Abstract
Lack of a system for site-specific genetic manipulation has severely hindered studies on the molecular biology of all Bartonella species. We report the first site-specific mutagenesis and complementation for a Bartonella species. A highly transformable strain of B. bacilliformis, termed JB584, was isolated and found to exhibit a significant increase in transformation efficiency with the broad-host-range plasmid pBBR1MCS-2, relative to wild-type strains. Restriction analyses of genomic preparations with the methylation-sensitive restriction enzymes ClaI and StuI suggest that strain JB584 possesses a dcm methylase mutation that contributes to its enhanced transformability. A suicide plasmid, pUB1, which contains a polylinker, a pMB1 replicon, and a nptI kanamycin resistance cassette, was constructed. An internal 508-bp fragment of the B. bacilliformis flagellin gene (fla) was cloned into pUB1 to generate pUB508, a fla-targeting suicide vector. Introduction of pUB508 into JB584 by electroporation generated eight Kanr clones of B. bacilliformis. Characterization of one of these strains, termed JB585, indicated that allelic exchange between pUB508 and fla had occurred. Analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunoblotting, and electron microscopy showed that synthesis of flagellin encoded by fla and secretion/assembly of flagella were abolished. Complementation of fla in trans was accomplished with a pBBR1MCS recombinant containing the entire wild-type fla gene (pBBRFLAG). These data conclusively show that inactivation of fla results in a bald, nonmotile phenotype and that pMB1 and REP replicons make suitable B. bacilliformis suicide and shuttle vectors, respectively. When used in conjunction with the highly transformable strain JB584, this system for site-specific genetic manipulation and complementation provides a new venue for studying the molecular biology of B. bacilliformis.
The Bartonella genus comprises a unique group of intracellular bacteria that employ arthropod-mediated transmission and hemotrophy as common parasitic strategies. Recent taxonomic reclassifications have expanded the number of Bartonella species from one, B. bacilliformis, to 11 based on sequence homology and genetic relatedness. Five of these species are presently considered agents of emerging infectious disease in humans (B. bacilliformis, B. clarridgeiae, B. elizabethae, B. henselae, and B. quintana), and the diseases share the symptoms of bacteremia, hemolytic anemia, recurrent fever, and a variety of vascular lesions (for recent reviews, see references 5, 25, and 34).
B. bacilliformis is the etiologic agent of a biphasic disease that is indigenous to the Andes mountain region of South America. Oroya fever is commonly used to describe the first phase, which is characterized by an acute syndrome of fever, malaise, and severe hemolytic anemia (16, 37, 46). Humans exhibit the acute hematic phase of disease within 2 to 3 weeks following inoculation of Bartonella into the bloodstream by the bite of a nocturnal sandfly, Lutzomyia verrucarum (20). Subsequent erythrocyte invasion accompanies a severe hemolytic anemia that is responsible for the high (40 to 80%) mortality rate observed in the absence of antibiotic therapy (20, 24, 28). The disease has killed over 10,000 humans in recorded time (20, 46). The chronic secondary phase of the disease, termed verruga peruana, develops approximately 4 weeks after the primary phase and is characterized by angiomatous cutaneous eruptions (20). During this phase, the bacteria invade vascular endothelial cells (14, 15, 32) and subsequently stimulate the formation of new blood vessels (14), a common sequela of bartonelloses. Recent reports of several atypical monophasic (verruga peruana) cases of B. bacilliformis in previously disease-free lowland elevations are cause for concern (2, 4).
Although a conjugative system for random Tn5-based mutagenesis has been reported for B. henselae (12), no means of site-directed mutagenesis exists for Bartonella species. This lack has been a major impediment to elucidating the molecular biology of this expanding group of emerging bacterial pathogens and was the impetus for the present study. Here we describe a system for site-specific mutagenesis and complementation of B. bacilliformis. This is the first report of site-specific mutagenesis and complementation for any of the Bartonella species.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Escherichia coli strains used for propagation of plasmids were grown overnight at 37°C in Luria-Bertani medium with antibiotic supplements when required (11). The strains of B. bacilliformis and E. coli used or generated in this study are summarized in Table 1. B. bacilliformis was routinely grown on heart infusion agar (Difco, Detroit, Mich.) supplemented with 5% defibrinated sheep erythrocytes and 2.5% filter-sterile sheep serum (Quad Five, Ryegate, Mont.) at 30°C in a water-saturated atmosphere. Antibiotic supplements for B. bacilliformis included kanamycin sulfate (25 μg/ml), and chloramphenicol (1 μg/ml) (both from Sigma Chemical Co., St. Louis, Mo.) and were used individually or combined depending upon the experimental conditions. Plates were routinely cultured by transferring growth from the surface of a 5-day culture plate to a fresh plate, using an initial plating density of approximately 4,000 CFU/plate. Since no suitable liquid growth medium is currently available for B. bacilliformis, the growth phase of the bacterium at harvest could not be determined. However, to ensure that the bartonellae were actively growing, colonies were first observed at 3 days and subsequently harvested at 5 days postinoculation.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or Reference |
|---|---|---|
| Strains | ||
| B. bacilliformis | ||
| KC583 | Neotype strain | 10 |
| KC584 | Peruvian isolate, 1963 | 10 |
| HG584 | Highly passaged KC584 transformed with pEST (Kmr, dcm mutant) | 19 |
| JB584 | HG584 cured of pEST (Kms) | This study |
| JB585 | fla gene of JB584 interrupted by insertion of pUB508 (Kmr, fla) | This study |
| JB686 | JB585 complemented in trans by transformation with pBBRFLAG (Kmr Cmr, fla+) | This study |
| E. coli | ||
| DH5α | Host strain used for cloning and propagation of plasmids | 17 |
| Plasmids | ||
| pUC18,19 | Cloning vectors; RepEc, Apr | 47 |
| pBK-CMV | Genomic library cosmid; RepEc, Kmr | 42 |
| pBBR1MCS | B. bacilliformis shuttle vector; RepEc, RepBb, Cmr | 27 |
| pBBR1MCS-2 | B. bacilliformis shuttle vector; RepEc, RepBb, Kmr | 26 |
| pUCK18 | pUC18 containing Kmr cassette (nptI); RepEc, Apr, Kmr | 36 |
| pUCK19 | pUC19 containing ∼1,500-bp PstI fragment of pUCK18; RepEc, Apr, Kmr | This study |
| pUB1 | Suicide plasmid; pUCK19 with a 1,118-bp BglII fragment removed to delete bla; RepEc, Kmr | This study |
| pUB508 | Flagellin-targeting suicide plasmid; pUB1 containing internal 508-bp Kpn-BglII fragment of fla; RepEc, Kmr | This study |
| pAUL1 | pBK-CMV containing 3,800-bp B. bacilliformis Sau3AI fragment with intact fla gene; RepEc, Kmr | This study |
| pFLAG3 | pUC18 containing 1,400-bp HindIII-EcoRI fragment of pAUL1 with intact fla gene; RepEc, Apr | This study |
| pBBRFLAG | Complementation shuttle vector; pBBR1MCS containing 2,158-bp HindIII fragment of pAUL1 with fla gene; RepEc, RepBb, Cmr | This study |
For testing the motility of B. bacilliformis strains, a Bartonella motility medium was devised. The defibrinated sheep erythrocyte supplement of standard Bartonella growth medium was replaced with sheep erythrocyte lysate (8) at a 5% final concentration to yield a translucent medium suitable for visual scoring, and the agar in the medium was reduced to 0.2% (wt/vol). Plates were poured and dried for 48 h at 22°C and dried for an additional 1 h at 45°C prior to inoculation, to reduce moisture on the surface of the agar.
Preparation and manipulation of DNA.
Chromosomal DNA from B. bacilliformis was prepared with CTAB (cetyltrimethylammonium bromide) by the methods of Ausubel et al. (6). Plasmid DNA for cloning was isolated from E. coli by the alkali lysis procedure of Birnboim and Doly (9). Plasmid DNA for electroporation experimentation was prepared with a Midi-Prep kit (Qiagen, Chatsworth, Calif.) or a Perfect Prep kit (5 PRIME-3 PRIME, Boulder, Colo.) as specified by the manufacturer. Cloning was accomplished by purifying individual DNA fragments from ethidium bromide-stained agarose gels with either a GeneClean kit (Bio 101, Inc., La Jolla, Calif.) or a QIAquick kit (Qiagen). Ligation and transformation of DNA into E. coli DH5α was done by standard procedures (38). The plasmids used in this study are summarized in Table 1.
DNA hybridization analysis.
Genomic DNA from B. bacilliformis strains and plasmid DNA were isolated, digested to completion with appropriate restriction enzymes, and resolved on ethidium bromide-stained 1% (wt/vol) agarose gels. The gels were blotted onto a supported nitrocellulose membrane (pore size, 0.45-μm; Schleicher & Schuell, Keene, N.H.) by the method of Southern (41) and baked for 1 h at 80°C. DNA probes were made by random primer extension with [α-32P]dCTP (New England Nuclear, Boston, Mass.) and were used to probe blots overnight at 50°C as previously described (7). The blots were then washed at high stringency and exposed for 1 h to X-ray film (X-Omat XAR-5; Eastman Kodak Co., Rochester, N.Y.) as previously described (7) to visualize hybridized DNA fragments.
Electroporation.
Approximately seven plates of 5-day-cultured B. bacilliformis cells were harvested into 1 ml of heart infusion broth at 4°C. The cells were subsequently washed four times with 1 ml of ice-cold 10% (vol/vol) glycerol in water with intermittent centrifugations at 2,090 × g for 20 min at 4°C. The final bacterial concentration was measured with a Petroff-Hauser counter and adjusted to 1010 cells/ml with 10% glycerol. Electrotransformation was performed with a gene pulser with 0.2-cm cuvettes (Bio-Rad Laboratories, Hercules, Calif.) that had been chilled on ice for at least 15 min. In general, a 44-μl bacterial suspension (1010 cells/ml) was combined with 1 to 2 μl of DNA (1 to 44 μg/μl) and electroporated with an exponential-decay waveform set at a field strength of 12.5 kV/cm, a pulse time of 5 ms, and capacitance held constant at 25 μF as previously described for Bartonella (19, 36). Immediately following electroporation, cells were removed from the cuvette by being resuspended in 1 ml of ice-cold sterile recovery broth (heart infusion broth containing 0.5% [wt/vol] bovine serum albumin, 5% [vol/vol] sheep erythrocyte lysate [8] and 5 mM l-methionine). The suspension was then transferred to a 15-ml sterile tube and incubated for 14 h at 30°C in a water-saturated atmosphere. This incubation period corresponds to approximately two B. bacilliformis generation times (8) and was used to allow antibiotic resistance marker expression. Transformants were isolated by being plated on standard Bartonella growth medium supplemented with kanamycin and/or chloramphenicol, when required for selection. Antibiotic-resistant colonies usually appeared after 6 to 7 days of incubation at 30°C.
PCR and oligonucleotides.
PCR amplification was achieved with a GeneAmp 2400 thermocycler (Perkin-Elmer, Norwalk, Conn.) by procedures developed by Mullis and Faloona (35). Reaction mixtures contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 200 μM each deoxynucleoside triphosphate, 4 mM MgCl2, 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer), 1 to 100 ng of template DNA, and 0.1 μg of each primer. The reaction proceeded for 30 cycles of 1 min at 94°C, 1 min at 50 to 60°C (depending on the calculated primer melting temperature), and 1 min at 72°C with an initial 5-min denaturation at 94°C and a final 7-min extension at 72°C. Single-stranded oligonucleotide primers specific for the fla gene, FLA5′ (5′-AAGCTTTAGAGATTGTTTTGCAAA-3′) and FLA3′ (5′-AAATATTCTGGCTGCCCTGATTTGC-3′), and the kanamycin cassette, NPTI5′ (5′-AGCCACGTTGTGTCTCAAAATCTC-3′) and NPTI3′ (5′-CGTCCCGTCAAGTCAGCGTAATGC-3′), were synthesized by The University of Montana Murdock Molecular Biology Facility. The “junction” amplimer set was designed to detect the integration of the pUB508 suicide plasmid at the fla locus and consisted of primers NPTI5′ and FLA3′. The target loci for each of the primers are illustrated in Fig. 1.
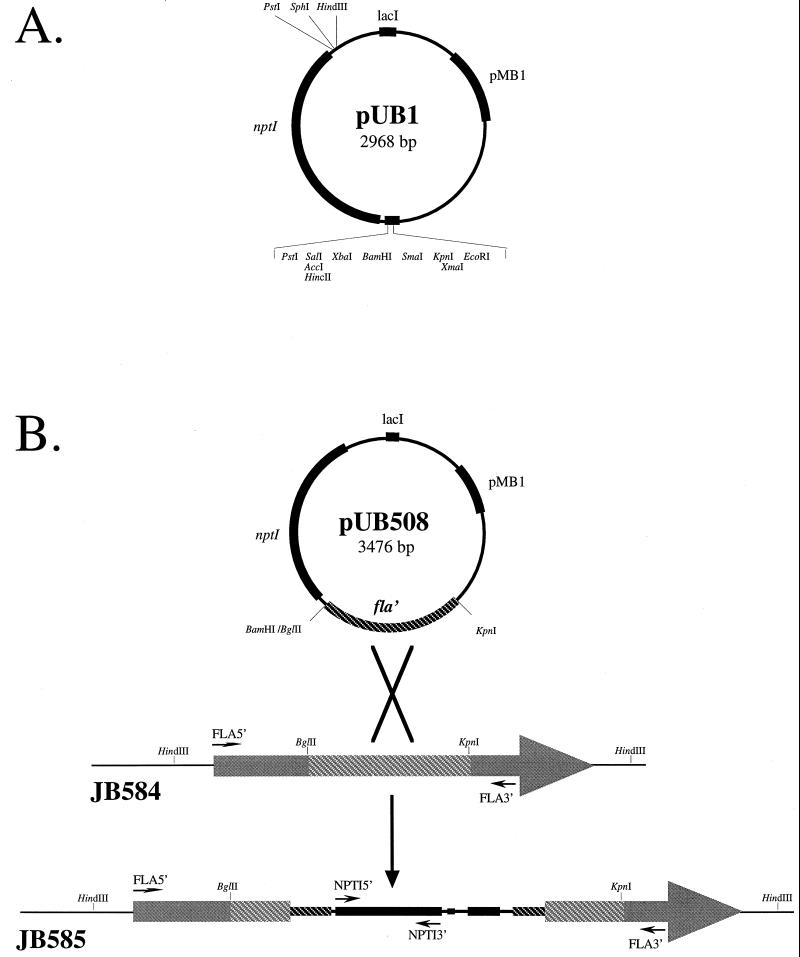
FIG. 1.
Illustration of suicide plasmid and schematic representation of site-specific fla disruption. (A) The B. bacilliformis suicide plasmid pUB1 harbors a multiple cloning site, the kanamycin resistance cassette neomycin phosphotransferase I (nptI), and the pMB1 replicon. (B) The flagellin gene-targeting suicide plasmid, derived from pUB1, is shown with the 508-bp BglII-KpnI internal fragment of fla (fla′) The transformable strain, JB584 (Kms, fla+), containing the wild-type 1,127-bp fla ORF, is shown. Homologous recombination resulted in site-specific insertion of pUB508 at the fla locus, generating strain JB585 (Kmr, fla). Note the position of the flagellin (FLA5′ and FLA3′) and kanamycin (NPTI5′ and NPTI3′) amplimers, indicated by the small arrows. (Figure not drawn to scale.)
SDS-PAGE and immunoblotting.
Whole-cell extracts of B. bacilliformis were prepared by boiling in sodium dodecyl sulfate (SDS) sample buffer for 10 min and were separated by SDS-polyacrylamide gel electrophoresis (PAGE) (12.5% [wt/vol] acrylamide), using procedures adapted from those of Laemmli (30). Approximately 20 μg of total extract protein was added per lane. Protein bands were visualized by staining with Coomassie brilliant blue (38). For immunoblots, separated proteins were electrophoretically transferred from gels to supported nitrocellulose membranes (pore size, 0.45 μm; Schleicher & Schuell) by the methods of Towbin et al. (43). Immunoblots were developed by using the rabbit anti-flagellin antiserum and procedures described by Scherer et al. (39).
Transmission electron microscopy (TEM).
B. bacilliformis was grown and harvested into 1 ml of heart infusion broth at 4°C. The cells were washed three times with 10% (vol/vol) glycerol at 4°C, with intermittent centrifugations at 2,090 × g for 20 min at 4°C, and finally resuspended in 10% glycerol. Aliquots of this suspension (15 μl) were placed on Formvar-coated 300-mesh copper-palladium grids (Electron Microscopy Sciences, Fort Washington, Pa.) and incubated for 5 min at 22°C. The grids were then stained with 2% uranyl acetate (pH 7.0) for 3 min, destained with 1 M ammonium acetate (pH 7.0) for 3 min, and washed with deionized water for 1 min. They were then air dried and observed at 75 kV with a 7100 transmission electron microscope (Hitachi, Mountain View, Calif.) located at The University of Montana Electron Microscopy Center.
Cloning the B. bacilliformis flagellin gene (fla).
The B. bacilliformis flagellin gene, fla, was chosen as the locus to develop a system of site-specific mutagenesis for two reasons. First, fla exists as a mapped, single-copy gene in B. bacilliformis (29), and mutations in fla are rarely lethal. Second, the phenotype associated with the gene is readily observable by TEM and easily scored by testing for motility.
fla was cloned, sequenced, and submitted to GenBank by another group (3). The fla gene was simultaneously isolated by our laboratory from a λZAP Express (Stratagene Cloning Systems, La Jolla, Calif.) expression library of B. bacilliformis by using rabbit anti-flagellin antiserum. A pBK-CMV cosmid clone containing the entire fla gene in a 3,800-bp Sau3AI fragment was subsequently excised from the λ clone as specified by the manufacturer (Stratagene) and termed pAUL1.
Purified pAUL1 was digested with HindIII, and the 2,158-bp fragment containing fla was isolated by agarose gel electrophoresis (1% [wt/vol] agarose) and purified by using a GeneClean II kit (Bio 101). Ligation of this fragment into the HindIII site of pUC18 resulted in pFLAG3, a source of fla DNA fragments for constructing the fla-targeting suicide plasmid.
Construction of suicide and shuttle/complementation plasmids.
The suicide plasmid pUB1 (Fig. 1A) was constructed in several steps. Previous studies showed that the pMB1 replicon was not recognized by the replicational machinery of Bartonella species (19, 36). Therefore, we reasoned that this replicon could be used to construct a suicide vector for Bartonella. To construct the plasmid, a ∼1,500-bp PstI fragment containing the nptI gene, encoding neomycin-kanamycin resistance, was subcloned from pUCK18 into pUC19, resulting in pUCK19. Subsequently, the β-lactamase (bla) gene of pUCK19 was deleted by removing a 1,118-bp BglII fragment and religated to generate pUB1 (Fig. 1A). To create a flagellin-specific suicide plasmid for insertional mutagenesis experiments, a 508-bp KpnI-BglII fragment from pFLAG3 containing an internal portion of fla was cloned into pUB1 to produce pUB508 (Fig. 1B).
A complementation shuttle plasmid, pBBRFLAG, was constructed by cloning the 2,158-bp HindIII fragment of pAUL1 into the broad-host-range vector pBBR1MCS. The resulting plasmid, pBBRFLAG, contains a chloramphenicol resistance cassette, the entire wild-type fla gene, and a REP replicon which is recognized by the replicational machinery of B. bacilliformis (see below).
RESULTS
Isolation of a highly transformable strain.
Grasseschi and Minnick previously reported mean transformation efficiencies of 7.8 × 105 following electroporation-mediated introduction of the cosmid pEST into B. bacilliformis KC584 (19). pEST is a cosmid harboring an RK2 origin of replication and a neomycin phosphotransferase I (nptI) gene encoding kanamycin resistance (36). We attempted to repeat these experiments with B. bacilliformis KC584 but were unable to isolate stable transformants. Electroporation-mediated introduction of a variety of constructs designed to disrupt genes in wild-type B. bacilliformis KC583 and KC584 were also unsuccessful. These trials included introduction of a variety of linear (double-stranded and single-stranded) and circular gene-targeting constructs designed to mutagenize two separate loci, fla and gyrB (7). A number of methods designed to either alleviate restriction (13, 21–23, 40, 44) or promote homologous recombination (1, 18, 31) were used in conjunction with the various gene-targeting constructs without success (data not shown).
The discrepancy between the reported transformation efficiencies and those found at the onset of this study suggested that one or more spontaneous mutations probably altered the genetic background of the KC584 strain used by Grasseschi and Minnick (19), resulting in a highly transformable strain of B. bacilliformis. To test this hypothesis, we obtained a frozen stock of the Kanr pEST-containing strain of B. bacilliformis (herein termed HG584) and cured this strain of the pEST cosmid by three passages, for a total of 15 days, in the absence of kanamycin sulfate. Six randomly selected clones were then subcultured independently and tested for sensitivity to kanamycin sulfate (25 μg/ml). All six clones exhibited wild-type sensitivities to kanamycin, and two of these Kans clones were cultivated and their genomic DNA was isolated. DNA hybridization analyses with a 32P-radiolabeled pEST probe did not detect pEST in the genome (data not shown), suggesting that the cosmid was cured from these two strains. Further verification by using PCR and nptI amplimers (NPTI5′ and NPTI3′) confirmed the absence of the Kanr marker (see Fig. 2, lane 2). One of these cured, Kans clones was termed JB584.
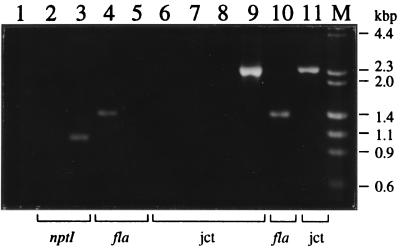
FIG. 2.
Electrophoretic analysis of PCR products derived from B. bacilliformis fla mutant and trans-complemented strains. PCR products generated by amplification of chromosomal DNA obtained from parent and recombinant strains by using three amplimer sets, nptI (NPTI5′ and NPTI3′), fla (FLA5′ and FLA3′), and junction (jct) (NPTI5′ and FLA3′) were respectively used to detect the kanamycin cassette, the flagellin gene, and the junction between fla and the inserted pUB508. Bars below the gel indicate the amplimer set used in each reaction. Amplimer sets and the respective DNA templates used in this analysis are as follows: lane 1 (NPTI5′ and NPTI3′, Fla5′ and Fla3′; no template), lane 2 (NPTI5′ and NPTI3′; JB584), lane 3 (NPTI5′ and NPTI3′; JB585), lane 4 (FLA5′ and FLA3′; JB584), lane 5 (FLA5′ and FLA3′; JB585), lane 6 (NPTI5′ and FLA3′; JB584), lane 7 (NPTI5′ and FLA3′; pUB508), lane 8 (NPTI5′ and FLA3′; pUB508 + JB584), lane 9 (NPTI5′ and FLA3′; JB585), lane 10 (FLA5′ and FLA3′ JB686), lane 11 (NPTI5′ and FLA3′ JB686). PCR products were resolved by agarose gel electrophoresis (1% agarose) and visualized by ethidium bromide staining. DNA size markers (lane M) are shown to the right.
The transformation efficiency of JB584 was subsequently assessed by electroporation-mediated introduction of pBBR1MCS-2. This plasmid was chosen because it is more stably maintained than the cosmid pEST and the multiple cloning site of pBBR1MCS plasmids enabled efficient cloning for complementation analyses. JB584 demonstrated a transformation efficiency of 5.2 × 103 transformants per μg of pBBR1MCS-2, which corresponds to one transformant per 8.4 × 104 cells. This value is within the lower range of efficiency reported by Grasseschi and Minnick (19); however, the relatively higher DNA concentrations (1 μg) and the different replicon used in the present study may account for the lower relative efficiency. Electroporation lethality, previously estimated at 31% (19), was not considered here or in the previous study when calculating the transformation efficiency of B. bacilliformis. Here, our focus was not on optimizing pBBR1MCS-2 electrotransformation but simply on isolating a strain that would efficiently serve as a host for allelic exchange experiments. Taken as a whole, these results demonstrated that the genetic background of the KC584 host strain used by Grasseschi and Minnick (19) was distinct from wild-type KC584 and suggested that JB584 would be an efficient host for allelic exchange experiments.
We hypothesized that the increased transformability of JB584 was due to one or more spontaneous mutations in the restriction-modification system, whereby restriction of introduced foreign DNA was reduced to a level that permitted plasmid replication and maintenance. To test this hypothesis, we compared genomic digests of KC583, KC584, JB584, HG584, JB585, and JB686 by using the methyl-sensitive restriction enzymes StuI (dcm sensitive) and ClaI (dam sensitive), with BamHI (methyl insensitive) as a positive control. Subsequent agarose gel electrophoresis revealed that StuI was able to digest genomic DNA from strains HG584, JB584, JB585, and JB686 but was unable to digest DNA from KC583 or KC584. These data strongly suggest that a dcm methylase is active in wild-type strains KC583 and KC584 and that the sites of methylation overlap the StuI recognition sequence. In contrast, ClaI and BamHI digestion of genomic DNA was evident in of all of the strains mentioned and there was no apparent difference in activity among any of the strains. There was no significant difference in the growth rate, colony morphology, or overt phenotypes between strains KC584 and JB584, suggesting that JB584 was suitable for use as a host strain for mutagenesis experiments. These results, combined with the previously mentioned descrepancies between the transformation efficiencies, strongly suggest that loss of dcm methylase activity occurred during the multiple passages of the KC584 strain used in the Grasseschi and Minnick (19) study and also suggest that JB584 and HG584 have essentially the same restriction-modification system genetic alterations and the curing event itself did not alter this genetic background. Because this highly passaged KC584 strain was no longer viable, we were resigned to curing the previously transformed strain (HG584) and using the resulting strain (JB584) for our subsequent manipulation studies.
Characterization of Kanr mutants by PCR.
Eight kanamycin-resistant clones were isolated following electroporation-mediated introduction of pUB508 into JB584. Three amplimer sets, designated nptI, fla, and junction, were used to characterize the genotype of the eight Kanr clones (refer to Fig. 1B for a schematic representation of amplimer target loci).
First, the nptI amplimer set (NPTI5′ and NPTI3′) generated a product in all eight Kanr strains, consistent with the expected 983-bp size (data not shown). This verified that the Kanr phenotype exhibited by these strains was not a result of natural mutation. The transformation efficiency was approximately 3.3 transformants per μg of pUB508, corresponding to one stable integrant per 1.6 × 109 cells. Figure 2 shows the product generated by the nptI amplimer set from one of these Kanr transformants, termed strain JB585 (Fig. 2, lane 3), and its absence from the parent strain, JB584 (lane 2).
Second, the fla amplimer set (FLA5′ and FLA3′) generated the expected 1,304-bp flagellin PCR product with the parent strain JB584 (Fig. 2, lane 4) and, in contrast, was absent from the Kanr strain JB585 (lane 5). This showed that the homologous recombination event occurred at the fla locus in JB585. Of the eight Kanr strains, two were analyzed with the fla amplimer set, and neither generated a product. We were unable to generate the expected ∼4.2-kb product resulting from amplification of the entire mutagenized locus spanning the inserted pUB508, even when the elongation time was increased to 2.5 min.
Finally, the junction amplimer set (NPTI5′ and FLA3′) was used to verify the hypothesized chromosomal fusion between the inserted nptI and the mutagenized fla. The ∼2,300-bp product generated from JB585 (Fig. 2, lane 9), as well as its absence from the parent strain (lane 6), confirms that the insertion of pUB508 occurred at fla as illustrated in Fig. 1B. In addition, as controls, pUB508 (lane 7) and a mixture of pUB508 and JB584 DNA (lane 8) were amplified with the junction amplimer set to further substantiate these results, since neither reaction produced an amplicon.
Characterization of the complemented mutant by PCR.
To develop a Bartonella system for in trans complementation, we restored the wild-type flagellin phenotype to a fla mutant. Plasmid pBBRFLAG, containing fla, a chloramphenicol resistance cassette, and a REP origin, was introduced into JB585 by electroporation. Transformants were selected on medium containing kanamycin sulfate (25 μg/ml) and chloramphenicol (1 μg/ml) and resulted in the isolation of strain JB686. Initial confirmation of in trans complementation was accomplished by electrophoretic analysis of PCR products. The fla amplimer set (FLA5′ and FLA3′) was used to detect the presence of the plasmid-located fla gene, which was absent in the fla mutant strain JB585 (Fig. 2, lane 5) but was present in both JB584 and the complemented mutant JB686 (lanes 4 and 10, respectively). Finally, the ∼2,300-bp product generated by the junction amplimer set (NPTI5′ and FLA3′) demonstrated that the chromosomal fla mutation was still present in the complemented mutant strain, JB686 (lane 11).
Characterization by DNA hybridization.
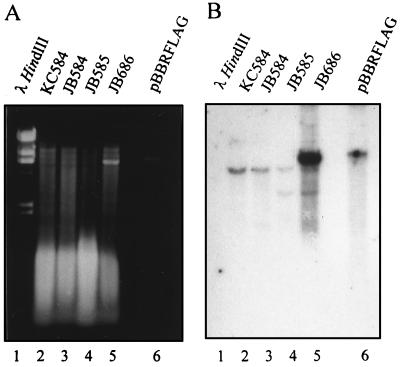
To further substantiate the genotype of the mutant and trans-complemented strains, high-stringency DNA hybridizations were performed (Fig. 3). Southern blots probed with the 32P-labeled wild-type fla PCR product (generated with the FLA5′ and FLA3′ amplimers) produced a distinct two-band hybridization pattern in ClaI-digested genomic DNA in strains containing the disrupted fla gene, i.e., JB585 (Fig. 3B, lane 4) and JB686 (lane 5). In addition, the pBBRFLAG complementation plasmid is clearly visible as a separate genetic element (Fig. 3A, lane 5).
FIG. 3.
Detection of the flagellin gene in wild-type, mutant, and trans-complemented strains of B. bacilliformis by DNA hybridization. (A) Ethidium bromide-stained agarose gel (1% [wt/vol] agarose) containing λ HindIII size standards (lane 1), ClaI-digested chromosomal DNA from KC584 (lane 2), JB584 (lane 3), JB585 (lane 4), and JB686 (lane 5), and ClaI-digested pBBRFLAG (lane 6). (B) Corresponding Southern blot following hybridization with the fla probe. Note the distinct two-band hybridization pattern in strains containing a disrupted fla gene (strains JB585 and JB686) and the presence of pBBRFLAG in the trans-complemented strain, JB686.
Analysis of flagellin production in generated strains by SDS-PAGE and immunoblotting.
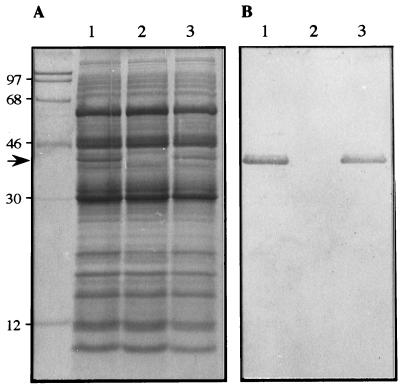
SDS-PAGE and immunoblotting were used to determine the effect of the pUB508 insertion on the synthesis of flagellin in the mutant strain, JB585, and the complemented strain, JB686 (Fig. 4). The wild-type 1,127-bp B. bacilliformis fla open reading frame (ORF) encodes a 42-kDa polypeptide (39). When whole-cell lysates were visualized by SDS-PAGE, the flagellin polypeptide was clearly synthesized in strain JB584 (Fig. 4A, lane 1). In contrast, the fla mutant, JB585 (lane 2), lacked the 42-kDa flagellin polypeptide, suggesting that flagellin synthesis had been disrupted. Finally, the 42-kDa flagellin polypeptide was clearly evident in the trans-complemented strain (lane 3), indicating that fla expression and synthesis was occurring from the plasmid locus. Immunoblot analysis was subsequently performed by reacting whole-cell lysates with rabbit anti-flagellin polyclonal antiserum. The immunoblot confirmed the presence of the flagellin polypeptide in strains JB584 and JB686 (Fig. 4B, lanes 1 and 3, respectively) and also demonstrated that flagellin synthesis was completely abolished in the mutant strain JB585 (lane 2). A truncated flagellin product was not observed in the mutant strain analyzed by this procedure.
FIG. 4.
Analysis of flagellin synthesis in the B. bacilliformis fla mutant and complemented mutant by SDS-PAGE and immunoblotting. (A) Whole-cell extracts of parent, flagellin mutant, and trans-complemented mutant strains were separated by SDS-PAGE (12.5% polyacrylamide) and stained with Coomassie brilliant blue. The 42-kDa flagellin polypeptide is present in the parent strain JB584 (lane 1) and is absent from the kanamycin-resistant flagellin mutant strain JB585 (lane 2). Flagellin synthesis is detected in trans from the complementation plasmid pBBRFLAG in strain JB686 (lane 3). (B) Corresponding immunoblot reacted with rabbit anti-flagellin polyclonal antiserum, indicating that flagellin synthesis is restored in the trans-complemented strain. Molecular mass standards are indicated to the left (in kilodaltons), and the 42-kDa flagellin polypeptide is marked by the arrow.
Ultrastructural characterization of strains by TEM.
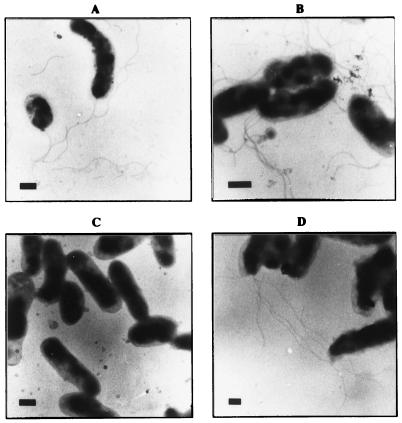
TEM was used to visualize the secretion and assembly of flagella in each of the strains (Fig. 5). Electron micrographs showed that the wild-type strain, KC584 (Fig. 5A), and the transformable strain, JB584 (Fig. 5B), maintained the normal synthesis, secretion, and assembly of flagellin polypeptides, resulting in the wild-type lophotrichous flagella. Second, as evidenced by the lack of flagellar filaments, the flagellin ORF of strain JB585 had been insertionally disrupted and the synthesis, secretion, and assembly of the flagellin polypeptide into filaments was abolished (Fig. 5C). Finally, not only did the complemented mutant, strain JB686, synthesize flagellin polypeptides from the plasmid locus, but also these polypeptides were secreted and assembled (Fig. 5D) as in the wild-type strain (Fig. 5A). Therefore, the flagellar phenotypes of the strains were consistent with the genotypes. There was no significant difference in the number of flagellar filaments or wavelength of the flagellar filaments, including in flagellated strains of JB584 containing pBBRFLAG or pBBR1MCS (data not shown).
FIG. 5.
Ultrastructure of generated strains as shown by TEM. Transmission electron micrographs were prepared by staining 5-day-old cultures of B. bacilliformis with 2% uranyl acetate. Flagella are observed the wild-type strain (KC584) (A) and the transformable strain (JB584) (B) but are absent from the fla mutant strain (JB585) (C) Polypeptide synthesis, secretion, and assembly of the flagella is restored in the trans-complemented strain (JB686) (D). Bars, 0.5 μm.
Motility phenotypes of generated strains.
Initially, phase-contrast microscopy was used to examine wet mounts of each strain. By using this method, a loss of motility was observed for strain JB585, whereas strains KC584, JB584, and JB686 exhibited indistinguishable motility phenotypes. Motility was subsequently assessed by the ability of the bacterium to spread within Bartonella motility medium. To develop the assay, we tested a wild-type strain in motility medium with agar concentrations ranging from 0.2 to 0.8% by stabbing the medium with an inoculation needle from 5-day-old cultures. Incubation for 7 days at 30°C demonstrated that agarose concentrations above 0.6% inhibited motility. However, at agarose concentrations of 0.4 and 0.2%, motility produced a uniform halo of growth within the medium away from the site of inoculation. The strains generated in this study were subsequently tested in the same way by using Bartonella motility medium containing 0.2% agar. Multiple inoculations with each of the strains consistently generated indistinguishable and uniform halos of growth for strains KC584, JB584, and JB686, indicating that neither the modified genotype resulting in enhanced transformability (JB584) nor fla expression from an extrachromosomal locus (JB686) has a detectable effect on motility. In contrast, the mutant strain, JB585, was nonmotile and did not produce a halo (data not shown).
In conclusion, site-directed insertion of pUB508 at the fla locus of JB584 generated the fla mutant, JB585, which lacks flagellin expression and is nonmotile. Subsequent electroporation-mediated introduction of pBBRFLAG into strain JB585 generated the trans-complemented strain, JB686, which has a motility phenotype that is indistinguishable from KC584.
DISCUSSION
The ability to genetically manipulate an organism is essential for a better understanding of its molecular biology. In vivo genetic manipulation of a bacterium generally consists of two fundamental techniques, transformation and mutagenesis. These techniques utilize constructs consisting of circular plasmids or linear fragments of DNA that possess genetic elements specific to the manipulation desired. These DNA constructs can be introduced into the bacterium by natural (conjugation and transduction) or artificial (electroporation and chemical) methods. In this study, we used electroporation to introduce a variety of plasmids (both replicative and nonreplicative) and linear DNA fragments and were able to demonstrate plasmid transformation, site-directed mutagenesis, and complementation in trans within B. bacilliformis.
Plasmid transformation was first demonstrated in Bartonella by electrotransformation of the cosmid pEST into B. (previously Rochalimaea) quintana (36) and was subsequently accomplished in B. bacilliformis by Grasseschi and Minnick using the same cosmid (19). In both studies, the Bartonella species recognized the RK2 origin of replication but did not recognize pMB1, ColE1, or F origins. These observations, combined with our results, suggested that pUB1 (harboring the pMB1 origin) could be used as a suicide plasmid for all Bartonella species. In the course of our research, we determined that the most consistent replicon for high transformation efficiencies in B. bacilliformis was the broad-host-range vector pBBR1MCS (27) and its derivatives (26), which contain the REP origin of replication.
Random mutagenesis by using chemical methods, UV light, or transposons is designed to generate nonspecific mutations that are subsequently selected by phenotypic or biochemical means. The major drawbacks to random mutagenesis are the difficulty in mutant isolation and the generation of secondary nonspecific mutations. These anomalous secondary mutations are of special concern when assessing the pathogenesis of an organism, since the virulence potential of a specific gene and gene product are in question relative to a wild-type background. Dehio and Meyer recently reported successful conjugation between E. coli and B. henselae as a means of plasmid transfer and delivery of Tn5 transposons on suicide plasmids for random gene inactivation (12).
Site-specific mutagenesis, the focus of this study, consists of two general methods, replacement recombination and insertional recombination, wherein single mutations are introduced at a specific genomic locus. Replacement recombination involves linear fragments of DNA designed so that during homologous recombination two crossover events result in the replacement of a target locus with the construct. We attempted replacement recombination with both single- and double-stranded linear DNA targeting two loci, gyrB (7) and fla, without success. Even when the highly transformable strain, JB584, was used as the host, replacement recombination was not achieved. However, insertional recombination with a circular segment of DNA, where a single homologous recombination event inserts the entire element into the chromosome, was successful in demonstrating site-specific genetic manipulation, as described in Results.
After several initial attempts to mutagenize the flagellin gene failed, we realized that there were in vivo barriers impeding homologous recombination in B. bacilliformis. This prompted us to try alternative methods for alleviating restriction-modification systems as well as biochemical and metabolic manipulations previously shown to increase the likelihood of homologous recombination. Although numerous manipulations were attempted, the generation of strain JB584 was the critical step toward successful mutagenesis. It is likely that a spontaneous mutation occured in a multiply-passaged KC584 strain used by Grasseschi and Minnick (19), allowing them to obtain exaggerated “wild-type” transformation efficiencies. By curing this Kanr HG584 strain of pEST, which resulted in JB584, we obtained a strain with a significant increase in transformation efficiencies, which encouraged us to use this strain as the parent for successful mutagenesis experiments.
In addition to being sequence specific, restriction enzymes such as StuI and ClaI are methylation sensitive, where restriction occurs only in the absence of methylation, enabling the organism to recognize self and nonself. While the genotype of JB584 is not fully known, genomic DNA extracted from this strain differed from that of the wild-type strain in that it was StuI sensitive, indicating a different methylation pattern and suggesting that a dcm methylase gene had been mutated. Since the altered methylation pattern did not result in lethal self-restriction of JB584, we speculate that a second mutation in the cognate restriction enzyme had occurred and that a lack of restriction enzyme activity explained the enhanced transformability of this strain. Alternatively, and more probably, a single, spontaneous mutation event involving the deletion of closely linked restriction and modification genes would also explain the phenotype of JB584. It is likely that wild-type strains of B. bacilliformis retain both an active dcm methylase and the cognate restriction enzyme and that they digest foreign DNA, but strains such as JB584 have lost the cognate restriction enzyme to ensure survival and do not digest foreign DNA. However, a full characterization of the specific restriction endonuclease mutation(s) in strain JB584 enabling higher transformation efficiencies remains to be determined.
The hybridization data generated in this study suggest that the B. bacilliformis flagellar filament is encoded by a single flagellin gene, whereas many bacteria possess multisubunit flagella. Furthermore, the data produced in this study conclusively show that inactivation of a single fla gene completely abolishes synthesis of the flagellum and generates a nonmotile and nonflagellated (bald) strain. The lophotrichous flagella of B. bacilliformis and the high degree of motility that they impart have been implicated as virulence determinants in several reports (8, 33, 39, 45). The strains generated in this study provide the molecular means to assess Koch’s postulates and thus more fully define the role of flagella in the pathogenesis of B. bacilliformis. Using the highly transformable strain, JB584, the pBBR1MCS shuttle vectors, and the fla-specific suicide plasmid, we have demonstrated site-specific mutagenesis and complementation for the first time in any Bartonella species. Two additional loci (ialB and the 16S–23S rDNA intergenic spacer) have subsequently been manipulated in our laboratory by using this system.
The Bartonella species are notoriously difficult to manipulate genetically. We have developed a system for electroporation-mediated site-specific mutagenesis and complementation for B. bacilliformis. The pBBR1MCS series of broad-host-range vectors were shown to be useful shuttle vectors for B. bacilliformis, and recent work in our laboratory with B. quintana shows that the REP origin is functional in other Bartonella species. We have constructed a suicide vector (pUB1) with a polylinker, which is ideally suited for manipulating any desired target gene in B. bacilliformis. As stated above, a spontaneous or natural mutation was apparently present in the host strain used by Grasseschi and Minnick (HG584) (19) and resulted in the inflated transformation efficiencies that they reported. The data produced in this study suggest that a strain of Bartonella with enhanced transformability can be isolated by selecting clones that are capable of maintaining a replicative plasmid, such as pBBR1MCS-2. Subsequent curing of these potential methylase restriction mutants can result in a well-defined, transformable host strain for subsequent mutagenesis experiments.
ACKNOWLEDGMENTS
We thank Joan Strange, Jim Driver, and Paul Dexter for their excellent technical assistance and Deborah Kane and Kay Crull (American Red Cross) for their continued generosity in supplying us with human erythrocytes. We are also grateful to Michael Kovach and Dennis Reschke for generous donations of plasmids and to Tracy Pfeifer, Scott Samuels, George Card, Pat Ball, Tim Gsell, Bill Holben, and Laura Smitherman for enthusiastic assistance and advice.
This work was supported by Public Health Service grant AI34050 from the National Institutes of Health (NIAID).
REFERENCES
- 1.Abbott P J. Stimulation of recombination between homologous sequences on carcinogen-treated plasmid DNA and chromosomal DNA by induction of the SOS response in Escherichia coli K12. Mol Gen Genet. 1985;201:129–132. doi: 10.1007/BF00397998. [DOI] [PubMed] [Google Scholar]
- 2.Alexander B. A review of bartonellosis in Ecuador and Colombia. Am J Trop Med Hyg. 1995;52:354–359. doi: 10.4269/ajtmh.1995.52.354. [DOI] [PubMed] [Google Scholar]
- 3.Arevalo-Jimenez J I, Williams M, Marks K, Ihler G M. Genbank accession no. L20677. 1993. Unpublished data. [Google Scholar]
- 4.Amano Y, Rumbea J, Knobloch J, Olson J, Kron M. Bartonellosis in Ecuador: serosurvey and current status of cutaneous verrucous disease. Am J Trop Med Hyg. 1997;57:174–179. doi: 10.4269/ajtmh.1997.57.174. [DOI] [PubMed] [Google Scholar]
- 5.Anderson B E, Newman M A. Bartonella species as emerging human pathogens. Clin Microbiol Rev. 1997;10:203–219. doi: 10.1128/cmr.10.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Strahl K. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 7.Battisti J M, Smitherman L S, Samuels D S, Minnick M F. Mutations in Bartonella bacilliformis gyrB confer resistance to coumermycin A1. Antimicrob Agents Chemother. 1998;42:2906–2913. doi: 10.1128/aac.42.11.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson L A, Kar S, McLaughlin G, Ihler G M. Entry of Bartonella bacilliformis into erythrocytes. Infect Immun. 1986;54:347–353. doi: 10.1128/iai.54.2.347-353.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner D J, O’Connor S P, Hollis D G, Weaver R E, Steigerwalt A G. Molecular characterization and proposal of a neotype strain for Bartonella bacilliformis. J Clin Microbiol. 1991;29:1299–1302. doi: 10.1128/jcm.29.7.1299-1302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 12.Dehio C, Meyer M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J Bacteriol. 1997;179:538–540. doi: 10.1128/jb.179.2.538-540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dzelzkalns V A, Bogorad L. Stable transformation of the cyanobacterium Synechocystis sp. PCC 6803 induced by UV irradiation. J Bacteriol. 1986;165:964–971. doi: 10.1128/jb.165.3.964-971.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia F U, Wojta J, Broadly K N, Davidson J M, Hoover R L. Bartonella bacilliformis stimulates endothelial cells in vitro and is angiogenic in vivo. Am J Pathol. 1990;136:1125–1135. [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia F U, Wojta J, Hoover R L. Interactions between live Bartonella bacilliformis and endothelial cells. J Infect Dis. 1992;165:1138–1141. doi: 10.1093/infdis/165.6.1138. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Caceres U, Garcia F U. Bartonellosis: an immunodepressive disease and the life of Daniel Alcides Carrion. Am J Clin Pathol. 1991;95:S58–S66. [PubMed] [Google Scholar]
- 17.Gibco-BRL. Gibco-BRL manual. Gaithersburg, Md: Gibco-BRL; 1991. [Google Scholar]
- 18.Gigliani F, Ciotta C, Del Grosso M F, Battaglia P A. pR plasmid replication provides evidence that single-stranded DNA induces the SOS system in vivo. Mol Gen Genet. 1993;238:333–338. doi: 10.1007/BF00291991. [DOI] [PubMed] [Google Scholar]
- 19.Grasseschi H A, Minnick M F. Transformation of Bartonella bacilliformis by electroporation. Can J Microbiol. 1994;40:782–786. doi: 10.1139/m94-123. [DOI] [PubMed] [Google Scholar]
- 20.Gray G C, Johnson A A, Thornton S A, Smith W A, Knobloch J, Kelley P W, Escudero L O, Huayda M A, Wignall F S. An epidemic of Oroya fever in the Peruvian Andes. Am J Trop Med Hyg. 1990;42:215–221. doi: 10.4269/ajtmh.1990.42.215. [DOI] [PubMed] [Google Scholar]
- 21.Hiom K, Thomas S M, Sedgwick S G. Different mechanisms for SOS induced alleviation of DNA restriction in Escherichia coli. Biochimie. 1991;73:399–405. doi: 10.1016/0300-9084(91)90106-b. [DOI] [PubMed] [Google Scholar]
- 22.Hiom K J, Sedgwick S G. Alleviation of EcoK DNA restriction in Escherichia coli and involvement of umuDC activity. Mol Gen Genet. 1992;231:265–275. doi: 10.1007/BF00279800. [DOI] [PubMed] [Google Scholar]
- 23.Holloway C T, Greene R C, Su Ching-Hsiang. Regulation of S-adenosylmethionine synthase in Escherichia coli. J Bacteriol. 1970;104:734–747. doi: 10.1128/jb.104.2.734-747.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurtaldo A, Musso J P, Merino C. La anemia en la enfermadad de Carrion (verruga peruana) Ann Fac Med Lima. 1938;28:154–168. [Google Scholar]
- 25.Koehler J E. Bartonella infections. Adv Pediatr Infect Dis. 1996;11:1–27. [PubMed] [Google Scholar]
- 26.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop R M, Jr, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 27.Kovach M E, Phillips R W, Elzer P H, Roop R M, Jr, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800. [PubMed] [Google Scholar]
- 28.Kreier J P, Ristic M. The biology of hemotrophic bacteria. Annu Rev Microbiol. 1981;35:325–338. doi: 10.1146/annurev.mi.35.100181.001545. [DOI] [PubMed] [Google Scholar]
- 29.Krueger C M, Marks K L, Ihler G M. Physical map of the Bartonella bacilliformis genome. J Bacteriol. 1995;177:7271–7274. doi: 10.1128/jb.177.24.7271-7274.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Lu C, Echols H. RecA protein and SOS. Correlation of mutagenesis phenotype with binding of mutant RecA proteins to duplex DNA and LexA cleavage. J Mol Biol. 1987;196:497–504. doi: 10.1016/0022-2836(87)90027-1. [DOI] [PubMed] [Google Scholar]
- 32.McGinnis Hill E, Raji A, Valenzuela M S, Garcia F, Hoover R. Adhesion to and invasion of cultured human cells by Bartonella bacilliformis. Infect Immun. 1992;60:4051–4058. doi: 10.1128/iai.60.10.4051-4058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mernaugh G, Ihler G M. Deformation factor: an extracellular protein synthesized by Bartonella bacilliformis that deforms erythrocyte membranes. Infect Immun. 1992;60:937–943. doi: 10.1128/iai.60.3.937-943.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minnick, M. F. Bartonella species. In M. Sussman (ed.), Molecular medical microbiology, in press. Academic Press, Ltd., London, United Kingdom.
- 35.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 36.Reschke D K, Frazier M E, Mallavia L P. Transformation of Rochalimaea quintana, a member of the family Rickettsiaceae. J Bacteriol. 1990;172:5130–5134. doi: 10.1128/jb.172.9.5130-5134.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynafarje C, Ramos J. The hemolytic anemia of human bartonellosis. Blood. 1961;17:562–578. [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Scherer D C, DeBuron-Connors I, Minnick M F. Characterization of Bartonella bacilliformis flagella and effect of antiflagellin antibodies on invasion of human erythrocytes. Infect Immun. 1993;61:4962–4971. doi: 10.1128/iai.61.12.4962-4971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi W, Zusman D R. Methionine inhibits developmental aggregation of Myxococcus xanthus by blocking the biosynthesis of S-adenosylmethionine. J Bacteriol. 1995;177:5346–5349. doi: 10.1128/jb.177.18.5346-5349.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 42.Stratagene Cloning Systems. Stratagene manual. Cloning Systems, La: Stratagene; 1993. Jolla, Calif. [Google Scholar]
- 43.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vericat J A, Guerrero R, Barbe J. Increase in plasmid transformation efficiency in SOS-induced Escherichia coli cells. Mol Gen Genet. 1988;211:526–530. doi: 10.1007/BF00425711. [DOI] [PubMed] [Google Scholar]
- 45.Walker T S, Winkler H H. Bartonella bacilliformis: colonial types and erythrocyte adherence. Infect Immun. 1981;31:480–486. doi: 10.1128/iai.31.1.480-486.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinman D, Dubos R J, Hirsch J G, editors. Bacterial and mycotic infections of man. 4th ed. Philadelphia, Pa: J. B. Lippincott Co.; 1965. The bartonella group; pp. 775–785. [Google Scholar]
- 47.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]