment in MPNs are predicted to reduce its ability to support hematopoiesis. Indeed, extramedullary hematopoiesis and splenomegaly are common in MPNs.
There is evidence implicating inflammatory mediators in the development of myelofibrosis. In particular, increased production of TGF-β produced by megakaryocytes and monocytes is found in most patients with MPNs (15–19). Using a thrombopoietin (TPO) overexpression model of MPN, Chagraoui et al. showed that deletion of Tgfb1 reduced but did not completely abrogate the development of myelofibrosis (20). Likewise, treatment with an inhibitor targeting TGFBR1 (ALK5) reduced myelofibrosis in mouse models of MPNs (21, 22). However, the role of TGF-β in the development of myelofibrosis remains controversial. In the aforementioned studies, although reduced, significant myelofibrosis remained despite inhibiting TGF-β signaling, raising the possibility that there are other mediators of myelofibrosis. Consistent with this possibility, Decker et al. showed that platelet-derived growth factor α (PDGFα) signaling in leptin receptor–positive (Lepr+) MSCs is required for the development of myelofibrosis (9). Moreover, Schneider et al. reported that CXCL4 signaling in Gli1+ MSCs contributes to myelofibrosis (10, 23). The cell of origin responsible for myelofibrosis is also controversial. As noted above, prior studies have implicated Lepr+ or Gli1+ MSCs as the target of inflammatory signaling leading to myelofibrosis (9, 10, 23). On the other hand, there are studies suggesting that monocyte-derived fibrocytes (24, 25) or endothelial cells (26) are the cell of origin.
In this study, we tested the hypothesis that TGF-β signaling in MSCs contributes to the development of myelofibrosis and hematopoietic niche disruption in MPNs. We show that genetic ablation of TGF-β signaling in MSCs, but not osteolineage cells, abrogates myelofibrosis. This effect is mediated by noncanonical c-Jun N-terminal kinase (JNK) signaling, and treatment of mice with a JNK inhibitor prevents the development of myelofibrosis. However, loss of TGF-β signaling in MSCs does not rescue the impaired hematopoietic niche function in MPNs, with persistent decreases in bone marrow cellularity, HSPC number, and Cxcl12 and Kitlg expression, and development of extramedullary hematopoiesis.
Results
TGF-β signaling in MSCs is required for the efficient induction of myelofibrosis by MPLW515L or Jak2V617F in mice.
Prior single-cell RNA sequencing studies of murine bone marrow MSCs show that Tgfbr2 is broadly expressed in MSCs, including Lepr+ perivascular stromal cells and osteolineage cells (27–31). To investigate the contribution of TGF-β signaling in MSCs to the pathogenesis of MPN, we used Osx-Cre Tgfbr2fl/fl mice as transplant recipients of hematopoietic cells carrying MPLW515L or Jak2V617F. The doxycycline-regulated Osx-Cre transgene has been shown to target osteoblasts, osteocytes, adipocytes, and perivascular stromal cells (32). Of note, our group recently showed that basal hematopoiesis is normal in Osx-Cre Tgfbr2fl/fl mice in which expression of Cre recombinase was activated at birth by removal of doxycycline, suggesting that TGF-β signaling in MSCs is dispensable for maintenance of the hematopoietic niche under basal conditions (33). To confirm efficient deletion of Tgfbr2, we generated Osx-Cre Tgfbr2fl/fl Ai9 mice, in which stromal cells targeted by Osx-Cre express TdTomato. Although Tgfbr2 mRNA expression in sorted TdTomato+ bone marrow MSCs was significantly reduced in Osx-Cre Tgfbr2fl/fl mice, Cre-mediated excision was incomplete, with Tgfbr2 mRNA levels reduced by approximately 66% compared with control cells (Figure 1A). Thus, to increase the percentage of Tgfbr2-deleted stromal cells, we generated Osx-Cre Tgfbr2fl/– Ai9 mice, in which 1 allele of Tgfbr2 is constitutively deleted. Tgfbr2 mRNA in TdTomato+ stromal cells was reduced to less than 10% of control cells in these mice. Moreover, single-cell RNA sequencing of sorted lineage– (CD45–CD3–B220–Gr-1–CD11b–Ter119–), PDGF receptor β+ (PDGFRβ+) cells from Osx-Cre Ai9 mice confirmed that the majority (>90%) of CXCL12+Lepr+ stromal cells expressed tdTomato (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI154092DS1).
Figure 1. TGF-β signaling in Osx-Cre–targeted mesenchymal stromal cells is not required for the development of the myeloproliferative phenotype by MPLW515L.
(A) Tgfbr2 mRNA expression in sorted lineage–tdTomato+ bone marrow MSCs relative to β-actin mRNA. (B) Schematic of experimental design. (C) Tgfb1 mRNA expression relative to β-actin in total bone marrow. (D) White blood cell (WBC) count, (E) red blood cell (RBC) count, (F) hematocrit (HCT), and (G) platelet (PLT) count 4–8 weeks after transplantation and (H) Kaplan-Meier estimates of survival. EV, empty vector; Tx, transplant. Data presented as the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by 1-way ANOVA (A and D–G) or Student’s t test (C).
We first used the MPLW515L retroviral model of MPN in which c-Kit+ cells from WT mice transduced with retrovirus expressing MPLW515L and GFP are transplanted into congenic Tgfbr2fl/fl or Osx-Cre Tgfbr2fl/– mice; we also included Tgfbr2fl/– mice to control for global Tgfbr2 haploinsufficiency (Figure 1B). Of note, Tgfb1 mRNA expression in total bone marrow was modestly but significantly elevated in this model (Figure 1C), with an increase in TGF-β+ megakaryocytes (Supplemental Figure 2). As reported previously, transplantation of MPLW515L HSPCs into Tgfbr2fl/fl recipients resulted in a fatal MPN characterized by leukocytosis and modest erythrocytosis and thrombocytosis (Figure 1, D–H). A similar hematopoietic phenotype was observed in Osx-Cre Tgfbr2fl/– recipients and animal survival was not altered. These data suggest that TGF-β signaling in MSCs is not required for the development of the lethal myeloproliferative hematopoietic phenotype induced by MPLW515L.
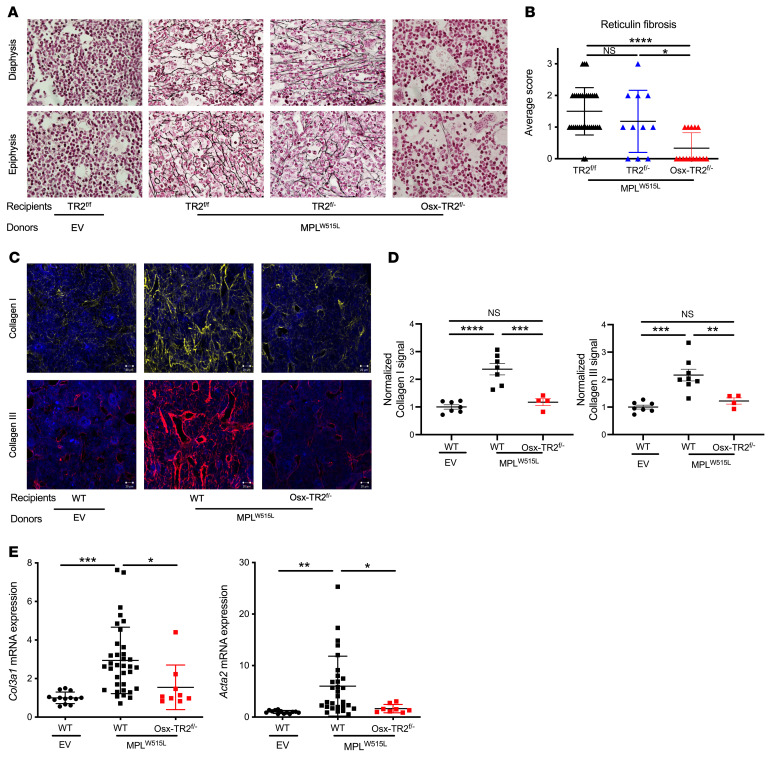
We next investigated the myelofibrosis phenotype induced by MPLW515L. As reported previously, transplantation of MPLW515L-transduced HSPCs induced a moderate to severe reticulin fibrosis in the bone marrow of both Tgfbr2fl/fl and Tgfbr2fl/– recipients (Figure 2, A and B). In contrast, reticulin fibrosis was significantly abrogated in Osx-Cre Tgfbr2fl/– recipients. Since we observed that Tgfbr2fl/fl and Tgfbr2fl/– recipients have similar phenotypes, we subsequently pooled data from these 2 groups as WT controls. To further characterize the myelofibrosis phenotype, we assessed collagen I and III deposition in the bone marrow. In WT recipients transplanted with MPLW515L-transduced HSPCs, a marked increase in perivascular collagen I and III was observed, which was abrogated in Osx-Cre Tgfbr2fl/– recipients (Figure 2, C and D). Increased α-smooth muscle actin (Acta2) expression has been reported in myelofibrosis (10). Indeed, in the MPLW515L transplantation model, Acta2 and Col3a1 (collagen III) mRNAs were modestly increased, whereas no elevation was observed in Osx-Cre Tgfbr2fl/– recipients (Figure 2E). Interestingly, modest splenic reticulin fibrosis was induced after transplantation of MPLW515L-transduced HSPCs in both WT and Osx-Cre Tgfbr2fl/– recipients (Supplemental Figure 3). Of note, micro-CT analysis showed that osteosclerosis did not develop in any of the cohorts (Supplemental Figure 4).
Figure 2. TGF-β signaling in Osx-Cre–targeted mesenchymal stromal cells is essential for the development of myelofibrosis by MPLW515L.
(A) Representative photomicrographs of femur sections stained for reticulin (original magnification, ×60). (B) Average score of fibrosis grading in the diaphysis and epiphysis. (C) Representative photomicrographs of femur sections stained for collagen I (yellow) or collagen III (red); nuclei were stained blue with DAPI. Scale bar: 20 μm. (D) Normalized fluorescence intensity for collagen I and III. (E) mRNA expression levels of collagen 3 (Col3a1) and α-smooth muscle actin (Acta2) relative to β-actin mRNA in total bone marrow. EV, empty vector; TR2fl/fl, Tgfbr2fl/fl; TR2fl/–, Tgfbr2fl/–; Osx-TR2fl/–, Osx-Cre Tgfbr2fl/–. Mice were analyzed 4–8 weeks after transplantation. Data presented as the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by Kruskal-Wallis test (B) or 1-way ANOVA (D and E).
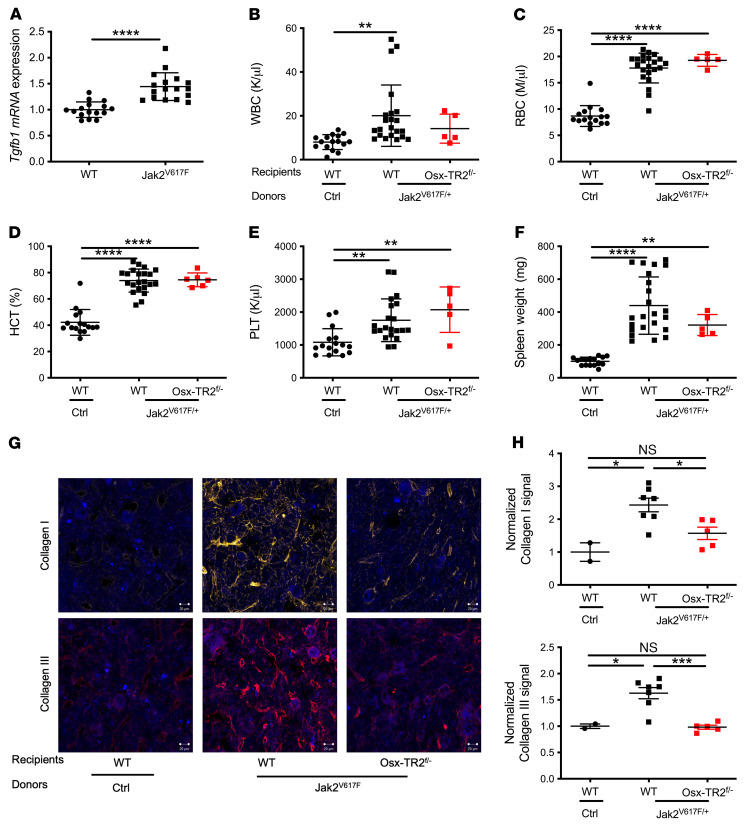
Similar results were observed in a Jak2V617F transgenic model of MPN. Bone marrow from Jak2V617F mice was transplanted into Osx-Cre Tgfbr2fl/– or control recipients. Increased Tgfb1 mRNA expression and TGF-β+ megakaryocytes were observed (Figure 3A and Supplemental Figure 2). The hematopoietic phenotype was similar in all mice, including erythrocytosis, leukocytosis, thrombocytosis, and splenomegaly (Figure 3, B–F). As reported previously (34), the degree of myelofibrosis in Jak2V617F mice was less severe than that observed in the MPLW515L model, with no consistent reticulin fibrosis seen. However, increased collagen I and III deposition in the bone marrow was observed in control, but not Osx-Cre Tgfbr2fl/–, recipients (Figure 3, G and H). Altogether, these data show that TGF-β signaling in MSCs is required for the development of myelofibrosis induced by both MPLW515L and Jak2V617F.
Figure 3. TGF-β signaling in Osx-Cre–targeted mesenchymal stromal cells is essential for the development of myelofibrosis by Jak2V617F.
(A) Tgfb1 mRNA expression relative to β-actin mRNA in total bone marrow. (B) White blood cell (WBC) count, (C) red blood cell (RBC) count, (D) hematocrit (HCT), and (E) platelet (PLT) count and (F) spleen weight 5 months after tamoxifen-induced Jak2V617F expression. (G) Representative photomicrographs of femur sections stained for collagen I (yellow) or collagen III (red); nuclei were stained blue with DAPI. Scale bar: 20 μm. (H) Normalized fluorescence intensity for collagen I and III. Osx-TR2fl/–, Osx-Cre Tgfbr2fl/–. Data presented as the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by Student’s t test (A) or 1-way ANOVA (B–E and G).
Abrogation of TGF-β signaling in MSCs does not rescue the defective hematopoietic niche in MPLW515L mice.
Decreased stromal cell expression of key niche factors, Cxcl12 and Kitlg, has been reported in several mouse models of MPNs (9–11). Consistent with these reports, we show that Cxcl12 mRNA expression in the bone marrow of control recipients transplanted with MPLW515L-transduced HSPCs was markedly decreased (Figure 4A). Surprisingly, a similar decrease in Cxcl12 mRNA expression was observed in Osx-Cre Tgfbr2fl/– recipients. Likewise, a modest but significant decrease in Kitlg mRNA expression in both control and Osx-Cre Tgfbr2fl/– recipients was observed (Figure 4B). Of note, we and others previously reported that loss of Cxcl12 or Kitlg expression in bone marrow MSCs is sufficient to disrupt hematopoietic niches, resulting in a decrease in bone marrow cellularity and HSC number and development of extramedullary hematopoiesis (1–3). Indeed, similar decreases in bone marrow cellularity and HSC number were observed in control and Osx-Cre Tgfbr2fl/– recipients of MPLW515L-transduced HSPCs (Figure 4, C and D). Moreover, splenomegaly and significant increases in HSCs and erythroid progenitors in the spleen were observed in both control and Osx-Cre Tgfbr2fl/– recipients (Figure 4, E–H, and Supplemental Figure 5). Collectively, these data suggest that, despite the absence of myelofibrosis, loss of TGF-β signaling in MSCs does not rescue the defective hematopoietic niche induced by MPLW515L.
Figure 4. TGF-β signaling in Osx-Cre–targeted mesenchymal stromal cells is not required for suppression of niche factor gene expression or development of extramedullary hematopoiesis by MPLW515L.
(A and B) mRNA expression of Cxcl12 (A) and Kitlg (B) in total bone marrow relative to β-actin mRNA. (C) Bone marrow cellularity per pelvis and (D) number of phenotypic HSCs (lineage–Sca1+c-kit+CD150+CD48–) in the bone marrow. (E) Spleen weight and (F) HSC number in spleen. (G and H) Number of erythroid progenitors in bone marrow (G) and spleen (H). I, proerythroblast; II, basophilic erythroblasts; III, polychromatic erythroblasts; IV, orthochromatic erythroblasts; EV, empty vector; Osx-TR2fl/–, Osx-Cre Tgfbr2fl/–. Data presented as the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by 1-way ANOVA.
Deletion of Tgfbr2 in osteolineage cells does not affect MPLW515L-induced myelofibrosis.
Osx-Cre targets a broad range of MSCs, including osteolineage cells, perivascular CXCL12-abundent reticular (CAR) cells, and adipocytes (32). To further identify the stromal cell population responsible for myelofibrosis, we used a Dmp1-Cre transgene that targets osteoblasts and approximately 30% of CAR cells, which we previously showed are enriched for osteolineage genes (35). Efficient deletion of Tgfbr2 was confirmed in TdTomato+ stromal cells isolated from Dmp1-Cre Tgfbr2fl/fl Ai9 mice (Supplemental Figure 6A). As expected, similar alterations in peripheral blood counts, splenomegaly, and bone marrow cellularity were observed in control and Dmp1-Cre Tgfbr2fl/fl recipients of MPLW515L-transduced HSPCs (Supplemental Figure 6, B–F). Interestingly, a similar degree of myelofibrosis, as measured by reticulin staining and Col3a1 mRNA expression, was observed in control and Dmp1-Cre Tgfbr2fl/fl recipients (Supplemental Figure 6, G–I). These data show that TGF-β signaling in osteolineage cells is not required for the development of myelofibrosis.
Canonical (SMAD4-dependent) TGF-β signaling in MSCs is not required for MPLW515L-induced myelofibrosis.
TGF-β signaling consists of SMAD-dependent canonical pathways and the SMAD-independent noncanonical pathways. To assess the role of canonical TGF-β signaling in the development of myelofibrosis, we generated Osx-Cre Smad4fl/fl mice. Cre-mediated recombination was induced postnatally by removing doxycycline chow at birth. We previously showed that deletion of Smad4 is very efficient in Osx-Cre–targeted cells, with essentially undetectable Smad4 mRNA (36). Of note, we reported that these mice have normal basal hematopoiesis, with no identifiable alteration in bone marrow MSCs (36). MPLW515L-transduced HSPCs were transplanted into Osx-Cre Smad4fl/fl recipients and similar degrees of leukocytosis, erythrocytosis, thrombocytosis, splenomegaly, and reduction in bone marrow cellularity were observed in control and MPLW515L-transduced HSPCs (Supplemental Figure 7). Surprisingly, the degree of myelofibrosis was similar in control and Osx-Cre Smad4fl/fl recipients, as measured by reticulin staining (Figure 5, A and B), collagen I and III immunofluorescence (Figure 5, C and D), and Col3a1 mRNA expression (Figure 5E). These data suggest that canonical (SMAD4-dependent) TGF-β1 signaling is not required for the development of myelofibrosis in the MPLW515L murine model of MPN.
Figure 5. Canonical (SMAD4-dependent) TGF-β signaling in mesenchymal stromal cells is not required for the induction of myelofibrosis by MPLW515L.
(A) Representative photomicrographs of femur sections stained for reticulin (original magnification, ×60). (B) Average score of fibrosis grading in the diaphysis and epiphysis. (C) Representative photomicrographs of femur sections stained for collagen I (yellow) or collagen III (red); nuclei were stained blue with DAPI. Scale bar: 20 μm. (D) Normalized fluorescence intensity for collagen I and III. (E) Col3a1 mRNA expression relative to β-actin mRNA in total bone marrow. EV, empty vector; Osx-Smad4fl/fl, Osx-Cre Smad4fl/fl. Mice were analyzed 4 weeks after transplantation. Data presented as the mean ± SEM. *P < 0.05; ***P < 0.001; ****P < 0.0001 by Kruskal-Wallis test (B) or 1-way ANOVA (D and E).
JNK activation by TGF-β contributes to the development of MPLW515L-induced myelofibrosis in mice.
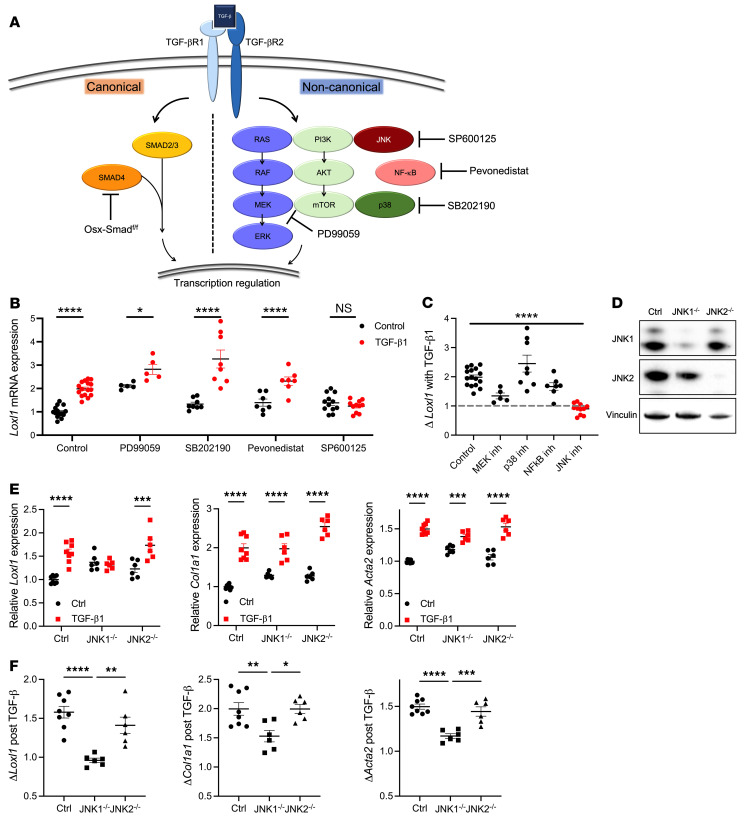
Noncanonical TGF-β signaling includes the activation of extracellular signal–regulated kinase (ERK), JNK, p38, PI3 kinase, and NF-κB pathways (Figure 6A and ref. 37). Treatment of cultured bone marrow MSCs with TGF-β1 resulted in increased expression of several fibrosis-associated genes, including Loxl1 (which encodes lysyl oxidase–like 1), Col1a1, and Acta2 (Figure 6B and Supplemental Figure 8, A–D). It also resulted in Smad2 and c-Jun phosphorylation, but not ERK1/2 phosphorylation (Supplemental Figure 8E). Consistent with this result, treatment with PD99059, a MEK inhibitor, resulted in a modest change in the basal expression of these genes, but it did not block their induction by TGF-β1 (Figure 6, B and C, and Supplemental Figure 8, A–D). Treatment with SB202190 (a p38 inhibitor) or pevonedistat (an NF-κB inhibitor) was associated with a significant decrease in basal Col1a1 and Acta2 mRNA expression, but also did not prevent TGF-β1–induced increases in the expression of these genes (Figure 6, B and C, and Supplemental Figure 8, A–D). In contrast, treatment with SP600125, a pan-JNK inhibitor, significantly reduced TGF-β1–induced increases in Loxl1, Col1a1, and Acta2 mRNA expression (Figure 6, B and C, and Supplemental Figure 8, A–D).
Figure 6. TGF-β contributes to fibrosis-related gene expression through noncanonical JNK signaling.
(A) TGF-β signaling pathways, with inhibitors used in this study included. (B) Relative Loxl1 mRNA in WT MSC cultures treated with TGF-β1 (10 ng/mL) and one of the following inhibitors: the MEK inhibitor PD99059 (20 μM), the p38 inhibitor SB202190 (20 μM), the NF-κB inhibitor pevonedistat (1 μM), or the JNK inhibitor SP600125 (20 μM). (C) Fold change (Δ) in Loxl1 mRNA expression induced by TGF-β1 in the presence of the indicated inhibitors. (D) Immunoblot showing JNK1 and JNK2 protein expression in control or Mapk8-deleted (JNK1–/–) or Mapk9-deleted (JNK2–/–) cells. (E) Relative Loxl1, Col1a1, and Acta2 mRNA in MSCs treated with TGF-β1 (10 ng/mL). (F) Fold change in Loxl1, Col1a1, and Acta2 mRNA expression induced by TGF-β1. Data presented as the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by 2-way ANOVA (B and E) or 1-way ANOVA (C and F).
JNK1 and JNK2, but not JNK3, are expressed in bone marrow MSCs (38). To determine whether JNK1 or JNK2 selectively contributes to TGF-β–induced myelofibrosis, we used CRISPR/Cas9 gene editing to delete Mapk8 (JNK1) or Mapk9 (JNK2) in MSCs. Both JNK1 and JNK2 were efficiently deleted (Figure 6D). Deletion of JNK1 completely abrogated TGF-β–induced Loxl1 expression and significantly reduced TGF-β–induced Col1a1 and Acta2 expression (Figure 6, E and F). However, deletion of JNK2 did not affect TGF-β–induced expression of fibrosis-related genes. Collectively, these data suggest that TGF-β1–induced activation of JNK1 is the major signal mediating myelofibrosis.
Finally, to assess the impact of JNK activation on the development of myelofibrosis, we treated WT mice transplanted with MPLW515L-transduced HSPCs with the pan-JNK inhibitor CC-930 (tanzisertib) twice daily for 4 weeks beginning 2 weeks after transplantation. Of note, this regimen was effective in blocking JNK signaling in lineage– bone marrow MSCs (Supplemental Figure 9). Treatment with CC-930 had no significant effect on MPLW515L-induced leukocytosis, erythrocytosis, thrombocytosis, or splenomegaly (Figure 7, A–D). However, treatment with CC-930 resulted in the nearly complete abrogation of myelofibrosis, as assessed by reticulin fibrosis, collagen I deposition, and Col3a1 mRNA expression (Figure 7, E–I). These data show that treatment with the pan-JNK inhibitor CC-930 prevents induction of myelofibrosis by MPLW515L in mice.
Figure 7. JNK inhibitor alleviates fibrosis phenotypes in MPLW515L-induced MPN.
WT mice transplanted with MPLW515L-transduced HSPCs were treated with pan-JNK inhibitor CC-930 (tanzisertib) twice daily for 4 weeks beginning 2 weeks after transplantation. (A) White blood cell (WBC) count, (B) red blood cell (RBC) count, (C) platelet (PLT) count, and (D) spleen weight. (E) Representative photomicrographs of femur sections stained for reticulin (original magnification, ×60). (F) Average score of fibrosis grading in the diaphysis and epiphysis. (G) Representative photomicrographs of femur sections stained for collagen I (yellow) or collagen III (red); nuclei were stained blue with DAPI. Sacle bar: 20 μm. (H) Normalized fluorescence intensity for collagen I. (I) Col3a1 mRNA expression relative to β-actin mRNA in total bone marrow. EV, empty vector; VE, vehicle-treated control. Data presented as the mean ± SEM. *P < 0.05, **P < 0.01, ****P < 0.0001 by 1-way ANOVA (A–D), Kruskal-Wallis test (F), or Student’s t test (H and I).
Discussion
In this study, we provide strong evidence that TGF-β signaling in Osx-Cre–targeted MSCs plays an essential role in the develop of myelofibrosis. These results are consistent with prior studies showing that genetic or pharmacologic inhibition of TGF-β signaling reduces reticulin fibrosis in TPO overexpression (20) or GATA1low (21) models of MPNs. In addition, Yue et al. showed that inhibition of TGF-β using galunisertib, an ALK5 (TGFBR1) inhibitor, reduces, but does not abrogate, myelofibrosis in the clinically relevant MPLW515L model of MPN. Of note, TGF-β signals through heterodimers of TGFBR2 with either TGFBR1 (ALK5) or ACVRL1 (ALK1), both of which are expressed at similar levels in bone marrow MSCs (28). Thus, the residual myelofibrosis in galunisertib-treated mice may be due to ALK1-dependent TGF-β signaling. Consistent with this conclusion, we show that genetic abrogation of all TGF-β signaling (by deleting Tgfbr2) in MSCs completely prevented the development of myelofibrosis in the majority of mice in both MPLW515L and Jak2V617F models of MPNs. Although requiring experimental validation, we suspect that the low-grade myelofibrosis observed in a minority of Osx-Cre Tgfbr2fl/– recipients is due to inefficient Cre-mediated excision of Tgfbr2.
Prior studies have implicated other inflammatory mediators in the pathogenesis of myelofibrosis. Decker et al. showed that reduction of PDGF signaling in MSCs by deletion of Pdgfra or by treatment with imatinib reduced myelofibrosis in the TPO overexpression model of MPN (9). However, treatment with PDGF did not induce fibrogenic gene expression in cultured bone marrow MSCs (Supplemental Figure 10). Whether PDGF signaling in MSCs provides a permissive signal required for TGF-β–induced fibrogenic gene expression will require further study. Schenider et al. reported that increased hematopoietic cell expression of Cxcl4 contributes to the development of myelofibrosis in the TPO overexpression MPN model (10, 23). However, the known receptors for CXCL4, CXCR3, and CCR1 are not expressed on bone marrow MSCs (28), suggesting that CXCL4 acts in an indirect fashion to induce myelofibrosis. Consistent with this conclusion, Schneider et al. showed that TPO-induced expression of TGF-β in Cxcl4–/– megakaryocytes was attenuated (23). Thus, CXCL4 may act upstream of TGF-β to contribute to myelofibrosis development.
Our data suggest that perivascular MSCs, but not osteoblast-lineage cells, are the key drivers of myelofibrosis. Specifically, we show that abrogation of TGF-β signaling in Osx-Cre–targeted, but not Dmp1-Cre–targeted stromal cells, prevents the development of myelofibrosis. These results are consistent with recent studies suggesting that Lepr+ or Gli1+ MSCs in the bone marrow are the key drivers of myelofibrosis (9, 10). Of note, constitutively active Lepr-Cre, similarly to postnatal active Osx-Cre, targets the majority of perivascular MSCs and osteoblasts (39, 40). Likewise, Gli1-Cre targets both perivascular and endosteal MSCs, including osteoblasts (41). Leimkuhler et al. recently reported results of single-cell RNA sequencing of bone marrow MSCs in a TPO overexpression model of MPN (11). They showed that adipogenic and osteogenic primitive subsets of Lepr+ MSCs expanded and upregulated expression of fibrosis-related genes. Of note, we previously showed that Dmp1-Cre targets an osteogenic subset of CAR (or Lepr+) stromal cells (35). Together, these data suggest that the adipogenic subset of Lepr+ MSCs is the cell of origin for myelofibrosis.
Our data show that noncanonical JNK-dependent TGF-β signaling is responsible for the induction of myelofibrosis. Deletion of Smad4 in bone marrow MSCs does not prevent the induction of myelofibrosis by MPLW515L. In contrast, treatment with a pan-JNK inhibitor abrogated the development of myelofibrosis by MPLW515L. This observation is consistent with a prior study showing that expression of c-Jun, a downstream target of JNK, is increased in all major human fibrotic conditions, including myelofibrosis (42). Another prior report also found activation of noncanonical TGF-β signaling, including JUN, in patients with myelofibrosis (43). Of note, there is evidence of cross-talk between canonical and noncanonical TGF-β signaling. Of particular relevance are data showing that JNK can phosphorylate and activate SMAD3 (44, 45). Whether the loss of SMAD3 activity contributes to suppression of myelofibrosis after JNK inhibitor treatment will require further study.
Our data suggest that treatment with JNK inhibitors may have therapeutic benefit in patients with myelofibrosis, although there are several important caveats. First, it is not clear whether JNK inhibitors are able to reverse myelofibrosis after it is established. Second, our data suggest that reversal of myelofibrosis may not improve hematopoietic niche function, raising questions about whether this treatment would significantly impact cytopenias present in many patients with MPN. Finally, a clinical trial of CC-930 (the pan-JNK inhibitor used in our study) in patients with idiopathic pulmonary fibrosis (NCT01203943) was terminated due to an unfavorable benefit/risk profile. Of note, preclinical studies suggesting a selective role for JNK1 in pulmonary fibrosis have led to an ongoing clinical trial of CC-90001, a JNK1-selective inhibitor, in patients with idiopathic pulmonary fibrosis (NCT03142191; ref. 46). Likewise, our in vitro data suggest that JNK1 is the major mediator of fibrosis-related gene expression in mouse bone marrow MSCs. Whether treatment with a JNK1-selective inhibitor would prevent or attenuate myelofibrosis will require further study.
In contrast to myelofibrosis, our data show that TGF-β signaling in MSCs is not required for the disruption of hematopoietic niches in MPNs. Specifically, the loss of TGF-β signaling in MSCs did not prevent the suppression of key niche factors, Cxcl12 and Kitlg, nor did it prevent the development of extramedullary hematopoiesis or loss of bone marrow HSCs. Prior studies have established the importance of Cxcl12 and Kitlg expression in MSCs in the maintenance of a functioning hematopoietic niche. Indeed, deletion of these genes in Osx-Cre–targeted (or Lepr-Cre–targeted) stromal cells results in HSPC mobilization and development of extramedullary hematopoiesis (1–3). Based on these observations, we propose that suppression of MSC Cxcl12 and/or Kitlg expression plays a key role in the disruption of the hematopoietic niche and development of extramedullary hematopoiesis, although we acknowledge that additional data from patients with MPNs is needed to confirm. These data also show for the first time to our knowledge that the signals that induce a fibrogenic program in bone marrow MSCs are distinct from those that suppress Cxcl12 and Kitlg expression. Our data show that the fibrogenic program is dependent on TGF-β signaling, while the signals that regulate niche factor expression remain unknown.
In summary, in this study, we show that noncanonical JNK-dependent TGF-β signaling in perivascular MSCs is responsible for the development of myelofibrosis but not hematopoietic niche disruption. JNK is a druggable target that may have therapeutic benefit in the treatment and/or prevention of myelofibrosis in patients with MPNs.
Methods
Mice.
Tgfbr2fl/fl (B6;129-Tgfbr2tm1Karl/J; ref. 47), Osx-Cre (B6.Cg-Tg Sp7-tTA,tetO-EGFP/cre1Amc/J; ref. 48), Ai9 [B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J; ref. 49], UBC-CreERT2 (B6.Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb/1J; ref. 50), Jak2V617F [B6N.129S6(SJL)-Jak2tm1.2Ble/AmlyJ; ref. 34], and Dmp1-Cre [B6N.FVB-Tg(Dmp1-cre)1Jqfe/BwdJ; ref. 51] mice were obtained from The Jackson Laboratory. Tgfbr2fl/– mice were generated by breeding Tgfbr2fl/fl with E2A-Cre [B6.FVB-Tg(EIIa-cre)C5379Lmgd/J; ref. 52] mice. Smad4fl/fl mice were a gift from Fanxin Long, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA. All mice were back-crossed onto the C57BL/6 background and were maintained under standard pathogen-free conditions. All mice used in this study were 6 to 12 weeks old. Both male and female mice were used equally in these studies. To induce recombination postnatally, Osx-Cre Tgfbrfl/– or Osx-Cre Smad4fl/fl breeders were maintained on 200 ppm doxycycline chow, which was removed after pups were born.
MPLW515L MPN model.
c-Kit+ HSPCs from WT mice were enriched using murine CD117 (c-Kit) microbeads and the autoMACS Pro Separator system, per the manufacturer’s instructions (Miltenyi Biotec) and cultured overnight at 37°C with 5% CO2 in stem cell media (MEM-α with 10% FBS, 100 U penicillin and streptomycin, 10 ng/mL murine TPO [mTPO], 10 ng/mL mIL-3, 10 ng/mL mFLT3L, and 100 ng/mL mSCF). To generate retrovirus, the retroviral packaging vector pCL-Eco was cotransfected with either MSCV-MPLW515L-IRES-GFP or MSCV-IRES-GFP (empty vector) into 293T cells using calcium phosphate precipitation. Freshly generated retrovirus was added to HSPC cultures containing polybrene (6 mg/mL). Cells were spin-infected at 1200g at 30°C for 90 minutes and then cultured overnight. Five hundred thousand transduced c-Kit+ cells were injected retro-orbitally into recipients, which had been irradiated with two 600-cGy doses, 16 hours apart. Recipient mice were placed on prophylactic antibiotics (trimethoprim-sulfamethoxazole) for 2 weeks following transplantation. Mice were sacrificed for analysis when at least one of the mice in the same transplanted cohort became moribund, which was around 4 to 8 weeks after transplantation. Of note, at time of terminal analysis, more than 95% of hematopoietic cells were GFP+.
Jak2V617F MPN model.
Donor bone marrow cells were collected from 6- to 8-week-old UBC-CreERT2 Jak2V617F or UBC-CreERT2 mice, and 2 million bone marrow cells were injected retro-orbitally into lethally irradiated recipients. Six weeks after transplantation, mice were treated with 3 mg tamoxifen in corn oil daily for 5 days via oral gavage to induce Cre recombinase activity. Mice were sacrificed for analysis 5 months after tamoxifen induction.
Immunostaining of femur sections.
Mouse femurs were fixed in 10% formalin for 24 to 48 hours at 4°C. Bones were then decalcified in 14% EDTA, pH 7.4, for 10 to 14 days at 4°C. Following incubation in 30% sucrose for 24 hours at 4°C to dehydrate, bones were embedded in optimal cutting temperature compound (OTC, Sakura Finetek). These tissue blocks were cut into 12-μm sections using a CryoJane system (Leica Biosystems). For immunostaining, the slides were blocked with 5% donkey serum in TBST for 1 hour at room temperature. Slides were then incubated with primary antibody overnight at 4°C, followed by incubation with secondary antibody for 1 hour at room temperature. The following antibodies were used: rabbit anti–collagen I (Abcam, ab34710), rabbit anti–collagen III (Abcam, ab7778), goat anti–TGF-β1 (LAP) (R&D Systems, AB-246-NA), PE–anti-CD41 (BD Biosciences, MWReg30), and Alexa Fluor 647–conjugated donkey anti–rabbit IgG (H+L) (Thermo Fisher Scientific). Finally, slides were mounted with ProLong Gold antifade reagent with DAPI (Life Technologies). Images were acquired with an LSM 700 microscope (Carl Zeiss). Fluorescence intensity was quantified using ImageJ (NIH).
Femur and spleen histology.
Mouse femurs and spleens were fixed in 10% formalin for 24 hours at 4°C. Femurs were then decalcified in 14% EDTA, pH 7.4, for 10–14 days at 4°C. Femurs and spleens were dehydrated via incubation in 30%, 50%, and 70% ethanol for 15 minutes each. Processed bones and spleens were further embedded in paraffin and sectioned into 5-μm sections and stained for reticulin fibers by the Musculoskeletal Research Histology Core at Washington University.
Histologic analysis of femur and spleen tissue.
The degree of fibrosis was graded in blinded fashion by 2 independent hematopathologists using adjusted WHO criteria (53). Scores were assigned based on the degree of fibrosis as well as the percentage of the femur that was fibrotic: 0, no fibrosis; 1, mild and patchy fibrosis; 2, moderate and patchy fibrosis; and 3, moderate to severe uniform fibrosis.
Quantitative real-time PCR.
To collect total bone marrow RNA, tibias from mice were either crushed (MPLW515L mice) with a mortar and pestle in 1 mL of TRIzol (Invitrogen) or flushed (Jak2V617F mice) with 1 mL of TRIzol. RNA was extracted following the manufacturer’s instructions. cDNA was prepared using iScript reverse transcription supermix (Bio-Rad). Quantitative real-time PCR (qRT-PCR) was performed using the TaqMan Universal RT Master Mix (Applied Biosystems). Data were collected on a StepOnePlus Real-Time PCR System (Applied Biosystems). TaqMan probe primers were all from Thermo Fisher Scientific: Actb VIC-MGB (Mm04394036_g1), Tgfb1 FAM-MGB (Mm01178820_m1), Tgfbr2 FAM-MGB (Mm03024091_m1), Col1a1 FAM-MGB (Mm00801666_g1), Col3a1 FAM-MGB (Mm01254476_m1), Acta2 FAM-MGB (Mm01546133_m1), and Loxl1 FAM-MGB (Mm01145738).
Flow cytometry.
Bone marrow and spleen were processed for flow cytometry as previously described (54). Cells were analyzed on a Gallios flow cytometer (Beckman Coulter). Data analysis was done using FlowJo version 10.7.1 software (Tree Star). The following antibodies were used for staining murine cells: anti–Gr-1 (clone RB6-8C5), anti-Ter119 (clone TER-119), anti-CD117 (clone 2B8), anti-CD48 (clone HM48-1), and anti-CD140b (PDGFRβ, clone APB5) from eBioscience; anti-CD45 (clone HI30), anti-CD11b (clone M1/70), anti-B220 (clone RA3-6B2), anti-CD3e (clone 145-2C11), anti-CD115 (clone AFS98), anti–Sca-1 (clone D7), anti-CD150 (clone TC15-12F12.2), and anti-CD71 (clone RI7217) from BioLegend.
Micro-CT analysis.
Mouse femurs were fixed in 10% formalin for 24 hours at 4°C and then dehydrated in 70% ethanol. Processed tissues were scanned at 10-μm voxel resolution using a Scanco μCT 40 by the Musculoskeletal Research Histology Core at Washington University.
Bone marrow MSC culture.
Total bone marrow cells from 3- to 4-week-old WT or Osx-Cre Smad4fl/fl mice were collected and cultured in MSC media (DMEM, 20% FBS, 100 U penicillin and streptomycin) overnight at 37°C with 5% CO2. Nonattached cells were removed the next day by gently aspirating the media. Cells were washed with PBS and fresh medium was added every day for another 2 days. Attached MSCs were expanded for 3 more days until they reached approximately 80% confluence. MSCs strongly attach to cell culture plates, so after trypsinization, cells were manually scraped to ensure efficient collection. For TGF-β1 and PDGF stimulation experiments, 200,000 MSCs were plated per well in 6-well plates and 10 ng/mL TGF-β1, PDGF-AA, PDGF-BB, or vehicle control (1% bovine serum albumin in PBS) was added every 24 hours for 3 days. For MAP kinase inhibitor treatments, 20 μM MEK inhibitor (PD99059), p38 inhibitor (SB202190), or JNK inhibitor (SP600125), or 1 μM NF-κB inhibitor (pevonedistat) or vehicle control (DMSO) were added every 24 hours for 3 days. Twenty-four hours after the last treatment, cells were lysed with TRIzol, and RNA was extracted for quantification using qRT-PCR analysis.
CRISPR/Cas9 gene editing.
Recombinant Cas9 protein (0.25 μL of 62 μM solution; Alt-R Streptococcus pyogenes Cas9 Nuclease V3, IDT), 1 μL of sgRNAs (50 μM), and 1 μL Buffer T (Neon, Invitrogen) were incubated at room temperature for 15 to 30 minutes. Cultured MSCs (100,000 cells) were washed with PBS, resuspended in Buffer T at 13,000 cells/μL, and then added to the Cas9/sgRNA mix. Cells were nucleofected with 1 pulse at 1500 V for 20 ms using the Neon transfection system (Invitrogen). Cells were allowed to expand in culture for 72 hours after nucleofection before stimulating with TGF-β1. For each gene, 3 different sgRNAs targeting the same exon were pooled to enhance editing efficiency. Sequences of the sgRNAs targeting exon 3 of Mapk8 (JNK1) were 5′-CATAAGAACTAGTTCTCGGT-3′, 5′-GGGTCTGATTCTGAAATGGC-3′, and 5′-TCGGTAGGCGCGCTTAGCAT-3′. Sequences of the sgRNAs targeting exon 3 of Mapk9 (JNK2) were 5′-TAAGAGGACGAGTTCACGGT-3′, 5′-TACAGTTCTTGGGATAAATG-3′, and 5′-AGGCTCTCTTTGCGTGCGTT-3′.
Immunoblotting.
MSCs were lysed with RIPA buffer (MilliporeSigma) per the manufacturer’s instructions, and proteins were denatured in SDS sample buffer with 9% β-mercaptoethanol at 100°C for 5 minutes. The following antibodies were used (all purchased from Cell Signaling Technology): anti-SMAD2 (catalog 5339), anti–phospho-SMAD2 (catalog 3104), anti–c-JUN (catalog 9165), anti–phospho-c-JUN (catalog 3270), anti-ERK1/2 (catalog 4695), anti–phospho-ERK1/2 (catalog 4370), anti-JNK1 (catalog 3708), anti-JNK2 (catalog 4672), and anti-vinculin (catalog 13901).
JNK inhibitor treatment in mice.
MPLW515L retrovirus–transduced HSPCs were transplanted into lethally irradiated WT recipients. Two weeks after transplantation, mice were treated with vehicle control (0.5% carboxymethyl cellulose, 0.25% Tween 80) or 50 mg/kg CC-930 (55) (MedChemExpress) twice daily via oral gavage for 4 weeks.
Single-cell RNA sequencing.
Mouse bone marrow from 1 male and 1 female Osx-Cre Ai9 mouse was crushed and dissociated with collagenase II (Worthington). Cells were sorted for lineage– and PDGFRβ+. Sorted cells were captured using 10× Genomics Chromium 3′ gel beads version 3.1. Gene expression libraries were sequenced on a NovaSeq S4 with the goal of achieving 50,000 reads per cell. A modified reference sequence was created to incorporate the Ai9 reporter sequence (positions 3897–6222; https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/using/count) into mm10 for mapping using Cell Ranger v6. Quality filters were applied to remove cells with mitochondrial transcripts greater than 10% and UMI counts greater than 50,000. Log normalization, PCA, dimensional reduction, and clustering were performed using the Seurat (v4.1.0) package in R (v4.1.1); https://www.r-project.org The single-cell RNA expression data have been submitted to NCBI’s Gene Expression Omnibus (GEO GSE200693).
Statistics.
Unpaired Student’s t test was used to evaluate the significance of differences between 2 groups. One-way ANOVA was used to evaluate the significance of differences between multiple groups. The Kruskal-Wallis test was used to evaluate the significance of differences of the categorical fibrosis grades. All data are presented as mean ± SEM. A P value of less than 0.05 was considered significant.
Study approval.
All animal studies were approved by the Institutional Animal Care and Use Committee at Washington University.
Authors contributions
JCY and DCL conceived and designed the experiments, analyzed the data, and wrote the manuscript. JCY performed the experiments. KAO performed and analyzed the single-cell RNA sequencing data. GAE generated the Osx-Cre Tgfbr2fl/fl mice. JRK generated the Osx-Cre Smad4fl/fl mice. SU performed the MSC culture experiments. EJD, TW, and HX performed the grading of fibrosis and captured images of bone marrow and spleen histology staining. DCL supervised the research.
Supplementary Material
Acknowledgments
We thank Amy Schmidt for technical assistance and Jackie Tucker-Davis for animal care. We thank Crystal Idleburg, Michael Brodt, and the Musculoskeletal Research Center Core for processing bone and spleen histology samples for reticulin staining and bones for micro-CT imaging and analysis. This work was supported by NIH grant R01 HL60772 (to DCL) and P01 CA101937-16.
Version 1. 04/19/2022
In-Press Preview
Version 2. 06/01/2022
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2022, Yao et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2022;132(11):e154092.https://doi.org/10.1172/JCI154092.
Contributor Information
Juo-Chin Yao, Email: juochinyao@wustl.edu.
Karolyn A. Oetjen, Email: koetjen@wustl.edu.
Tianjiao Wang, Email: tianjiao.wang@wustl.edu.
Haoliang Xu, Email: hlxu2006@gmail.com.
Grazia Abou-Ezzi, Email: graziaabouezzi@gmail.com.
Joseph R. Krambs, Email: jrkrambs@wustl.edu.
Salil Uttarwar, Email: suttarwar@wustl.edu.
Eric J. Duncavage, Email: duncavagee@wustl.edu.
Daniel C. Link, Email: danielclink@wustl.edu.
References
- 1.Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495(7440):231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenbaum A, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495(7440):227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding L, et al. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481(7382):457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao JC, Link DC. Concise review: the malignant hematopoietic stem cell niche. Stem Cells. 2017;35(1):3–8. doi: 10.1002/stem.2487. [DOI] [PubMed] [Google Scholar]
- 5.Tefferi A, Vardiman JW. Classification and diagnosis of myeloproliferative neoplasms: the 2008 World Health Organization criteria and point-of-care diagnostic algorithms. Leukemia. 2008;22(1):14–22. doi: 10.1038/sj.leu.2404955. [DOI] [PubMed] [Google Scholar]
- 6.Kota J, et al. Aberrant signal transduction pathways in myeloproliferative neoplasms. Leukemia. 2008;22(10):1828–1840. doi: 10.1038/leu.2008.236. [DOI] [PubMed] [Google Scholar]
- 7.Mehta J, et al. Epidemiology of myeloproliferative neoplasms in the United States. Leuk Lymphoma. 2014;55(3):595–600. doi: 10.3109/10428194.2013.813500. [DOI] [PubMed] [Google Scholar]
- 8.Tefferi A. Primary myelofibrosis: 2012 update on diagnosis, risk stratification, and management. Am J Hematol. 2011;86(12):1017–1026. doi: 10.1002/ajh.22210. [DOI] [PubMed] [Google Scholar]
- 9.Decker M, et al. Leptin-receptor-expressing bone marrow stromal cells are myofibroblasts in primary myelofibrosis. Nat Cell Biol. 2017;19(6):677–688. doi: 10.1038/ncb3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider RK, et al. Gli1+ mesenchymal stromal cells are a key driver of bone marrow fibrosis and an important cellular therapeutic target. Stem Cell. 2017;20(6):785–800. doi: 10.1016/j.stem.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leimkuhler NB, et al. Heterogeneous bone-marrow stromal progenitors drive myelofibrosis via a druggable alarmin axis. Cell Stem Cell. 2020;28(4):637–652. doi: 10.1016/j.stem.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, et al. Effects of chromatin-modifying agents on CD34+ cells from patients with idiopathic myelofibrosis. Cancer Res. 2007;67(13):6417–6424. doi: 10.1158/0008-5472.CAN-07-0572. [DOI] [PubMed] [Google Scholar]
- 13.Migliaccio AR, et al. Altered SDF-1/CXCR4 axis in patients with primary myelofibrosis and in the Gata1 low mouse model of the disease. Exp Hematol. 2008;36(2):158–171. doi: 10.1016/j.exphem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulsen LW, et al. A histomorphometric study of haematological disorders with respect to marrow fibrosis and osteosclerosis. APMIS. 1998;106(4):495–499. doi: 10.1111/j.1699-0463.1998.tb01377.x. [DOI] [PubMed] [Google Scholar]
- 15.Rameshwar P, et al. Monocyte adhesion in patients with bone marrow fibrosis is required for the production of fibrogenic cytokines. Potential role for interleukin-1 and TGF-beta. J Immunol. 1994;153(6):2819–2830. [PubMed] [Google Scholar]
- 16.Le Bousse-Kerdilès MC, et al. Differential expression of transforming growth factor-beta, basic fibroblast growth factor, and their receptors in CD34+ hematopoietic progenitor cells from patients with myelofibrosis and myeloid metaplasia. Blood. 1996;88(12):4534–4546. doi: 10.1182/blood.V88.12.4534.bloodjournal88124534. [DOI] [PubMed] [Google Scholar]
- 17.Martyré MC, et al. Elevated levels of basic fibroblast growth factor in megakaryocytes and platelets from patients with idiopathic myelofibrosis. Br J Haematol. 1997;97(2):441–448. doi: 10.1046/j.1365-2141.1997.292671.x. [DOI] [PubMed] [Google Scholar]
- 18.Campanelli R, et al. Evaluation of the bioactive and total transforming growth factor β1 levels in primary myelofibrosis. Cytokine. 2011;53(1):100–106. doi: 10.1016/j.cyto.2010.07.427. [DOI] [PubMed] [Google Scholar]
- 19.Malara A, et al. Megakaryocyte contribution to bone marrow fibrosis: many arrows in the quiver. Mediterr J Hematol Infect Dis. 2018;10(1):e2018068. doi: 10.4084/MJHID.2018.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chagraoui H, et al. Prominent role of TGF-beta 1 in thrombopoietin-induced myelofibrosis in mice. Blood. 2002;100(10):3495–3503. doi: 10.1182/blood-2002-04-1133. [DOI] [PubMed] [Google Scholar]
- 21.Zingariello M, et al. Characterization of the TGF-β1 signaling abnormalities in the Gata1low mouse model of myelofibrosis. Blood. 2013;121(17):3345–3363. doi: 10.1182/blood-2012-06-439661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue L, et al. Efficacy of ALK5 inhibition in myelofibrosis. JCI Insight. 2017;2(7):e90932. doi: 10.1172/jci.insight.90932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gleitz H, et al. Increased CXCL4 expression in hematopoietic cells links inflammation and progression of bone marrow fibrosis in MPN. Blood. 2020;136(18):2051–2064. doi: 10.1182/blood.2019004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verstovsek S, et al. Role of neoplastic monocyte-derived fibrocytes in primary myelofibrosis. J Exp Med. 2016;213(9):1723–1740. doi: 10.1084/jem.20160283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maekawa T, et al. Myeloproliferative leukemia protein activation directly induces fibrocyte differentiation to cause myelofibrosis. Leukemia. 2017;31(12):2709–2716. doi: 10.1038/leu.2017.112. [DOI] [PubMed] [Google Scholar]
- 26.Erba BG, et al. Endothelial-to-mesenchymal transition in bone marrow and spleen of primary myelofibrosis. Am J Pathol. 2017;187(8):1879–1892. doi: 10.1016/j.ajpath.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Baryawno N, et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177(7):1915–1932. doi: 10.1016/j.cell.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tikhonova AN, et al. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569(7755):222–228. doi: 10.1038/s41586-019-1104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baccin C, et al. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat Cell Biol. 2020;22(1):38–48. doi: 10.1038/s41556-019-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolock SL, et al. Mapping distinct bone marrow niche populations and their differentiation paths. Cell Rep. 2019;28(2):302–311. doi: 10.1016/j.celrep.2019.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong L, et al. Single cell transcriptomics identifies a unique adipose lineage cell population that regulates bone marrow environment. Elife. 2020;9:e54695. doi: 10.7554/eLife.54695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, et al. Osx-Cre targets multiple cell types besides osteoblast lineage in postnatal mice. PLoS One. 2014;9(1):e85161. doi: 10.1371/journal.pone.0085161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abou-Ezzi G, et al. TGF-β signaling plays an essential role in the lineage specification of mesenchymal stem/progenitor cells in fetal bone marrow. Stem Cell Reports. 2019;13(1):48–60. doi: 10.1016/j.stemcr.2019.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mullally A, et al. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell. 2010;17(6):584–596. doi: 10.1016/j.ccr.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Link DC. Targeting of mesenchymal stromal cells by Cre-recombinase transgenes commonly used to target osteoblast lineage cells. J Bone Miner Res. 2016;31(11):2001–2007. doi: 10.1002/jbmr.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krambs JR, et al. Canonical signaling by TGF family members in mesenchymal stromal cells is dispensable for hematopoietic niche maintenance under basal and stress conditions. PLoS One. 2020;15(5):e0233751. doi: 10.1371/journal.pone.0233751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulder KM. Role of Ras and MAPKs in TGFbeta signaling. Cytokine Growth Factor Rev. 2000;11(1–2):23–35. doi: 10.1016/s1359-6101(99)00026-x. [DOI] [PubMed] [Google Scholar]
- 38.Nakano R, et al. Biological properties of JNK3 and its function in neurons, astrocytes, pancreatic β-cells and cardiovascular cells. Cells. 2020;9(8):1802. doi: 10.3390/cells9081802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizoguchi T, et al. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev Cell. 2014;29(3):340–349. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou BO, et al. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15(2):154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Y, et al. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat Commun. 2017;8(1):2043. doi: 10.1038/s41467-017-02171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wernig G, et al. Unifying mechanism for different fibrotic diseases. Proc Natl Acad Sci U S A. 2017;114(18):4757–4762. doi: 10.1073/pnas.1621375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciaffoni F, et al. Activation of non-canonical TGF-β1 signaling indicates an autoimmune mechanism for bone marrow fibrosis in primary myelofibrosis. Blood Cells Mol Dis. 2015;54(3):234–241. doi: 10.1016/j.bcmd.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engel ME, et al. Interdependent SMAD and JNK signaling in transforming growth factor-beta-mediated transcription. J Biol Chem. 1999;274(52):37413–37420. doi: 10.1074/jbc.274.52.37413. [DOI] [PubMed] [Google Scholar]
- 45.Liu Q, et al. A crosstalk between the Smad and JNK signaling in the TGF-β-induced epithelial-mesenchymal transition in rat peritoneal mesothelial cells. PLoS One. 2012;7(2):e32009. doi: 10.1371/journal.pone.0032009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popmihajlov Z, et al. CC-90001, a c-Jun N-terminal kinase (JNK) inhibitor, in patients with pulmonary fibrosis: design of a phase 2, randomised, placebo-controlled trial. BMJ Open Respir Res. 2022;9(1):e002060. doi: 10.1136/bmjresp-2021-001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levéen P, et al. Induced disruption of the transforming growth factor beta type II receptor gene in mice causes a lethal inflammatory disorder that is transplantable. Blood. 2002;100(2):560–568. doi: 10.1182/blood.V100.2.560. [DOI] [PubMed] [Google Scholar]
- 48.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133(16):3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 49.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruzankina Y, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1(1):113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu Y, et al. DMP1-targeted Cre expression in odontoblasts and osteocytes. J Dent Res. 2007;86(4):320–325. doi: 10.1177/154405910708600404. [DOI] [PubMed] [Google Scholar]
- 52.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93(12):5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Passamonti F, Maffioli M. Update from the latest WHO classification of MPNs:a user’s manual. Hematology Am Soc Hematol Educ Program. 2016;2016(1):534–542. doi: 10.1182/asheducation-2016.1.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richards MK, et al. Pivotal role of granulocyte colony-stimulating factor in the development of progenitors in the common myeloid pathway. Blood. 2003;102(10):3562–3568. doi: 10.1182/blood-2003-02-0593. [DOI] [PubMed] [Google Scholar]
- 55.Plantevin Krenitsky V, et al. Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor. Bioorg Med Chem Lett. 2012;22(3):1433–1438. doi: 10.1016/j.bmcl.2011.12.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.